Abstract
An accurate, nonsurgical diagnostic test for brain tumors is currently unavailable, and the methods of monitoring disease progression are not fully reliable. MicroRNA profiling of biological fluids has recently emerged as a diagnostic tool for several pathologic conditions. Here we tested whether microRNA profiling of cerebrospinal fluid (CSF) enables detection of glioblastoma, discrimination between glioblastoma and metastatic brain tumors, and reflects disease activity. We determined CSF levels of several cancer-associated microRNAs for 118 patients diagnosed with different types of brain cancers and nonneoplastic neuropathologies by quantitative reverse transcription PCR analysis. The levels of miR-10b and miR-21 are found significantly increased in the CSF of patients with glioblastoma and brain metastasis of breast and lung cancer, compared with tumors in remission and a variety of nonneoplastic conditions. Members of the miR-200 family are highly elevated in the CSF of patients with brain metastases but not with any other pathologic conditions, allowing discrimination between glioblastoma and metastatic brain tumors. Quantification of as few as 7 microRNAs in CSF enables differential recognition of glioblastoma and metastatic brain cancer using computational machine learning tools (Support Vector Machine) with high accuracy (91%–99%) on a test set of samples. Furthermore, we show that disease activity and treatment response can be monitored by longitudinal microRNA profiles in the CSF of glioblastoma and non–small cell lung carcinoma patients. This study demonstrates that microRNA-based detection of brain malignancies can be reliably performed and that microRNAs in CSF can serve as biomarkers of treatment response in brain cancers.
Keywords: biomarkers, brain metastasis, cerebrospinal fluid, glioblastoma, leptomeningeal metastasis, microRNA
The most frequently occurring brain malignancies in adults are metastatic brain cancers (eg, from lung, breast), followed by glioblastoma (GBM). GBM is the most aggressive primary brain cancer, which generally has a poor prognosis, with median survival of about 14 months, despite intensive treatment.1 Currently, diagnosis of brain tumors is performed with histology of tumor tissue, if biopsy is available, and in some cases cytological analysis of cerebrospinal fluid (CSF) for the presence of cancer cells. CSF can be accessed readily for longitudinal disease monitoring during and after therapy. However, cytological analysis of CSF has low sensitivity, as it often appears negative when tumor progression is present.2 It is also nonquantitative and technically challenging; there is no routine way to subtype the malignancy and monitor molecular changes from CSF, indicating the need for more accurate and reliable biomarkers and methods. The aim of this study is to determine whether specific microRNAs (miRNAs) that are present in CSF can serve as biomarkers for particular brain malignancies and disease activity.
MiRNAs are small endogenous mediators of RNA interference and key regulatory components of many biological processes required for organism development, cell specialization, and homeostasis. Many miRNAs exhibit tissue-specific patterns of expression and are deregulated in various cancers, where they can be either oncogenic (oncomirs) or tumor-suppressive. The recent discovery of miRNAs in secreted membrane vesicles (exosomes),3,4 as well as in the blood serum5,6 and other body fluids,7 suggests that miRNAs play a role in intracellular communication in a both paracrine and endocrine manner. It has also opened a new exciting direction for study of miRNAs as biomarkers for diseases, and cancer diagnostics by miRNA profile in blood serum has become a quickly growing field.8
Several studies have reported detection of miRNA, among several biological fluids, in CSF,9–11 raising the possibility that miRNAs in CSF might serve as informative biomarkers of CNS diseases. Such a possibility, largely unexplored until now, is supported by the finding that different types of brain cancer have distinct signatures of miRNA expression, with some miRNA species abundant in cancer while undetectable in healthy brain.12–14 Since CSF is separated from blood circulation by the blood–brain barrier, it is conceivable that CSF might better retain a unique signature of miRNA expression specific for brain tumors.
Two recent studies demonstrated the usefulness of miRNA profiling in CSF for diagnostics of brain lymphoma and GBM.11,15 In our current study, we tested the levels of several candidate miRNAs in the CSF of patients with GBM and compared them with those of metastatic brain cancers and a variety of nonneoplastic CNS diseases. We found a strong association between the particular types of brain cancer and the presence of specific miRNAs in CSF. Using this approach, we were able to recognize GBM and metastatic brain cancers and to discriminate between them with about 95% accuracy. Our results demonstrate the utility of miRNA as a biomarker of high-grade brain malignancies and reveal their value for the development of diagnostic and prognostic tools, as well as for monitoring of CNS pathology in general.
Materials and Methods
Collection of Samples
CSF and brain tumor samples were obtained from the Department of Neurosciences, Translational Neuro-Oncology Laboratories, Moores UCSD Cancer Center, UC San Diego; the Department for Neurosurgery at Brigham and Women's Hospital; and the Department for Neurosurgery at Göttingen University Medical Center over the period of 2–5 years. All human materials were obtained and used in accordance with the policies of institutional review boards at UC San Diego, Brigham and Women's Hospital, and Göttingen University Medical Center. CSF samples were obtained either through lumbar puncture or through Ommaya reservoir during surgery, as indicated in Supplementary material, Table S1. Samples obtained intraoperatively were collected immediately before tumor resection. At least 1 mL of each CSF sample was cleared of cells and debris immediately after collection by brief centrifugation and stored in aliquots at −80°C. All tumor specimens were fresh-frozen on dry ice and stored at −80°C until tested.
