Abstract
Glioblastomas present as diffuse tumors with invasion into normal brain tissue and frequently recur or progress after radiation as focal masses because of glioma-initiating cells. The role of the urokinase-type plasminogen activator receptor (uPAR) and cathepsin B in stem-like phenotype has been extensively studied in several solid tumors. In the present study, we demonstrated that selection of glioma-initiating cells using CD133 expression leads to a specific enrichment of CD133+ cells in both U87 and 4910 cells. In addition, CD133+ cells exhibited a considerable amount of other stem cell markers, such as Nestin and Sox-2. Radiation treatment significantly enhanced uPAR and cathepsin B levels in glioma-initiating cells. To downregulate radiation-induced uPAR and cathepsin B expression, we used a bicistronic shRNA construct that simultaneously targets both uPAR and cathepsin B (pCU). Downregulation of uPAR and cathepsin B using pCU decreased radiation-enhanced uPAR and cathepsin B levels and caused DNA damage-induced apoptosis in glioma cell lines and glioma-initiating cells. The most striking finding of this study is that knockdown of uPAR and cathepsin B inhibited ongoing transcription by suppressing BrUTP incorporation at γH2AX foci. In addition, uPAR and cathepsin B gene silencing inversely regulated survivin and H2AX expression in both glioma cells and glioma-initiating cells. Pretreatment with pCU reduced radiation-enhanced expression of uPAR, cathepsin B, and survivin and enhanced DNA damage in pre-established glioma in nude mice. Taken together, our in vitro and in vivo findings suggest that uPAR and cathepsin B inhibition might serve as an adjunct to radiation therapy to target glioma-initiating cells and, therefore, for the treatment of glioma.
Keywords: apoptosis, glioma-initiating cells, γH2AX, radiation
Glioblastoma (GBM) is one of the most frequently occurring primary brain tumors and is characterized by aggressive clinical behavior and biologic heterogeneity. GBMs are the most lethal brain tumor, with a median survival of less than 12 months because of resistance to radiation and other treatments.1 GBMs present as diffuse tumors with invasion into normal brain tissue but frequently recur or progress after radiation as focal masses, which suggests that only a fraction of tumor cells is responsible for regrowth; these cells are known as glioma-initiating cells (GICs) or cancer stem cells (CSC).2 Despite controversial results CD133 is the most accredited stem cell marker that confers radioresistance to glioma.3–5 The human CD133 protein has recently gained attention as a cell-surface marker that can be used to isolate GICs.6
The histone variant H2AX is a major guardian of the genome and has been previously associated with cell death after cytotoxic therapy.7 In addition, depletion of H2AX has been demonstrated to cause genome instability and, consequently, carcinogenesis.8,9 Furthermore, H2AX knockout mice are prone to develop tumors.10 H2AX is involved in various molecular events, such as DNA damage response, chromatin remodeling, gene silencing, and apoptosis.11,12 In addition, phosphorylation of H2AX is reported to inhibit transcription by inhibiting assembly of the transcription complex without heterochromatin formation.13,14
Apoptosis is an evolutionarily conserved multistep cascade regulated by proteins that promote or counteract apoptotic cell death. Survivin, a unique member of inhibitors of apoptosis proteins (IAPs), has recently attracted many researchers because of its broad distribution, evolutionary conservation, and anti-apoptotic activity.15 Moreover, survivin has greater potential for inhibition of apoptosis than any other apoptotic inhibitor, including Bcl2.16 Recently, the anti-apoptotic function of survivin in relation to H2AX was reported in urothelial carcinoma.17
uPAR anchors uPA to the cell membrane, interacts with many potential ligands, and associates with several members of integrin family, which indicate its strong involvement in cell adhesion and migration.18 uPAR is also involved in the initiation of several intracellular signal-transduction pathways, including MEK–ERK and JAK–STAT pathways that involve cytoskeletal components and cytosolic and transmembrane kinases.19 Both uPAR and cathepsin B are known to be associated in close proximity to αVß3 integrins and have been implicated in their ability to initiate signaling events.20 uPAR and cathepsin B have been implicated in the maintenance of the malignant phenotype in glioma.21,22 Recent reports demonstrated that uPAR and cathepsin B are also involved in the maintenance of the stem-like phenotype in solid tumors.23,24 However, the role of uPAR and cathepsin B has not been reported in glioma stem cells. In the present study, uPAR and cathepsin B were targeted using small interfering RNA (siRNA) to understand the molecular mechanisms involved in the glioma stem-like phenotype. Our previous study has demonstrated that gene silencing using siRNA against uPAR and cathepsin B induced apoptosis in glioma.25
In the present study, we demonstrate that downregulation of uPAR and cathepsin B enhanced radiation-induced apoptosis in glioma cell lines (non-GICs) and GICs. Moreover, knockdown of uPAR and cathepsin B decreased survivin expression by inhibiting run-on transcription in both non-GICs and GICs. Our gene silencing experiments further demonstrate that uPAR and cathepsin B downregulation enhanced expression of γH2AX and H2AX by inhibiting c-Met signaling. Therefore, the activation of programmed cell death or apoptosis using siRNA against uPAR and cathepsin B in CSCs could be a promising strategy for the treatment of glioma.
Materials and Methods
Ethics Statement
The Institutional Animal Care and Use Committee of the University of Illinois College of Medicine at Peoria approved all surgical interventions and post-operative animal care. The consent was written and approved.
Cell Lines
The U87 glioma cell line was obtained from the American Type Culture Collection; 4910 glioma xenograft cells were kindly provided by Dr. David James (University of California-San Francisco, San Francisco, CA). U87 and 4910 cells were grown in DMEM medium and RPMI 1640 medium, respectively, and supplemented with 10% FBS and 1% penicillin/streptomycin. GICs were isolated from the U87 glioma cells and 4910 glioma xenograft cells (non-GICs) and cultured in DMEM/F-12 medium containing LIF (10 ng/mL), hEGF (20 ng/mL), bFGF (20 ng/mL), and N2 supplements.
CD133 Staining and FACS Sorting
For FACS analysis and purification, glioma cells enriched in DMEM/F-12 medium plus growth factors were dissociated, washed, and incubated with PE-conjugated CD133 antibody (Milteny Biotech) at a dilution of 1:10 in phosphate-buffered saline (PBS)–bovine serum albumin (BSA) for 30 min at 4°C. For the control, cells were incubated with an isotype IgG antibody. Dead cells were analyzed and excluded using trypan blue at 1:1000 (FL3 channel). Expression level analysis and sorting were done on FACScan and FACSAria, respectively (BD Bioscience). Purified cells were collected and cultured in DMEM/F-12 medium plus growth factors.
Secondary Neurosphere Formation Assay
After primary spheres formed and reached 100–200 cells, they were dissociated into single cells and plated onto a 96-well plate for the secondary neurosphere formation assay by limiting dilution as described previously.26 In brief, the cells in single cell suspension were diluted and plated at 1–2 cells/well. Cells were fed by changing half the medium every 2 days. The wells were scored for sphere formation after 14 days.