RNA Isolation and MiRNA Profiling
CSF samples were lyophilized and total RNA was extracted using the mirVana miRNA isolation kit (Ambion) according to the manufacturer's protocol. The amount of RNA extracted from the CSF samples was within 50–2500 ng/mL, consistent with previous findings.4 Total RNA from frozen tumor tissues was isolated using Trizol reagent (Invitrogen). The levels of individual miRNAs in CSF and tumors were determined by TaqMan miRNA assays from Applied Biosystems. Four nanograms of total RNA was used in 6 µL reverse transcription (RT) reactions with specific miRNA reverse transcriptionprobes, prior to TaqMan real time RT-PCR reactions that were performed in duplicates. MiR-24, which is relatively uniformly represented in CSF,15 was detected in all CSF samples and used as an internal control for normalization (Supplementary Fig. S1a). However, since miR-24 levels themselves were not uniform across the CSF samples, miR-125, an alternative abundant marker, was used for normalization, producing similar results (Supplementary Fig. S1b). The data normalized to miR-125b as well as nonnormalized data are presented in Supplementary Figs S2 and S3, respectively. We were unable to identify a miRNA marker less variable across the CSF samples and observed generally higher miRNA CSF levels in neoplastic cases relative to nonneoplastic controls. This trend may reflect a release of miRNA-containing microvesicles by cancer cells4 and/or destruction of the brain tissue in neoplastic conditions. MiRNA levels were calculated relative to corresponding miR-24 levels using standard calculations: 2ΔCt, where ΔCt = CtmiR-24 – CtmiR-X). All data are means of technical duplicates, and the standard errors of the means were calculated between duplicates.
Sample Classification and Data Analysis
A total of 118 patients of 2 neuro-oncology clinics and corresponding CSF samples were analyzed in this study. One hundred eight patients were classified into 6 groups based on clinical and pathologic diagnoses (including CSF cytology and tumor histology when applicable) and MRI findings (Table 1; the detailed patient characteristics are listed in Supplementary material, Table S1). The first control group, referred as “nonneoplastic,” included patients with various neurological conditions other than brain neoplasia. The patients in this group had no cancer at the time of CSF collection, and no previous history of CNS malignancies. The second group, “GBM,” included patients diagnosed with active GBM. GBM was referred to as clinically active when a primary enhancing tumor mass was apparent and growing by MRI as assessed by the neuro-oncologist (S.K.) at the time of CSF sample collection and was further classified as GBM by tumor tissue histology. The two groups called “Breast to Brain” and “Lung to Brain” comprised samples from patients with parenchymal brain metastasis from breast carcinoma and lung cancer (including small cell and non–small cell lung carcinoma), respectively. The presence of metastases in these patients was confirmed by MRI at the time of CSF collection. Two additional groups represented patients with documented leptomeningeal metastasis of these cancers (CSF- or MRI-positive disease). Seven additional patients not included in the groups described above were analyzed separately. These patients represent cases of remission of primary and metastatic brain tumors, as indicated by no detectable brain tumor at the time of CSF collection based on imaging features, clinical stability, and CSF cytology. The remaining 3 patients were analyzed in the longitudinal study.
Table 1.
Groups of patients included in this study
| Group | N | Clinical/pathology-based diagnosis |
|---|---|---|
| Control | 15 | Nonneoplastic neurological conditions: headache (4)a, trigeminal neuralgia, memory problem, gait difficulty, dementia, Parkinson disease, myelitis (2), normal pressure hydrocephalus, encephalitis, neuropathy, benign cerebellar lesion, Hodgkin disease with no CNS cancer |
| GBM | 19 | Glioblastoma multiforme (glioma grade IV) |
| Breast to brain | 16 | Breast cancer metastasis to brain |
| Breast LM | 26 | Breast cancer leptomeningeal metastasis |
| Lung to brain | 28 | Lung cancer metastasis to brain |
| Lung LM | 4 | Lung cancer leptomeningeal metastasis |
Abbreviation: N, number of patients per group.
aThe number of patients with a particular diagnosis, if more than one, is indicated in parentheses.
Statistical Analysis and Support Vector Machine–Based Data Classification
The differences in CSF miRNA levels between groups of samples were determined using Graph Pad Prism software by Wilcoxon signed rank test, and 2-tailed P-values were calculated.
Support Vector Machine (SVM) was implemented within a machine learning software package (weka, www.cs.waikato.ac.nz/ml/weka). In such an approach, miRNA levels from the samples are treated as independent variables and the type of cancer, if any, as a variable to be predicted. The SVM is trained and tested on such a dataset using a standard N-fold cross-validation process. In this process, the SVM is trained on all samples except one and tested on that holdout sample. The procedure is repeated as many times as there are samples in the dataset—hence, each sample forms the holdout set once and only once. We found the following choice of nondefault parameters working best: classifier, Sequential Minimal Optimization; kernel, radial basis function (RBF); and a complexity parameter one for all tasks, except breast versus lung metastasis, in which case it was 100. Threshold cycle values were used for the classification as-is, with no standardization or normalization, except that 1000 was used in place of a Ct value in cases of undetectable miRNA.
The Cancer Genome Atlas (TCGA) miRNA expression microarray data for GBM patients were downloaded from http://tcga-data.nci.nih.gov/tcga/homepage.htm. The fold difference in specific signals between GBM (n = 261) and normal brain (n = 10) tissue were calculated for each miRNA as described.4
Results
The aims of this pilot study were (1) to determine whether specific miRNAs could serve as CSF biomarkers of GBM, (2) to determine whether specific miRNAs in CSF could differentiate between GBM and other most common brain cancers, such as metastatic brain cancers, and (3) to examine whether miRNAs in CSF could serve as biomarkers of GBM and metastatic brain disease activity and of response to therapy.
MiR-10b Is Present and MiR-21 Is Elevated in CSF of Glioblastoma and Brain Metastasis Patients
To identify miRNA biomarkers for GBM, we used a candidate approach based on our previous miRNA profiling data.4,16,17 An additional analysis of miRNA expression in 261 GBM patients utilized the TCGA dataset and revealed a panel of miRNAs deregulated in GBM relative to normal brain tissues (Fig. 1A). Among them, miR-10b and miR-21 were the most strongly upregulated (Fig. 1A). Both miR-10b and miR-21 were also found to be highly elevated in GBM tumors analyzed in many independent studies.18–21 MiR-10b is a unique molecule, as it is the only known miRNA undetectable in normal brain while highly expressed in GBM.18,22 We have therefore chosen it as our top priority candidate. Expression of miR-10b is also associated with metastatic phenotypes of several solid cancers, including those of the breast and lung.23,24
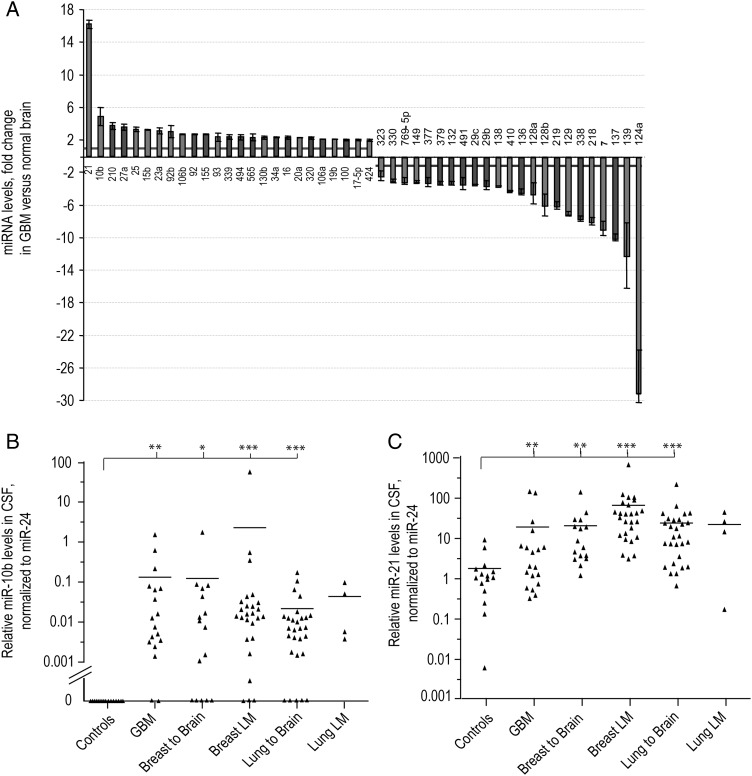
Fig. 1.
MiR-10b and miR-21 upregulation in GBM and their CSF levels in patients with GBM or metastatic brain cancer and nonneoplastic controls. (A) MiRNAs were deregulated in GBM more than 2-fold compared with normal brains. MiRNA levels were obtained by the analysis of TCGA miRNA microarray data; error bars represent standard deviation between individual probe sets present for each miRNA on the arrays. (B) MiR-10b and (C) miR-21 levels were examined by qRT-PCR in CSF samples of neurological patients. Relative levels were normalized to miR-24, as described in Materials and Methods and demonstrated for individual CSF samples. Horizontal lines indicate arithmetic mean of miRNA levels for each group of patients: “Controls” = nonneoplastic neuropathologic cases, “GBM” = glioblastoma cases, “Breast to Brain” and “Lung to Brain” = breast and lung cancer brain metastasis, “Breast LM” and “Lung LM” = breast and lung cancer leptomeningeal metastasis, respectively. Differences between group means have been determined by nonparametric Wilcoxon signed rank test, and the significance is indicated by asterisks: *P < .05, **P < .001, ***P < .0001. MiR-10b and miR-21 CSF levels normalized to miR-125b are presented in Supplementary Fig. S2A and S2B.
We examined miR-10b levels in the CSF samples of our study cohort patients and detected miR-10b–specific quantitative (q) RT-PCR product in the CSF of 17 of 19 GBM patients (89%, Fig. 1B). This is consistent with our previous finding of miR-10b expression in ∼90% of GBM tumors.17 MiR-10b was also detected in the CSF of 81% of patients with brain and leptomeningeal metastasis of both breast and lung cancer (Fig. 1B). In some instances of both GBM and metastatic brain cancer, miR-10b was present in CSF samples, while CSF cytology analysis produced negative results, suggesting that miR-10b may serve as a more sensitive CSF marker for brain cancer detection compared with CSF cytology.
None of the patients with various nonneoplastic neurological conditions showed detectable levels of miR-10b at 40 cycles of the qRT-PCR reaction. Raw qRT-PCR Ct values representing specific CSF levels of miR-10b, and other miRNAs are shown in Supplementary material, Table S2. Therefore, miR-10b in CSF is a highly indicative marker of high-grade primary and metastatic brain cancers.
We next assessed CSF levels of another candidate miRNA, miR-21, which is the most common miRNA elevated in GBM and other cancers25 and also most strongly upregulated in GBM compared with normal brain (Fig. 1A). We found that miR-21 levels were significantly increased in the CSF of most GBM and metastatic patients relative to its levels in the control CSF samples (Fig. 1C), suggesting that miR-21 may represent an additional CSF biomarker for both GBM and metastatic brain cancer.
The levels of 3 additional candidate miRNAs upregulated in GBM relative to normal brain, miR-15b, miR-17-5p, and miR-93 (Fig. 1A), were determined in a randomly selected set of several CSF samples. The levels of all 3 miRNAs were slightly higher in the CSF of GBM and metastatic brain cancer patients relative to the nonneoplastic controls; however, these differences did not reach significance and were abolished by data normalization to both miR-24 and miR-125b (Supplementary Fig. S4).
MiR-200 Family in the CSF Is Indicative of Brain Metastasis
MiR-10b is expressed in most extracranial tissues26,27 (Supplementary Fig. S5) and is abundant in blood serum.28 However, it is not expressed in brain and not detectable in the CSF of noncancer patients. Therefore, miR-10b and other miRNAs seem unlikely to pass the blood–brain barrier under nonneoplastic conditions, and miRNAs in CSF might therefore reflect a unique miRNA signature of brain. On the other hand, miR-10b is highly expressed in breast and lung tissues, and its presence in the CSF of lung and breast cancer patients with CNS metastasis indicates that metastatic cells bring their signature miRNAs to CSF. Based on these data, we have searched for other miRNA CSF biomarkers that could enable discrimination between GBM and metastatic brain tumors. Such miRNAs should be highly expressed in a primary carcinoma or tissues of its origin (eg, lung or breast) but not in brain or GBM.