Culture and Transfection Conditions
Unless otherwise mentioned, all cultures were performed in 100-mm culture plates. All transfections were performed using FuGene HD transfection reagent according to the manufacturer's protocol (Roche Applied Science). U87 and 4910 cells (non-GICs or GICs) were transfected with scrambled vector (pSV), a bicistronic shRNA construct targeting uPAR and cathepsin B (pCU) or siRNA against H2AX (pH2AX) (SC: 62464; Santa Cruz Biotechnology) as described elsewhere.27 At 48 h (non-GICs) or 24 h (GICs) after transfection, they were treated with 10 Gy radiation and incubated for another 24 or 48 h in CO2 incubator. for overexpression of H2AX, non-GICs or GICs were transfected with a plasmid expressing full-length human cDNA clone of H2AX (FLH; SC 125294; OriGene Technologies) for 24 h and treated with 10 Gy radiation for another 24 h. Met Inhibitor-PHA-665752 hydrate (10µM) (PHA) was used in the present study.
Immunofluorescent Assay
For immunofluorescent staining of undifferentiated neurospheres, spheres were plated onto 4-well chamber slides in DMEM/F-12 medium containing 10% FBS for 2 h. Cells were then fixed with 4% paraformaldehyde and stained with anti-CD133, anti-Nestin, anti-Sox-2, and anti-Tuj-1 antibodies and analyzed with a confocal microscope (Olympus BX61 Fluoview). Overlay of images was done using SPOT Advanced software (for Windows, version 4.0.8).
FACS analysis
For identification of stem cell markers, U87 and 4910 GICs were dissociated, washed with PBS, pelleted at 1000 rpm for 5 min, and resuspended at a concentration of 1 × 106 cells/mL in PBS. Cells were then incubated with HRP-conjugated or fluorescein-conjugated CD133, Nestin, Sox-2, and Tuj-1 antibodies for 1 h on ice; pelleted; and washed 3 times with PBS to remove excess primary antibody. Isotype IgG was used as a control. Cells that were incubated with HRP-conjugated primary antibodies were then resuspended in 1 mL of PBS containing Alexa Fluor-labeled secondary antibody and incubated for 1 h on ice. After 3 washes with PBS, cells were analyzed on a Counter EPICS XL AB6064 flow cytometer (Beckman Counter). For cell cycle analysis, U87 and 4910 non-GICs and GICs were washed with PBS, pelleted at 1000 rpm for 5 min, and resuspended in 50 µg/mL of propidium iodide at a concentration of 1 × 106 cells/mL. After 30 min of incubation at 4°C, the DNA content was analyzed using a flow cytometer.
TUNEL Assay and Run-On Transcription Assay
TUNEL assay was performed as described previously25 using an In Situ Cell Death Detection Kit (Roche Diagnostics). In run-on transcription experiments, BrUTP incorporation into cell nuclei for the detection of RNA synthesis was performed using a scratch labeling method.13 Here, GICs and non-GICs were plated onto 4-well chamber slides and treated with pCU and radiation as described previously, and 150 µL of medium containing BrUTP (SC: 70443, Santa Cruz Biotechnology) at a final concentration of 5 mM were added to each well. With the tip of a hypodermic needle, a series of parallel scratches were made about 0.5 mm apart in 2 directions: from top to bottom and from left to right. After 30 min, cells were washed with PBS, fixed with ice-cold 4% paraformaldehyde solution in PBS for 10 min, and placed in 70% ethanol at 4°C overnight. After fixing, cells were incubated with a mixture of anti-BrdU antibody plus anti-phospho H2AX antibodies for 1 h, subsequently counterstained with Alexa Fluor-conjugated secondary antibodies, and analyzed under a confocal microscope (Olympus BX61 Fluoview). Overlay of images was done using SPOT Advanced software (for Windows, version 4.0.8).
Western Blotting and Immunoprecipitation
For Western blot analysis, equal amounts of protein fractionated on SDS-PAGE were immunoblotted with primary antibodies and subsequently incubated with species-specific, HRP-conjugated secondary antibodies (Santa Cruz Biotechnology). Signals were detected using the ECL Western blotting detection system (Pierce). Unless otherwise mentioned, all primary antibodies and species-specific secondary antibodies conjugated to HRP and Alexa Fluor used in this study were obtained from Santa Cruz Biotechnology.
uPAR was immunoprecipitated from 500 µg of total protein using anti-uPAR antibody and protein A coupled with G agarose beads (20 µg). The precipitates were washed 5 times with lysis buffer and once with PBS. The pellet was then resuspended in 1X sample buffer (50 mM Tris [pH, 6.8], 100 mM bromophenol blue, and 10% glycerol), incubated at 90°C for 10 min before electrophoresis to release the proteins from the beads, and immunoblotted for p-Met.
Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from the transfected cells using TRIzol reagent (Invitrogen) according to standard protocol. DNase-treated RNA was used as a template for reverse transcription followed by PCR analysis using specific primers as listed below. The PCR conditions were as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 45 s. GAPDH was used as an internal control.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| uPAR | 5′GCCTTACCGAGGTTGTGTGT3′ | 5′CATCCAGGCACTGTTCTTCA3′ |
| Cathepsin B | 5′GCTACAGCCCGACCTACAAA3′ | 5′CCAGTAGGGTGTGCCATTCT3′ |
| CD133 | 5′CCAAGTTCTACCTCATGTTTGG3′ | ACCAACAGGGAGATTGCAAAGC3′ |
| Sox-2 | 5′CACCTACAGCATGTCCTACTC3′ | 5′CATGCTGTTTCTTACTCTCCTC3′ |
| Nestin | 5′CGTTGGAACAGAGGTTGGAG3′ | 5′TCCTGAAAGCTGAGGGAAG3′ |
| TUJ-1 | 5′CTGCTGGCAGCTGGAGTGAG3′ | 5′CATAAATACTGCAGGAGGGC3′ |
| Survivin | 5′ACCACCGCATCTCTAC3′ | 5′TCCTCTATGGGGTCGT3′ |
| H2AX | 5′GGCCTCCAGTTCCCAGTG3′ | 5′TCAGCGGTGAGGTACTCCAG3′ |
| GAPDH | 5′CGGAGTCAACGGATTTGGTCGTAT3′ | 5′AGCCTTCTCCATGGTGGTGAAGAC3′ |
Immunohistochemical Analysis
Stereotactic implantation of U87 and 4910 non-GICs and GICs (1 × 105) was performed as described previously.28 Mice were divided into the following groups: control, radiation alone, pCU treatment alone, and radiation plus pCU. Radiation was given in 2 doses (5 Gy on days 8 and 10), and pCU treatment was given using Alzet mini-pumps at a flow rate of 0.25 µL/h. Mice were observed for 30 days. Once control animals showed chronic symptoms (about 30 days), the animals were euthanized by cardiac perfusion using 10% buffered formalin. The brains were removed, stored in 10% formalin, and embedded in paraffin according to standard protocols. Paraffin-embedded tumor sections were subjected to rehydration by passing through a series of xylene and 100% and 90% ethanol. Sections were stained with hematoxylin and eosin (H&E) to characterize tumor growth as described previously.25,29 In addition, tumor sections were stained for cathepsin B, uPAR, survivin, and H2AX and analyzed as described previously.
Results
Isolation and Identification of GICs
GBMs are heterogeneous, aggressive neoplasms containing GBM stem cells or GICs that exhibit the capacity for unlimited growth as multicellular spheres in medium and efficient tumor initiation in immunodeficient animals. Therefore, isolation and characterization of GICs have been considered to be extremely important for better understanding the biochemical/molecular pathways that support the stem-like tumor-initiating phenotype.