According to miRNA profiling across different tissues, miRNAs of the miR-200 family are good candidates fulfilling this criterion. All members of this family are highly expressed in lung and breast tissues and epithelial cancers, including lung and breast carcinomas, but are barely detectable in brain27,29 (see Supplementary Fig. S6). On the other hand, we have found that the miR-200 family, unlike miR-10b, is not expressed in GBM and other primary brain tumors, making it a putative biomarker for metastatic brain cancer (Fig. 2A).
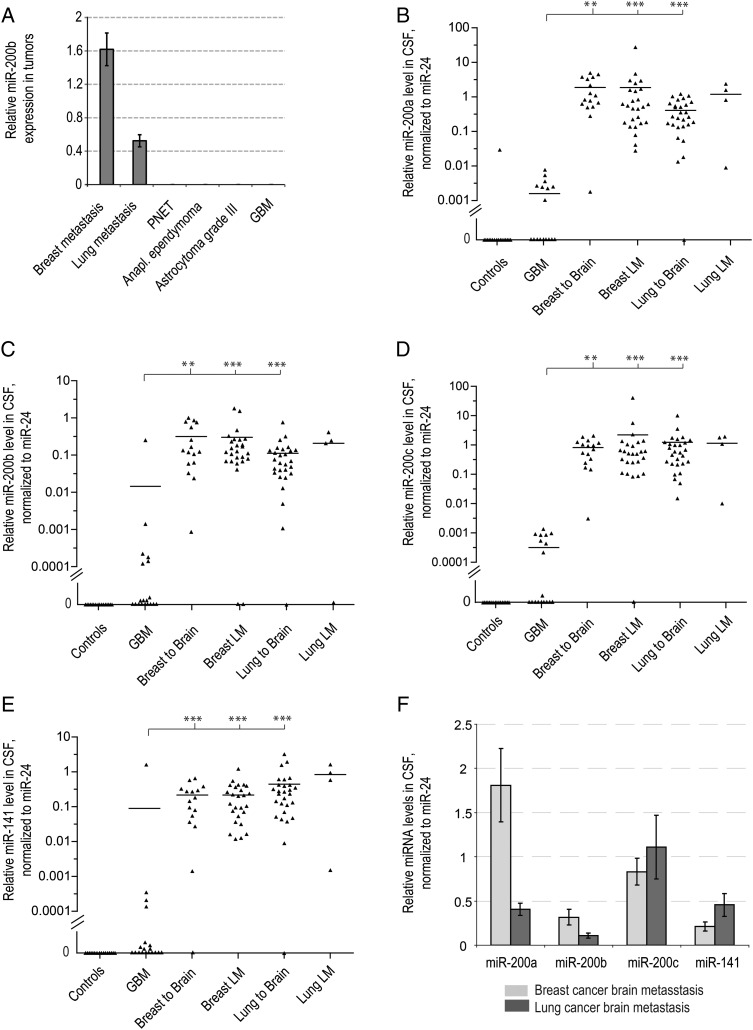
Fig. 2.
Detection of miRNAs of miR-200 family in metastatic brain cancer patients. (A) MiR-200b expression levels were examined by qRT-PCR in various primary and metastatic brain tumor tissue specimens and normalized to miR-24, as described in Materials and Methods. Error bars indicate standard errors between technical duplicates. PNET = primitive neuroectodermal brain tumor. MiR-200a (B), miR-200b (C), miR-200c (D), and miR-141 (E) levels were examined by qRT-PCR in the CSF samples of neurological patients. MiRNA levels are normalized to miR-24 and demonstrated for individual patients. There are 9 samples with undetectable miR-200 levels (values = 0) present in the GBM group. Horizontal lines indicate arithmetic mean for each group of samples. Differences between group means that reached statistical significance, as determined by a nonparametric Wilcoxon signed rank test, are indicated with asterisks: *P < .05, **P < .001, ***P < .0001. Corresponding values normalized to miR-125b are presented in Supplementary Fig. S2C–F. (F) The average levels of miR-200a/miR-200b and miR-141/miR-200c cluster miRNAs in CSF of metastatic brain cancer patients. The error bars represent the standard error of the mean for each group of patients.
To explore a potential of miRNA-200 to distinguish between GBM and metastatic brain cancer, we assessed the levels of 4 miR-200 family members, miR-200a, miR-200b, miR-200c, and miR-141, in the CSF of control, GBM, and metastatic brain cancer patients. Remarkably, all 4 miRNAs were highly expressed in the majority of CSF samples collected from the patients with brain and leptomeningeal metastasis, but not in the control or GBM cases (Fig. 2B–E). These data suggest that miR-200 levels might be used for discriminating between primary brain cancer and brain metastasis.
In an attempt to discriminate between metastasis from breast versus lung cancer, we assessed miR-195 levels in several randomly selected CSF samples, since circulating miR-195 was proposed as a differential biomarker of breast versus lung cancer.30 However, we found no significant difference in miR-195 levels in CSF of breast and lung cancer metastasis patients (Supplementary Fig. S7). Another miRNA, miR-1, is expressed at higher levels in breast versus lung tissue according to miRNA expression profiles,27 but we found miR-1 undetectable in the CSF of both the breast and lung cancer cohorts of patients. Breast and lung carcinomas express strikingly similar miRNA repertoire.26 However, we noted significantly higher amounts of miR-200a and miR-200b (2 miRNAs encoded as a cluster at chromosome 1p36.33) in the CSF of the patients with breast cancer relative to lung cancer, while CSF levels of miR-141 and -200c (coencoded at chromosome 12p13.31) were similar in breast and lung cancer cases (Fig. 2F). These data suggest that the ratios between miRNAs of 2 different miR-200 genomic clusters in CSF may be informative for discrimination between brain metastasis from breast versus lung cancer.
Computational Classification of High-Grade Brain Malignancies Based on CSF MiRNA Profiling
The relationships discovered between miRNA CSF levels and diagnostic outcomes are illustrated by a simple diagnostic tree (Fig. 3A). We next tested whether the samples could be classified more accurately (nonneoplastic control vs GBM vs metastasis) using a “machine-learning technique” based on the SVM concept. This technique was previously applied to a wide range of biological problems, including mRNA and miRNA expression data analysis in cancers.31–33
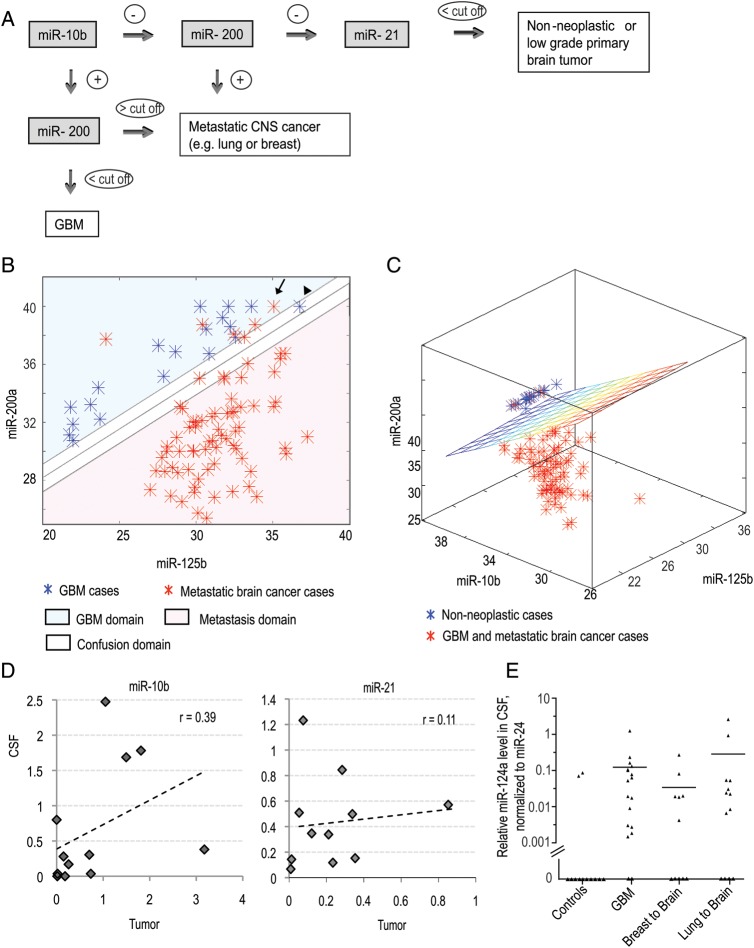
Fig. 3.
Computational classification of high-grade brain malignancies based on CSF miRNA profiling and the origin of miRNA in CSF. (A) Classification tree of brain cancer patients based on CSF miRNA biomarkers (miR-10b, -21, and -200). (B and C) Application of SVM with linear kernel to classification of specimens: (B) classification of GBM vs metastatic brain cancer on the basis of Ct levels of 2 miRNAs (miR-200a and miR-125b) in the CSF. The Ct values of corresponding miRNAs are plotted. A specimen is classified as GBM/metastasis depending on whether the Ct levels of the 2 miRNAs place it above/below the separation line. The line illustrates the automatically generated classifier. The arrow depicts a misclassified sample and the arrowhead depicts a “confused” sample. (C) Classification of GBM and brain metastasis versus nonneoplastic control cases on the basis of Ct levels of 3 miRNAs (miR-10b, miR-200a, and miR-125b) in the CSF. A specimen is classified to a particular group depending on whether its Ct levels place it below/above the separation plane. The plane illustrates the automatically generated classifier. (D) The relation between miR-10b and miR-21 levels in brain tumors and matching CSF samples collected from the same patients. The Pearson coefficients (r) of linear regression between 2 data sets were calculated for each miRNA. (E) Relative miR-124a levels were determined in CSF of a randomly selected cohort of control and brain cancer patients by qRT-PCR analysis. Relative levels were normalized to miR-24, as described in Materials and Methods and demonstrated for individual CSF samples. Horizontal lines indicate arithmetic mean of miRNA levels for each group of patients.
We applied various SVM algorithms for classification of the samples. In one case (GBM vs metastasis classification) a very simple linear classifier provided discrimination with about 95% accuracy. The levels of 2 miRNAs, miR-200a and miR-125b, were used in this case as independent variables, and a linear function of these 2 Ct levels was employed as a classifier, with the coefficients calculated in the process of the classifier training (Fig. 3B).
Another case that allows for a similar interpretation is the classification of GBM and brain metastasis versus nonneoplastic controls. In that case we constructed a linear classifier that uses Ct levels of 3 miRNAs—miR-10b, miR-200a, and miR-125b—as features. Accordingly, a 2-dimensional plane in the space spawned by the levels of these 3 miRNAs separates the space into the 2 domains as demonstrated in Fig. 3C.
Similarly, we tested various SVM classifiers and found that the RBF kernel provides the best separation between all classes of samples. The best classification accuracy is achieved using the levels of 7 miRNAs—miR-10b, miR-21, miR-125b, miR-141, miR-200a, miR-200b, and miR-200c—as independent variables. This analysis revealed that different types of cancer are distinguished from each other as well as from nonneoplastic controls with the average cross-validation accuracy of about 90% (Table 2). Thus, the SVM incorrectly predicted the class of about 1 of 10 previously unseen samples. This analysis suggests a possibility of computational differential diagnostics of brain cancers using miRNA profiling.
Table 2.
Accuracies of classification of brain tumors by SVM analysis
| Comparison | Instances classified in the test sets, n (%) |
|
|---|---|---|
| Correctly | Incorrectly | |
| GBM vs nonneoplastic controls | 31 (91.2) | 3 (8.8) |
| Metastasis vs nonneoplastic controls | 88 (98.9) | 1 (1.1) |
| GBM and metastasis vs nonneoplastic controls | 105 (97.2) | 3 (2.8) |
| GBM vs metastasis | 89 (95.7) | 4 (4.3) |
| GBM vs non-GBM (all others) | 102 (94.5) | 6 (5.5) |
| Metastasis vs nonmetastasis (all others) | 100 (92.6) | 8 (7.4) |
| Breast vs lung metastasis | 51 (68.9) | 23 (31.1) |
An automatically generated classifier (SVM with RBF kernel) is used for the classification. Cross-validation approach in which the specimens are split into the train and test groups is used for calculating the accuracies. Used as input data for the analysis were levels of miR-10b, miR-21, miR-125b, miR-141, miR-200a, miR-200b, and miR-200c in CSF.