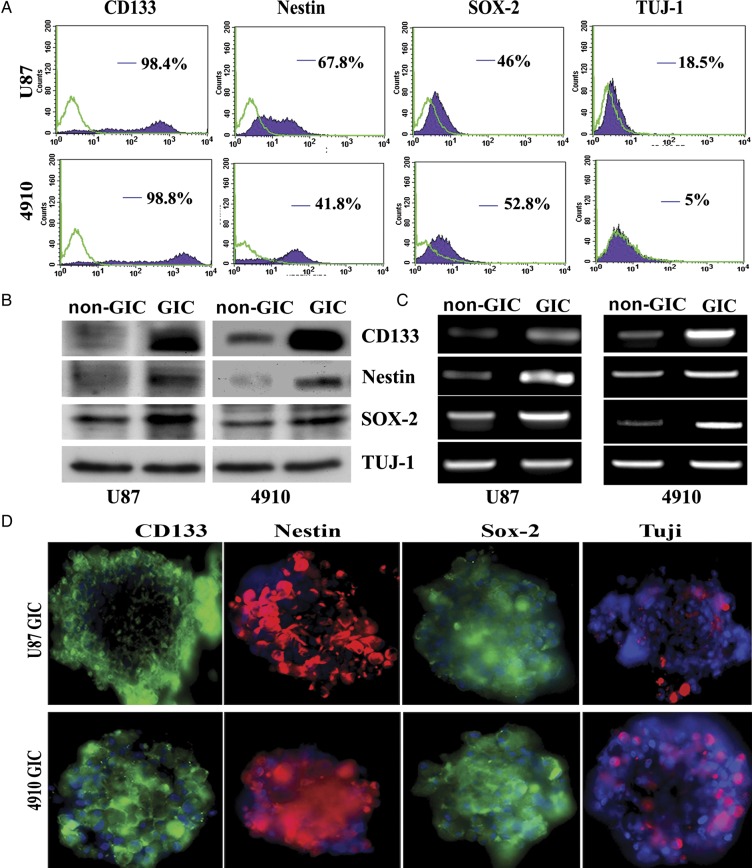
To determine whether there is a population of cells with the unique characteristics of stem cells and distinct from normal glioma cells, we studied U87 glioma cells and 4910 glioma xenograft cells. Primary cultures were grown as monolayers attached to the bottom of flasks in a medium containing FBS. We switched the glioma cells into culture conditions known to be permissive for stem cell proliferation,26 and neurosphere-like colonies with 100–200 cells appeared in 1 week (Supplementary Fig. S1A). The percentage of CD133+ cells were evaluated in primary glioma cells and freshly dissociated glioma sphere cultures using flow cytometry (Supplementary Fig. S1B). The results show that U87 and 4910 glioma cells enriched in DMEM/F-12 medium plus growth factors contained 12% and 18% CD133+ cells, respectively, whereas glioma cells cultured in their respective growth medium exhibited less than 4% CD133+ cells. Furthermore, isolated GICs of U87 and 4910 cells showed 98.4% and 98.8% expression of CD133, respectively, when subjected to FACS analysis (Fig. 1A). CD133+ cells also exhibited a considerable amount of other stem cell markers, including Nestin (67.8% in U87 and 41.8% in 4910 GICs) and Sox-2 (46% in U87 and 52.8% in 4910 GICs), at the cell-surface level. However, expression of Tuj-1, a lineage marker, was significantly less in both U87 (18.5%) and 4910 (5%) GICs. At the protein and mRNA levels, significant enrichment of CD133, Nestin, and Sox-2 was found in U87 and 4910 CD133+ fractions, compared with non-GICs; however, there was no significant change in the expression of Tuj-1 (Fig. 1B–C). Immunofluorescence assay further confirmed the expression of CD133, Nestin, Sox-2, and Tuj-1 in both U87 and 4910 glioma spheres (Fig. 1D). Of importance, the expression of CD133 was stable for 5 in vitro passages (data not shown).
Fig. 1.
Characterization of CD133-positive cells from the U87 glioma primary cell line and 4910 glioma xenograft cells. (A) Cell-surface expression of CD133, Nestin, Sox-2, and Tuj-1 in CD133-positive glioma cells. U87 and 4910 GICs (CD133+) were grown in DMEM/F-12 serum-free medium with growth factors as described in Materials and Methods. Glioma spheres were dissociated into single cells, stained with specific monoclonal antibodies (mAbs) to CD133, Nestin, Sox-2, and Tuj-1, isotype control matched mAbs and used for flow cytometric analysis. The results are given as the percentage of positive cells in the total population. In the histogram, the purple line represents staining with positive mAB, and the green line represents the isotype control matched mAb. (B) Expression of stem cell markers at the protein level in non-GICs and GICs. Cell lysates were prepared from non-GICs and dissociated GICs, and the expression of stem cell markers was determined by Western blot analysis using specific antibodies. (C) Expression of stem cell markers at mRNA level in non-GICs and GICs. Total RNA was extracted from both non-GICs and GICs, and mRNA expression of stem cell markers was measured by RT-PCR. (D) Immunofluorescence staining on GBM spheres. GBM spheres were stained by stem cell markers CD133 (green), Nestin (red), Sox-2 (red), and Tuj-1 (green) and counterstained with species-specific Alexa Fluor secondary antibodies. Cells were located by counterstaining with DAPI (blue).
Effect of Radiation on Cell Cycle Distribution of U87 and 4910 Non-GICs and GICs
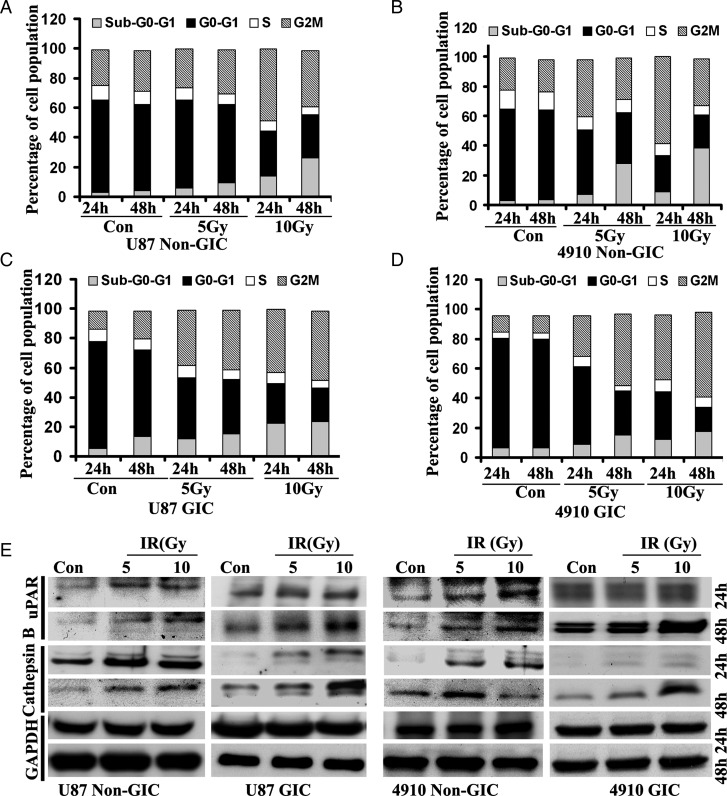
Stem cells contribute to glioma radioresistance by increasing DNA repair capacity through preferential activation of the DNA damage response.30 To compare the intrinsic radioresistance of GICs with that of non-GICs, it is necessary to perform cell-cycle analysis under normal conditions for each cell type. Distribution through the cell cycle could account for differences in radioresistance with cells in the G2-M phase over time.31 After radiation treatment, both U87 and 4910 non-GICs and GICs were arrested in the G2-M phase of the cell cycle. The results depicted in Fig. 2A and B show that 10 Gy radiation-induced G2-M arrest in U87 and 4910 non-GICs 24 h after treatment. However, G2-M arrest was significantly higher after 48 h in both U87 and 4910 GICs (Fig. 2C–D). These results indicate that GICs are more radioresistant than are normal glioma cells.