The Origin of MiRNA in CSF
MiRNAs detected in the CSF of brain cancer patients may originate from brain tumor cells, from surrounding brain tissues, or from extracranial tissues due to blood–brain barrier disruption associated with tumors and treatments (eg, radiation). To discriminate between these possibilities we determined miR-10b and miR-21 expression levels in tumor biopsies obtained during brain surgery and corresponding CSF samples from the same patients. To avoid contamination by tumor tissues, the CSF samples were obtained intraoperatively before tumor resection. We observed a trend toward positive correlation between miR-10b expression level in the brain tumor and corresponding CSF specimens, and no such trend for miR-21 (Fig. 3D). Of note, miR-10b is expressed in tumors but not in normal brain tissues, while miR-21 is elevated in tumors but is also present in normal brain.16,22 Taking these expression patterns into account, our data suggest that miRNA composition of the CSF is established by tumor cells as well as by the cells of surrounding brain tissues. Another abundant CNS-specific miRNA, miR-124a, which is absent in metastatic and GBM tumors34 (Fig. 1A), was present in the CSF of many GBM and metastatic cases (Fig. 3E), confirming that miRNA is partially released to CSF through the destruction of the brain tissues. Further, we found miR-200 species detectable in the CSF of approximately 50% of GBM patients, but none of the control nonneoplastic patients, suggesting some amount of transudation of miRNA from circulation (Supplementary Fig. S3C–F). The CSF levels of miR-200 were significantly lower in GBM patients than in patients with miR-200–expressing tumors (eg, breast and lung metastasis) and became negligible following data normalization (Fig. 2B–E and Supplementary Fig. S2C–F). These data further indicate that the major miRNA supply of CSF is provided by the brain and tumor tissues.
MiRNAs in CSF of Brain Cancer Patients as Markers of Disease Activity
To examine whether CSF levels of miRNAs reflect disease status/activity, we studied miRNA in CSF of active GBM and metastatic brain cancer versus tumor remission cases. The disease was considered in remission if, following treatment, there was no evidence of tumor mass detected by MRI and if CSF cytological analysis was negative. Neither miR-10b nor miR-200 family members were detected after 40 cycles of qRT-PCR reaction in CSF samples in any remission case (Supplementary material, Table S3, Fig. S8). MiR-21 levels were significantly lower in cancer remission cases compared with active GBM and metastatic brain cancer cases before treatment (Supplementary Fig. S8B). These data suggest that miRNAs analyzed in this study may reflect the activity of brain tumors.
To further test whether the CSF levels of specific miRNAs reflect disease status/activity and responsiveness to therapy, miRNA levels were determined in the CSF of lung cancer and GBM patients longitudinally during the course of erlotinib treatment. MiRNA analysis was accompanied by MRI, CSF cytology, and clinical monitoring of the disease status. A patient with non–small cell lung carcinoma (patient A) developed parenchymal and leptomeningeal disease during the course of treatment and medication adjustment (Fig. 4A). Erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor, was given orally at the dose of 150 mg daily and increased at time of progression to 600 mg every 4 days and further to 900 mg (at 41 weeks) to achieve higher brain/CSF concentration,35 followed by a prolonged remission. The levels of both miR-10b and miR-200 members in CSF of this patient are concordant with the MRI results, rising during relapse and returning to background levels after the increase of erlotinib dosage (significant reduction by 45 weeks; Fig. 4A).
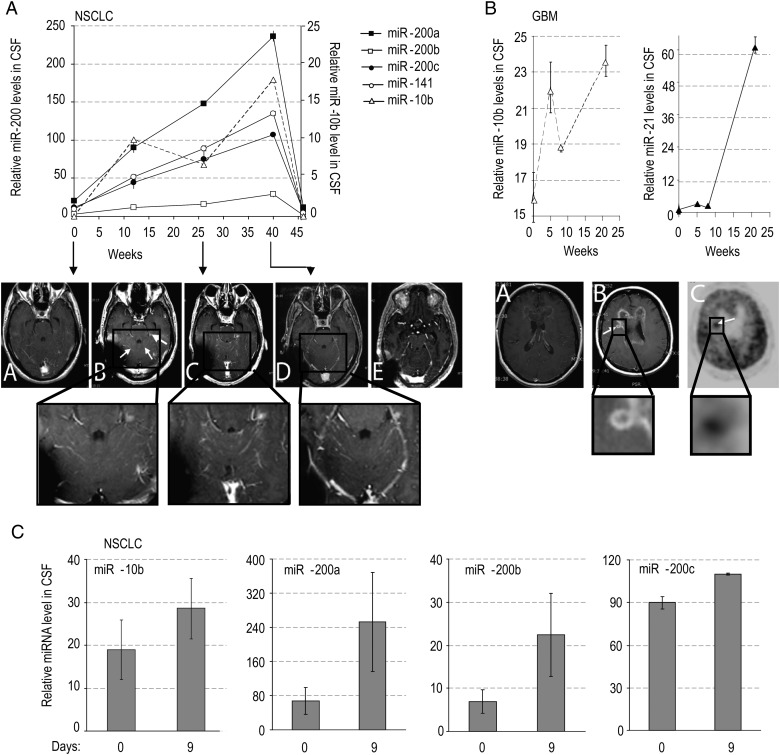
Fig. 4.