Fig. 2.
Effect of radiation on cell-cycle progression and on uPAR and cathepsin B expression in GICs. (A–D) Radiation-induced changes in the cell-cycle distribution of GICs. U87 and 4910 GIM cell lines and GICs were treated with different doses of radiation for 24 and 48 h, stained with propidium iodide, and DNA content was measured by flow cytometry. Histograms represent the percent of cells in sub G0-G1, G0-G1, S, and G2-M phases. The data represent 1 of 2 independent experiments (n = 2). (E) Radiation increased expression of uPAR and cathepsin B in GICs. Cell lysates from 0, 5, and 10 Gy radiation-treated non-GICs and GICs were collected and analyzed for expression of uPAR and cathepsin B using specific antibodies. The experiments were repeated 3 times and representative blots are shown. GAPDH was used as a loading control.
Effect of Radiation on Expression of uPAR and Cathepsin B
Our previous findings suggest that radiation promotes the invasive potential of cancer cells, which is associated with increased expression of cathepsin B and uPAR.32 Therefore, we investigated the expression pattern of uPAR and cathepsin B in irradiated U87 and 4910 non-GICs and GICs using Western blot analysis. The expression levels of uPAR and cathepsin B exhibited a dose-dependent increase in both U87 and 4910 non-GICs (Fig. 2E). The expression levels of uPAR and cathepsin B were not altered in 24 h but increased in a dose-dependent manner within 48 h of treatment in both U87 and 4910 GICs (Fig. 2E). Additional experiments were performed at 24 and 48 h after radiation treatment in non-GICs and GICs, respectively.
Effect of pCU on Radiation-Induced DNA Damage
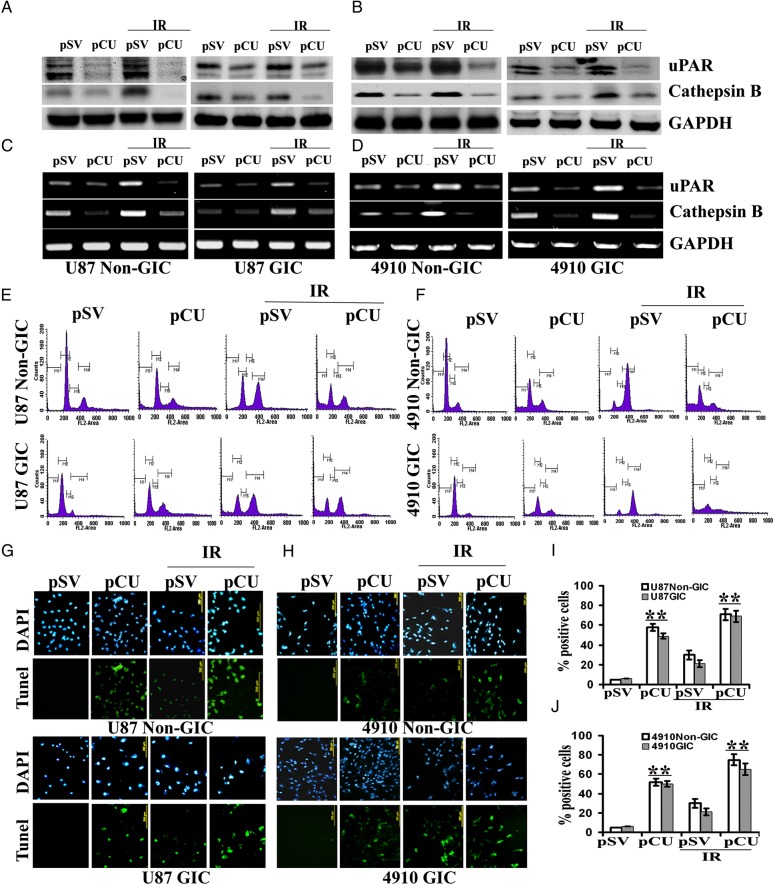
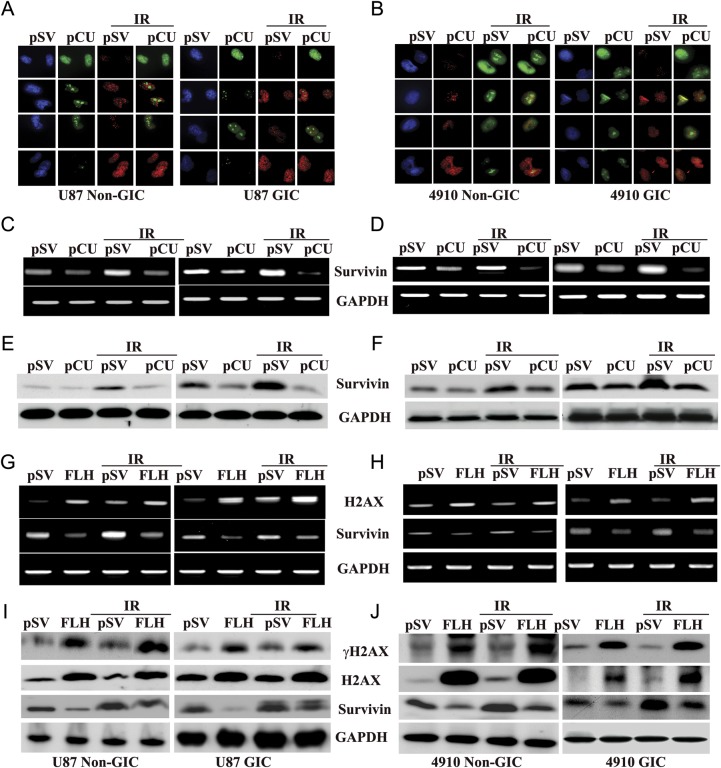
Radiation induces arrest at cellular interphase checkpoints to allow the cells to repair DNA strand breaks before continuing the cell cycle, or it induces apoptosis if DNA repair is not possible.33 Because uPAR and cathepsin B play key roles in stem-like phenotypes and initiation of signaling cascades related to DNA damage, we determined the potential of siRNA-mediated downregulation of uPAR and cathepsin B in sensitizing radiation-induced DNA damage in U87 and 4910 non-GICs and GICs. As expected, pCU treatment alone caused a significant downregulation of uPAR and cathepsin B both at the transcriptional and translational levels. Moreover, siRNA-mediated targeting of uPAR and cathepsin B using pCU further reduced the radiation-induced expression of uPAR and cathepsin B at both transcriptional and translational levels (Fig. 3A–D).
Fig. 3.