CSF levels of miRNA markers in metastatic lung cancer and GBM patients during treatment with erlotinib. MiRNA levels were examined by qRT-PCR in the CSF samples of lung cancer (A and C) and GBM patients (B) during the time course of erlotinib treatment. The disease progression and drug response were concomitantly monitored by MRI, as follows. For Patient A: serial axial postgadolinium MRIs of the lung cancer patient's brain during the course of progression of disease and stability and improvement on MRI with escalating doses of erlotinib. A: At 0 weeks, while the patient was taking erlotinib, there was no leptomeningeal and parenchymal enhancement and CSF cytology was negative; B: at 3 weeks progression on erlotinib at 150 mg daily dosing with new cerebellar leptomeningeal enhancement (small arrows) and nodule (large arrow), erlotinib increased to 600 mg every 4 days at 9 weeks; C: at 29 weeks on showing stable leptomeningeal enhancement and nodule; D: at 40 weeks showing reduction in leptomeningeal enhancement and nodule, erlotinib increased to 900 mg every 4 days at 41 weeks; E: at 64 weeks, after 6 cycles of chemotherapy with carboplatinum and pemetrexed due to lung cancer progression, showing further reduction in leptomeningeal enhancement and nodule has disappeared. For Patient B: A: At 2 weeks for patient with recurrent GBM with prominent mass effect and enhancement felt to be radiation changes rather than tumor based on MRI spectroscopy and FDG-PET scan, on erlotinib at 600 mg every 4 days; B: at 26 weeks on treatment, showing progression on MRI with new lesion (arrow) concerning for tumor; C: at 27 weeks on treatment showing hypermetabolic area (arrow) on PET consistent with tumor and biopsy confirmed. For Patient C: had inadequate treatment due to functional status and rapidly progressed over a few weeks, which was reflected by an increase in levels of miR-200 family members in a short interval.
Patient B (GBM, postradiation; Fig. 4B) initially exhibited negative cytological CSF analysis, and his status was interpreted as pseudoprogression based on MRI spectroscopy and fluorodeoxyglucose (FDG)-PET negative enhancing disease. CSF level of miR-21 in this patient was initially comparable to the levels in the control (nonneoplastic) group, while miR-10b was significantly elevated. A dramatic increase in miR-21 levels and further increase in miR-10b levels at a later time (25 weeks) indicated disease progression that was confirmed by MRI, PET-CT, and repeat biopsy of the lesion. Neither miRNA was behaving individually as a simple biomarker of tumor burden in this case; nevertheless, these data are suggestive of high sensitivity of miRNA as biomarkers. A further extended study is needed to evaluate CSF miRNAs as markers of GBM progression versus pseudoprogression. Patient C (Fig. 4C) had inadequate treatment due to functional status and rapidly progressed over a few weeks, which was reflected by an increase in levels of miR-200 family members.
Altogether, these data indicate for the first time that CSF miRNA levels may serve as biomarkers of brain cancer progression and response to therapy.
Discussion
In this study, we developed a new approach for detection and monitoring of different types of brain cancer by examining specific miRNAs in CSF. We have chosen a few candidate miRNAs based on prior knowledge about their expression profiles in normal brain versus various brain cancers and other tissues. Moreover, important pleiotropic roles of our top candidates miR-10b and miR-21 in gliomagenesis are well established.17,18,22,36,37 Similarly, miR-200 function in epithelial-to-mesenchymal transition and metastasis of epithelial tumors has been intensively investigated.38
MiR-10b was our top priority candidate because it is the only known miRNA that is absent in normal brain but expressed in GBM.18,22 We detected miR-10b in CSF of the majority (89%) of GBM patients but not in the patients with noncancer neuropathology. Consistently, undetectable miR-10b levels in CSF of healthy individuals were observed in the study of others.10 Since miR-10b is highly expressed in various extracranial tissues, the absence of miR-10b in the CSF of control patients indicates that it may not pass the blood–brain barrier under nonneoplastic conditions. If the blood–brain barrier is not permeable for at least some miRNAs, that would create a unique advantage for miRNA profiling of CSF as a diagnostic/monitoring tool for brain conditions. Profiling of other biofluids would lack this specificity of the CSF signature.
Since miR-10b is robustly expressed in breast and lung cancers and is detected in the CSF of patients with brain metastasis from these organs, we hypothesized that metastatic cells transport their miRNA signature to CNS, which could be used for discrimination between primary and metastatic brain tumors. On the basis of this hypothesis, we searched for miRNA markers that are absent in brain and GBM but abundant in breast and lung tissues, such as the miR-200 family. Indeed, we found that the presence of 4 different miR-200 family members in CSF is highly specific for metastatic brain tumors but not GBM. Furthermore, the ratio between different miR-200 members in the CSF appeared informative of the origin of metastasis (breast vs lung cancer). Breast and lung cancers are the most frequent causes of brain metastasis, accounting for ∼50% of total brain tumors. We anticipate the demonstrated recognition of these tumors by miRNA CSF signature to be extended to other types of brain cancer, primary as well as metastatic, such as melanoma, kidney, and colon cancer. The unique miRNA signatures of these tissues may efficiently serve as CSF biomarkers.
The SVM-based computational analysis of our data revealed that a well-established computational algorithm could be used for “automated diagnostics” on the basis of acquired qRT-PCR numerical data. The results demonstrate that the power of miRNA-based assessment is about 91%–99%, depending on the task, when the detection levels of 7 miRNAs (miR-10b, miR-21, miR-141, miR-200a, miR-200b, miR-200c, and miR-125b) are used as input data. The levels of several other miRNAs analyzed on a small pilot set of samples (eg, miR-1 and miR-195) did not demonstrate reliable differences between the groups and therefore were not further studied. However, we suggest that finding additional miRNA biomarkers and the follow-up analysis of larger cohorts of patients may further increase the accuracy of the approach. Particularly, another miRNA, miR-15b, was recently shown to be elevated in the CSF of patients with GBM.15 Although our limited pilot study of several GBM patients has not confirmed this finding, a larger-scale study may be needed to validate the utility of this miRNA as a biomarker of brain cancer.