Simultaneous downregulation of uPAR and cathepsin B with radiation-enhanced accumulation of cells in the sub-G0/G1 phase. (A–B) U87 and 4910 non-GICs and GICs were transfected with pSV and pCU with or without radiation as described in Materials and Methods. uPAR and cathepsin B expression levels were determined by Western blotting. (C–D) Expression of uPAR and cathepsin B at the mRNA level. Total RNA was extracted from both non-GICs and GICs, and mRNA expression levels of uPAR and cathepsin B were determined by RT-PCR. (E–F) Distribution of cells in different phases of cell cycle. Non-GICs and GICs transfected with pSV and pCU with or without radiation were trypsinized and stained with propidium iodide as per standard protocols. Changes in cell-cycle phases were determined by measuring cellular DNA content using a flow cytometer. Histograms represent the percent of cells in sub G0-G1, G0-G1, S, and G2-M phases. (G–H) Cells were stained for apoptosis using TdT-mediated dUTP nick end-labeling (TUNEL) assay. Data shown are representative of 3 experiments. (I–J) Quantification of apoptotic cells expressed as percent of DAPI-stained cells. Bars represent the mean ± SD of 3 experiments (**P < 0.001).
To test whether pCU treatment had an effect on radiation-induced DNA damage, we determined cellular DNA content using flow cytometry after treatment of both non-GICs and GICs with pSV and pCU alone or in combination with radiation. Analysis of the histograms (Fig. 3E–F) indicates that pCU treatment alone increased the apoptotic population by 15-, 6-, 22-, and 8-fold in U87 non-GICs, U87 GICs, 4910 non-GICs, and 4910 GICs, respectively, compared with the control. The combination treatment of radiation and pCU increased the apoptotic population by 22-, 10-, 26-, and 12-fold in U87 non-GICs, U87 GICs, 4910 non-GICs, and 4910 GICs, respectively, compared with the control.
To further confirm the effect of uPAR and cathepsin B downregulation on radiation-induced DNA damage, we performed the TUNEL assay. The results show that pCU either alone or in combination with radiation significantly increased the number of TUNEL-positive cells in both U87 and 4910 non-GICs and GICs (Fig. 3G–H). Quantification revealed that 58% of pCU-treated U87 non-GICs and 42% of pCU-treated U87 GICs cells were TUNEL positive. In addition, the combination treatment with radiation further increased the number of TUNEL-positive cells to 71% and 69% in U87 non-GICs and GICs, respectively, compared with DAPI-stained cells (100%) (Fig. 3I). The percentage of TUNEL-positive cells was approximately 50% with pCU alone and 75% with pCU and radiation in 4910 non-GICs; the percentage of TUNEL-positive cells was 50% and 65% with pCU alone and with radiation, respectively, in 4910 GICs (Fig. 3J).
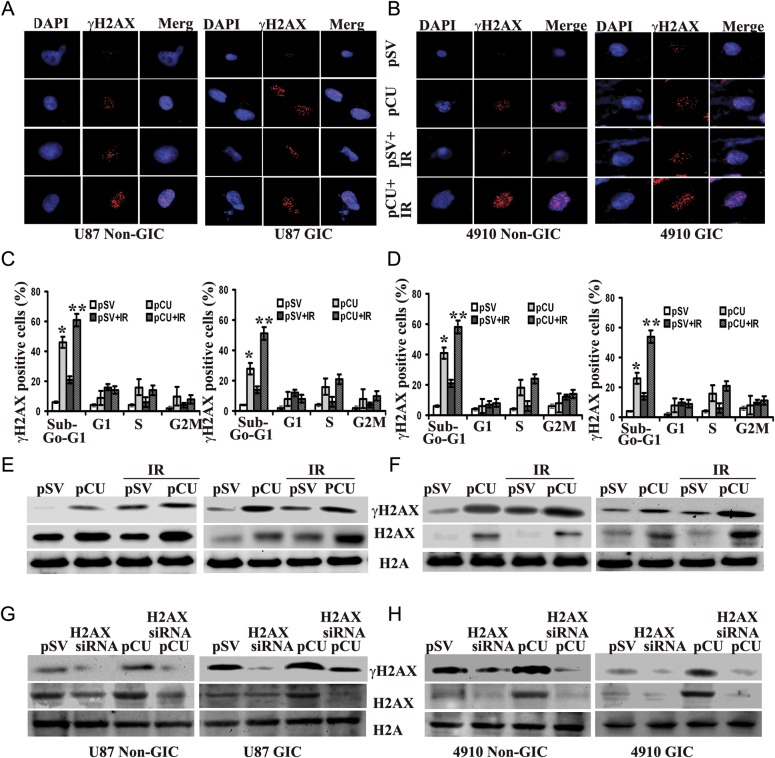
γH2AX is one of the earliest DNA double-strand break markers, and the quantity of γH2AX-bound DNA usually reflects the extent of DNA damage.34 To check the DNA damage response, we assessed the γH2AX foci by immunofluorescence staining. The results depicted in Fig. 4A and B show that pCU treatment alone or in combination with radiation significantly increased the γH2AX foci in both U87 and 4910 non-GICs and GICs.
Fig. 4.
Effect of pCU on radiation-induced DNA damage. (A–B) Induction and dispersal of γH2AX foci in U87 and 4910 non-GICs and GICs. pSV- and pCU-transfected cells were treated with or without radiation, stained with γH2AX antibody, and counterstained with species-specific Alexa Fluor secondary antibodies. Cells were located by counterstaining with DAPI (blue) and observed for fluorescence. (C–D). FACS analysis of gated γH2AX expression. Cells were stained with propidium iodide and γH2AX antibody, counterstained with species-specific Alexa Fluor antibody, and analyzed by flow cytometry. After gating, expression of γH2AX was divided into sub G0-G1, G1, S, and G2-M phases, and percent expression is expressed in bar graphs. 10 000 cells were analyzed during each process. Error bars indicate ± SD (*P < 0.05 and **P < 0.001). (E–F) Expression of total and phospho-H2AX expression in U87 and 4910 non-GICs and GICs. Cell lysates from pCU-treated and pCU + radiation-treated non-GICs and GICs were collected and analyzed for expression of H2AX and γH2AX using specific antibodies. The experiments were repeated 3 times, and representative blots are shown. GAPDH was used as a loading control. (G–H) Effect of H2AX siRNA on pCU-induced expression of H2AX and γH2AX. GICs and non-GICs were co-transfected with pCU and H2AX siRNA for 72 h. Cell lysates were prepared and analyzed for expression of H2AX and γH2AX by Western blotting. The experiments were repeated 3 times, and representative blots are shown.
To further evaluate the expression of γH2AX, FACS analysis and Western blotting were performed. When γH2AX expression and DNA content was gated and divided into sub G0-G1, G0-G1, S, and G2-M phases by FACS analysis, γH2AX expression increased in the sub G0-G1 phase of pCU-treated cells and pCU + radiation-treated cells, compared with the pSV control (Fig. 4C–D). Western blot analysis confirmed that pCU alone or pCU plus radiation overexpressed phosphorylated H2AX (Ser-139). Unexpectedly, overexpression of γH2AX was accompanied by an increase in total H2AX protein levels (Fig. 4E–F). However, core Histone H2A was not altered in any of the cell lines used in this study (data not shown). To further confirm our results, both U87 and 4910 non-GICs and GICs were cotransfected with pCU and siRNA against H2AX. The results show that basal expression and pCU-induced expression levels of both total and phosphorylated H2AX were significantly decreased, compared with controls (Fig. 4G–H). These results indicate that uPAR and cathepsin B downregulation enhanced the expression of both total and phosphorylated H2AX.
uPAR and Cathepsin B Downregulation Caused a Transcriptional Block that Sensitized Glioma Cells to Apoptosis
Chromatin aggregation induced by excess histone has previously been shown to abrogate transcription.35 To determine whether ongoing transcription is affected in uPAR and cathepsin B-depleted cells, we performed a run-on transcription assay by following in vivo incorporation of 5-bromouridine triphosphate (BrUTP). The results of the present study show that incorporation of BrUTP (nascent RNA) was observed throughout chromosome territories in pSV-treated cells. However, suppression of BrUTP incorporation at γH2AX foci was noticed in pCU-treated non-GICs and GICs and even more significantly in pCU + radiation-treated cells (Fig. 5A–B). We also examined whether the distribution of nascent RNA sites was affected by pCU-induced DNA damage using antibodies against γH2AX. We found a strong negative correlation between γH2AX and BrUTP incorporation in pSV-, pCU-, pSV + radiation-, and pCU + radiation-treated U87 and 4910 non-GICs and GICs.
Fig. 5.
Effect of pCU on run-on transcription. (A–B) Simultaneous visualization of transcription (green) and γH2AX (red) in pCU-treated U87 and 4910 non-GICs and GICs with and without radiation. Cells were transfected with pCU alone or pCU plus radiation and incubated with BrUTP at the final concentration of 5 mM. Cells were fixed and stained with anti-BrdU antibody and anti-phospho H2AX antibodies for 1 h and counterstained with species-specific Alexa Fluor-conjugated secondary antibodies for 1 h. Before mounting, cells were treated with DAPI and analyzed under a confocal microscope (Olympus BX61 Fluoview). Overlay of images was done using SPOT Advanced software (Windows version 4.0.8). (C–D) Expression of survivin mRNA. Total RNA was extracted from both non-GICs and GICs, and mRNA expression of survivin was determined by RT-PCR. GAPDH was used as a loading control. (E–F) Expression of survivin protein. Cell lysates from pCU-treated U87 and 4910 non-GICs and GICs with and without radiation were analyzed for expression of survivin by Western blotting. (G–H) Expression of survivin mRNA after transfection with full-length H2AX (FLH). Total RNA was extracted from both non-GICs and GICs, and mRNA expression of H2AX and survivin was determined by RT-PCR. (I–J) Lysates from FLH-treated cells with or without radiation were analyzed for γH2AX, H2AX and survivin proteins by Western blotting.
Studies focusing on radioresistance of glioma cells have demonstrated the potential influence of specific proteins, including survivin.36 To directly analyze whether abrogation of gene transcription stimulates apoptosis, we determined the expression of survivin at both the mRNA and protein levels by RT-PCR and Western blot analyses. Analysis of RT-PCR data revealed that survivin mRNA expression predominantly decreased with pCU in both U87 and 4910 non-GICs and GICs, compared with respective pSV controls (Fig. 5C–D). In GICs, expression of survivin significantly increased with radiation treatment as compared to non-GICs, which indicates that GICs are more radioresistant than are non-GICs. However, radiation plus pCU significantly decreased the expression of survivin in both non-GICs and GICs, compared with pCU alone. Similarly, expression of survivin at protein levels also decreased with pCU treatment alone or in combination with radiation (Fig. 4E–F). From the aforementioned results, we can conclude that uPAR and cathepsin B downregulation sensitized non-GICs and GICs by inhibiting expression of survivin. To test whether H2AX directly regulates survivin expression, non-GICs and GICs were transiently transfected with a H2AX-overexpressing plasmid with or without radiation, and expression levels of H2AX and survivin were determined by RT-PCR and Western blot analyses. Increased expression of H2AX in U87 and 4910 non-GICs and GICs at both transcriptional and translational levels indicates the efficiency of the H2AX-overexpressing plasmid (Fig. 5G–J). Further, expression of survivin was negatively correlated with the overexpression of H2AX at both the transcriptional and translational levels (Fig. 5G–J). These results indicate that survivin might be regulated by the histone variant H2AX.
c-Met Signaling Is Regulated by uPAR and Cathepsin B
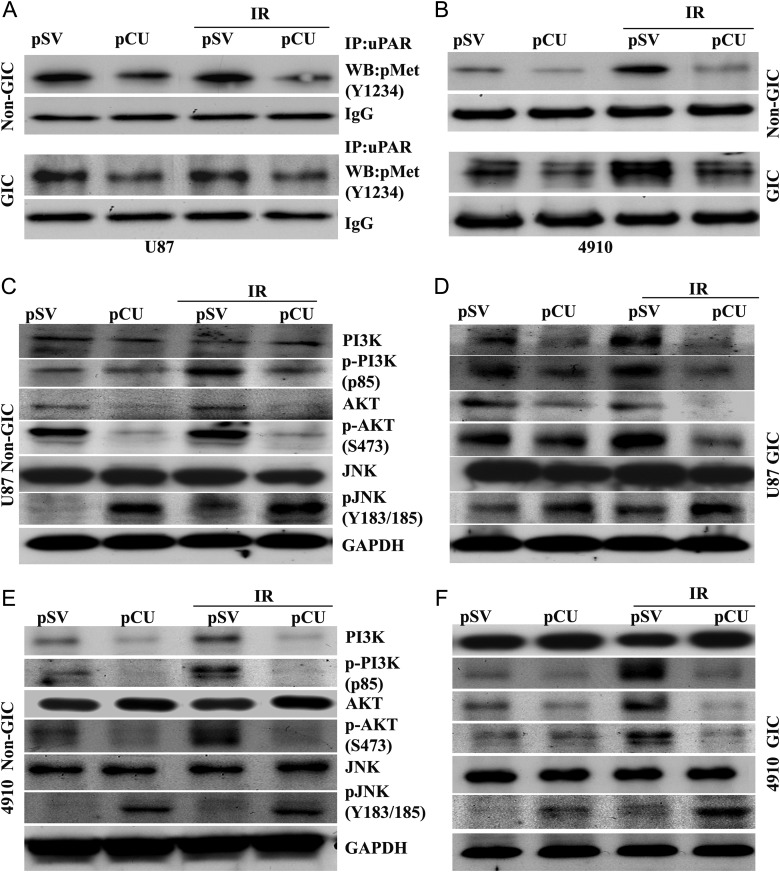
Hepatocyte growth factor, c-Met, and its receptor are associated with glioma invasion and a glioma stem-like phenotype.37,38 To test whether the regulation of H2AX by uPAR and cathepsin B is mediated by c-Met, we determined the expression of phospho-Met (Y1234), an active form of c-Met, in uPAR and cathepsin B-depleted non-GICs and GICs by Western blotting. The results showed that expression of phospho-Met significantly decreased with pCU treatment alone and with radiation, compared with pSV controls in both U87 and 4910 non-GICs and GICs (data not shown). uPAR is a glycosylphosphatidylinositol (GPI)-anchored plasmamembrane protein and relies on transmembrane co-receptors for intracellular signaling, including receptor tyrosine kinases.37 Therefore, we determined the interaction of uPAR with c-Met in uPAR and cathepsin B-depleted cells by co-immunoprecipitation. The results indicated that bulk of phospho-Met was co-precipitated with uPAR in pSV- and pSV + radiation-treated U87 and 4910 non-GICs and GICs (controls). However, phospho-Met levels in uPAR coprecipitates were reduced in pCU-treated cells and, more significantly, in pCU + radiation-treated cells, which indicates a strong interaction between uPAR and c-Met (Fig. 6A–B). Because c-Met signaling is associated with PI3K, AKT, and JNK in glioma stem cells, we also determined their expression levels in uPAR and cathepsin B-depleted cells. The results show that expression of total JNK was not altered among the treated and untreated U87 and 4910 non-GICs and GICs (Fig. 6C–F). However, phosphorylated PI3K and AKT were reduced, and p-JNK was enhanced in pCU-treated and pCU + radiation-treated cells. Furthermore, Western blot analysis of c-Met inhibitor-treated cells showed increased expression of γH2AX and decreased the expression of p-AKT (Supplementary Figs. S2A–B). These results indicate that uPAR and cathepsin B might regulate c-Met/PI3K/Akt/JNK signaling in glioma cells.