Remarkably, characteristic tumor miRNAs, miR-10b and miR-200, were not detected in the CSF of cancer remission cases, strongly suggesting that these miRNAs may also reflect disease status/activity. Further, the correlation between miR-10b/miR-200 CSF content and MRI data observed for GBM and lung cancer patients in a time course of erlotinib treatment is the first indication that miRNAs in CSF may serve as biomarkers of brain cancer progression and responsiveness to treatment. However, because these observations are based on individual cases, larger prospective studies have to be done to validate them.
In this study the patient control group did not include subjects with enhancement of nonneoplastic origin. Enhancing lesions may influence CSF composition through their own local pathology and associated phenomenon of transudation. A prospective study on such subjects will be important to determine the full and varied utility of discovered miRNA markers for discrimination between enhancing lesions of neoplastic and nonneoplastic nature (eg, cancer versu ischemia, hemorrhage, or pseudoprogression). In this regard, further validation of miRNAs as markers of tumor progression versus pseudoprogression will be especially valuable.
There are several advantages of using miRNAs as biomarkers for brain cancer compared with other biomarkers, such as mRNA and proteins. MiRNAs are highly stable, and their detection by qRT-PCR is quantitative, extremely sensitive (requiring only nanograms of starting RNA material), highly specific, and reproducible. The fact that the majority of CSF samples used in this study were obtained through lumbar puncture suggests that this relatively simple medical procedure is sufficient for miRNA analysis and can potentially provide an accurate diagnosis of brain cancers. While this manuscript was being prepared for submission, a separate report demonstrating the usefulness of miRNA profiling in CSF for the diagnosis of glioma was published,15 further underscoring the feasibility of this approach for clinical application. The use of CSF biomarkers would be of particular value in management of patients who are not surgical candidates due to tumor size, tumor location, or underlying medical condition. As the miRNA biomarkers reported here have been selected based on a number of independent profiling studies that utilized resected tumors as well as tumor biopsies,34 most likely these markers will also be useful for diagnostic evaluation and management of patients who are inoperable.
In summary, our analysis suggests that miRNA profiling of CSF allows detection of GBM and metastatic brain cancers and discrimination between these tumors with a high level of confidence. At the same time, the pilot longitudinal study results indicate that a CSF miRNA profiling–based technique might be developed for disease monitoring and management (eg, assessment of relapses and remissions, after treatment follow-up, and examination of chemo- and radiotherapy efficacy) which can help determine the most effective strategy of treatment.
Supplementary Material
Funding
This work was supported by National Institutes of Health grants [R01CA138734-01A1 to A.K., K08CA124804, ARRA 3P30CA023100-25S8 to S.K.]; Sontag Foundation [Distinguished Scientist Awards to A.K., Distinguished Scientist Award to S.K.]; and James S. McDonnell Foundation award to S.K. Clinical specimen banking and database are supported by generous donations from patients to S.K.
Supplementary Material
Acknowledgments
We thank Dr. Caifu Chen and Applied Biosystems for supplying miRNA RT and qPCR primers. We also thank Dr. Kai Sonntag and Dr. Erik Uhlmann for reviewing the manuscript.
Conflict of interest statement. None declared.
References
- 1.Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol. 2008;10(1):79–87. doi: 10.1215/15228517-2007-038. doi:10.1215/15228517-2007-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grewal J, Saria MG, Kesari S. Novel approaches to treating leptomeningeal metastases. J Neurooncol. 2012;106(2):225–234. doi: 10.1007/s11060-011-0686-2. doi:10.1007/s11060-011-0686-2. [DOI] [PubMed] [Google Scholar]
- 3.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. doi:10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 4.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. doi:10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. doi:10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 6.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. doi:10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 7.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. doi:10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. doi:10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 10.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. doi:10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117(11):3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 12.Birks DK, Barton VN, Donson AM, et al. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56(2):211–216. doi: 10.1002/pbc.22723. doi:10.1002/pbc.22723. [DOI] [PubMed] [Google Scholar]
- 13.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nass D, Rosenwald S, Meiri E, et al. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19(3):375–383. doi: 10.1111/j.1750-3639.2008.00184.x. doi:10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14(1):29–33. doi: 10.1093/neuonc/nor169. (2011) doi:10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. doi:10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 17.Gabriely G, Yi M, Narayan RS, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71(10):3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. doi:10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasayama T, Nishihara M, Kondoh T, et al. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasivefactors, uPAR and RhoC. Int J Cancer. 2009;125(6):1407–1413. doi: 10.1002/ijc.24522. doi:10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 19.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. doi:10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 20.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. doi:10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 21.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. doi:10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. doi:10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219(2):214–221. doi: 10.1002/path.2586. doi:10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. doi:10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 25.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. doi:10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. doi:10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56(12):1871–1879. doi: 10.1373/clinchem.2010.147553. doi:10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. doi:10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heneghan HM, Miller N, Kelly R, et al. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15(7):673–682. doi: 10.1634/theoncologist.2010-0103. doi:10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Li HR, Fan JB, et al. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics. 2006;7:202. doi: 10.1186/1471-2105-7-202. doi:10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. doi:10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 33.Keller A, Leidinger P, Borries A, et al. miRNAs in lung cancer - studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. doi:10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonntag KC, Woo TU, Krichevsky AM. Converging miRNA functions in diverse brain disorders: A case for miR-124 and miR-126 [published online ahead of print December 8, 2001] Exp Neurol. doi: 10.1016/j.expneurol.2011.11.035. doi:10.1016/j.expneurol.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackman DM, Holmes AJ, Lindeman N, et al. Response and resistance in a non-small-cell lung cancer patient with an epidermal growth factor receptor mutation and leptomeningeal metastases treated with high-dose gefitinib. J Clin Oncol. 2006;24(27):4517–4520. doi: 10.1200/JCO.2006.06.6126. doi:10.1200/JCO.2006.06.6126. [DOI] [PubMed] [Google Scholar]
- 36.Schramedei K, Morbt N, Pfeifer G, et al. MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene. 2011;30(26):2975–2985. doi: 10.1038/onc.2011.15. doi:10.1038/onc.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13(6):580–590. doi: 10.1093/neuonc/nor033. doi:10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. doi: 10.4161/rna.5.3.6558. doi:10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.