Fig. 6.
Downregulation of uPAR and cathepsin B altered c-Met signaling in GICs. (A–B) Interaction of uPAR with c-Met in U87 and 4910 non-GICs and GICs. An immunoprecipitation assay was performed to determine the effect of uPAR, and cathepsin B knockdown on interaction of uPAR with c-Met. Total cell lysates were subjected to immunoprecipitation with uPAR and co-immunoprecipitates of uPARs were analyzed by Western blotting using specific antibodies to c-Met (phospho-Met). Blots are representative of 3 experiments. (C–F) Effect of downregulation of uPAR and cathepsin B on PI3K, AKT, and JNK. Cell lysates were analyzed for expression of total and phosphorylated PI3K, AKT, and JNK in both pCU-treated and pCU + radiation-treated U87 and 4910 non-GICs and GICs by Western blotting. Blots are representative of 3 independent experiments.
Effect of uPAR and Cathepsin B Downregulation on DNA Damage of U87 and 4910 Non-GICs and GICs in an Orthotropic Model
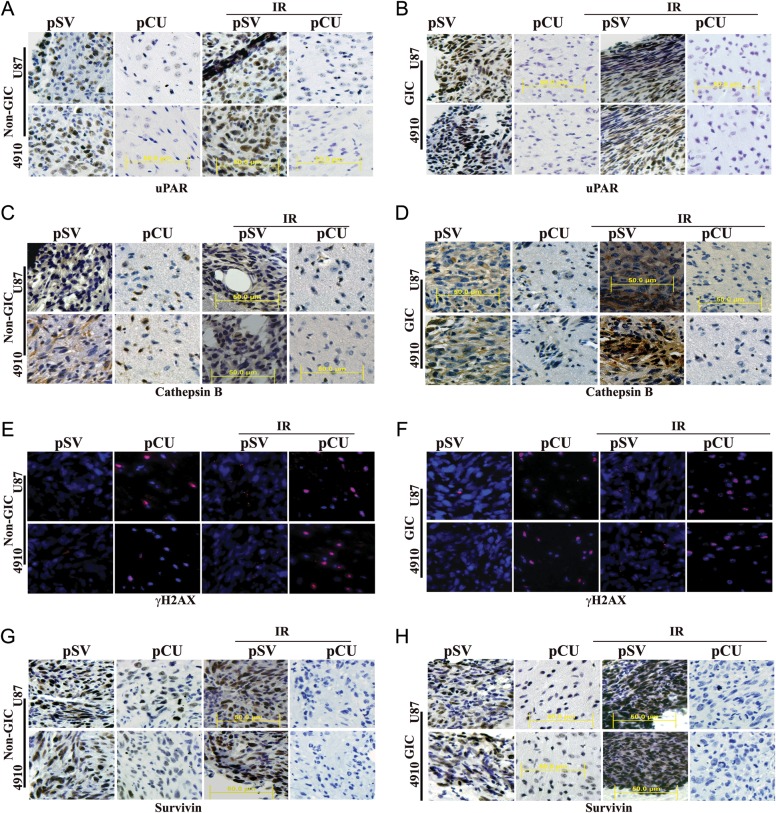
To determine the effect of uPAR and cathepsin B downregulation on DNA damage in an intracranial model, nude mice were intracranially implanted and treated as described in the Materials and Methods section. Tumor samples were taken, and paraffin-embeded sections were prepared for immunohistopathological examination. pSV- and pSV + radiation-treated brain sections had a large spread of tumor cells whereas pCU-treated and pCU + radiation-treated brain sections had a small number of tumor cells as illustrated by H&E staining (data not shown). Immunohistochemical analysis revealed decreased expression levels of uPAR and cathepsin B in pCU-treated and pCU + radiation-treated brain tissue sections of both U87 and 4910 non-GICs and GICs, compared with controls (Fig. 7A–D). To check DNA damage in vivo, we performed immunofluorescence staining of γH2AX on paraffin-embedded brain tissue sections. The results demonstrated significant γH2AX foci in pCU-treated and pCU + radiation-treated brain tissue sections, compared with control sections (Fig. 7E–F). Furthermore, our immunohistochemical analysis experiments also demonstrated that expression levels of survivin were significantly decreased in pCU-treated and pCU + radiation-treated tissue sections, compared with controls (Fig. 7G–H). These in vivo experiments confirm our in vitro data showing that uPAR and cathepsin B downregulation caused DNA damage-induced apoptosis in non-GICs and GICs.
Fig. 7.
pCU combined with radiation enhanced DNA damage-induced apoptosis in vivo. (A–D) U87 and 4910 non-GICs and GICs (1 × 105) were injected intracranially into anesthetized nude mice. Tumors were allowed to grow for 1 week, and pCU and radiation treatments were given as described in Materials and Methods. Once the control animals showed chronic symptoms (3–4 weeks), the brains were harvested and sectioned. After deparaffinization, sections were immunoprobed for uPAR and cathepsin B using specific antibodies. After staining nuclei with DAPI, the slides were mounted and visualized under a confocal microscope. Immunohistochemical analysis of brain sections used anti-uPAR (A–B) and anti-cathepsin B antibodies (C–D). Sections were photographed (40X). (E–F) Sections were immunoprobed for γH2AX using specific antibodies, followed by appropriate Alexa Fluor-conjugated secondary antibodies. After staining nuclei with DAPI, the slides were mounted and visualized under a confocal microscope. (G–H) Immunohistochemical analysis of survivin in pCU-treated and pCU + radiation-treated brain tissue sections.
Discussion
GBMs are the most lethal primary brain tumor, with a median survival of less than 12 months because of resistance to radiation and other treatments.1 Gliomas present as diffuse tumors with invasion into normal brain tissue but frequently recur after radiation, which suggests that only a fraction of tumor cells (GICs) is responsible for recurrence.2 This subpopulation often exhibits CD133 positivity and increases with radiation. Therefore, identification of CSCs (CD133+) provides a powerful tool for the investigation of the tumorigenic process and is crucial for developing targeted therapies.39 In the present study, we characterized the abundance of CD133+ cells, which often demonstrates radioresistance.3 We found that CD133+ glioma cells expressed higher levels of Nestin and Sox-2, suggesting that CD133+GICs may play an important role in the tumor's ability to resist chemotherapy and radiation.40 In addition, we found a similar expression pattern of Tuj-1 (β-Tubulin–III) in both non-GICs and GICs, indicating neuronal origin.
Although radiation therapy is one of the mainstays of glioma treatment, recent studies have demonstrated that radiation promotes malignant behavior of cancer cells.41,42 In the present study, we have observed that radiation treatment increased the G2-M cell population in a dose-dependent manner in non-GICs and in a time and dose-dependent manner in GICs, indicating the radioresistance of GICs. Radiation-sensitive tumor cells undergo cell-cycle arrest in vitro more readily than do radiation-resistant tumor cells.43
uPAR and cathepsin B are overexpressed in high-grade gliomas.21,22 Our previous findings have suggested that the expression levels of uPAR and cathepsin B correlate with the invasiveness of gliomas.44,45 For the most part, uPAR and cathepsin B have been studied as a gene product that controls invasion and metastasis of gliomas. However, the functions of uPAR and cathepsin B as a gene product involved in generating radioresistance of GICs refocuses attention on these molecules toward earlier steps in glioma development. In the present study, we noticed that uPAR and cathepsin B increased with radiation in a dose-dependent manner in non-GICs and in a dose and time-dependent manner in GICs. Thus, the observed radiation-induced increase in uPAR and cathepsin B activities in GICs is not only integral to radiation resistance but may also aid survival in a relatively aggressive setting.
Our previous report demonstrated that downregulation of uPAR and cathepsin B efficiently induced cell death in glioma cells.25 A recent report demonstrated that combined treatment with radiation and siRNA targeting potential molecules in cancer cells could enhance cell killing.46 In the current study, we have demonstrated that pCU treatment alone caused significant apoptosis and pCU combined with radiation further enhanced apoptosis in both U87 non-GICs and GICs as shown by FACS analysis. TUNEL assay further confirmed that pCU treatment alone or in combination with radiation-induced DNA fragmentation in both U87 and 4910 non-GICs and GICs. Thus, targeting uPAR and cathepsin B might sensitize radiation-induced DNA damage in glioma cells by affecting the uPAR/cathepsin B signaling pathway.
We further examined the mechanism by which downregulation of uPAR and cathepsin B induces radiosensitization in gliomas. Histone, the guardian of the genome, is subject to a wide variety of side chain modification that may be important in regulating DNA metabolism and chromatin structure.47 The most important post-translational modification linked to replication, transcription, and DNA damage is phosphorylation of H2AX.48 The most important clinical application of γH2AX measurement is to follow double-strand break levels induced by radiation and chemotherapy as a marker of treatment efficiency. H2AX is rapidly phosphorylated at Ser 136 after the induction of DNA double-strand breaks by ionization49 and genotoxic agents.12 Besides its association with double-strand breaks, γH2AX has been found to play a role in apoptosis.50 Therefore, assessment of H2AX phosphorylation as a reporter of DNA damage-induced apoptosis by pCU can be clinically useful.51 Massive accumulation of γH2AX foci 72 h after treatment, with pCU alone or with radiation, indicates irreparable DNA damage in non-GICs and GICs in the current study. Its appearance is concurrent with the initiation of DNA damage, suggesting that γH2AX formation is an early chromatin modification following initiation of DNA fragmentation during apoptosis.52 In the present study, we also observed that massive increase of γH2AX was accompanied by an increase in total H2AX in pCU-treated and pCU + radiation-treated GICs and non-GICs. This may be attributable to reciprocal regulation of histone synthesis and histone incorporation into chromatin, as it has previously been suggested.53 Both basal and pCU-induced expression of total and phosphorylated H2AX was reduced significantly by H2AX siRNA as compared to cells transfected with pSV, indicating that upregulation of H2AX is in part involved in pCU-induced apoptosis in non-GICs and GICs.
In the present study, downregulation of uPAR and cathepsin B sensitized glioma cells to DNA damage-induced apoptosis in a process that is associated with a disruption of ongoing gene transcription as evidenced by suppressed incorporation of BrUTP at nuclear segments containing γH2AX foci. Other studies previously found that massive formation of γH2AX-induced chromatin changes that inhibited assembly of transcription complexes without heterochromatin formation.13 As reported in previous studies, transcriptional shut down cannot be sustained for a prolonged period without affecting cell viability.53 Although it is a nonspecific mechanism, we cannot rule out possible direct pro-apoptotic mechanisms of H2AX.54 Therefore, evaluating anti-apoptotic protein survivin in relation to the γH2AX expression would help further clarify our findings.
In our cohort, γH2AX overexpression by pCU or pCU plus radiation also inhibited expression of survivin. However, radiation alone increased the expression of survivin in both non-GICs and GICs. In recent years, survivin has been highlighted in glioma, which is largely attributed to its radiation resistance.55,56 Of interest, overexpression of H2AX significantly reduced the expression of survivin at the protein and mRNA levels. Our findings suggest that uPAR and cathepsin B regulate the anti-apoptotic function of survivin at least in part through H2AX in glioma cells.
DNA damage-induced phosphorylation of H2AX at Ser 139 is mediated by members of the PI3K kinase group. Lu et al. reviewed that in addition to the PI3K group, JNK (a member of the stress-activated MAP kinase group) phosphorylates H2AX.54 In the present study, we were able to detect increased expression of p-JNK (Y183/185) and decreased expression of p-PI3K (p85) and p-AKT (S473) in pCU-treated and pCU + radiation-treated non-GICs and GICs. However, levels of total JNK were unchanged in pSV-, pCU-, and pCU + radiation-treated cells. However, we previously reported that downregulation of uPAR and cathepsin B decreased expression of p-JNK in SNB19 glioma cells.57 Earlier reports suggested that JNK involved in pro-survival and pro-apoptotic signaling depends on cell type and stress.58,59 Our previous reports also demonstrated that uPAR and cathepsin B regulate PI3K/AKT as a survival pathway in glioma.25,29
uPAR is GPI-anchored plasma membrane protein and relies on transmembrane co-receptors for transducing intracellular signals, including receptor tyrosine kinase.37 Functionally significant c-Met signaling has been demonstrated previously in glioma stem cells.37 For the first time, our co-immunoprecipitation experiments demonstrate that uPAR interacts with c-Met in normal glioma and GIC. Cathepsin B may also be involved in c-Met signaling indirectly through uPAR, at least in glioma cells, since simultaneous downregulation of uPAR and cathepsin B decreased the interaction of uPAR with c-Met.
Tumor tissue with a decreased tumor cell density and fragmented DNA is indicative of apoptosis. Our in vivo studies show that the simultaneous downregulation of uPAR and cathepsin B caused DNA damage-induced apoptosis as shown by massive and large γH2AX foci. In addition, decreased expression of survivin in pCU-treated and in pCU + radiation-treated cells further confirmed the DNA damage-induced apoptosis in non-GICs and GICs. Therefore, our findings suggest that uPAR and cathepsin B inhibition might serve as an adjunct to radiation therapy to target GICs and for the treatment of glioma.
Supplementary Material
Funding
This work was supported by the National Cancer Institute (CA116708 to J.S.R.). Contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
Acknowledgement
We thank Noorjehan Ali for technical assistance, Shellee Abraham for manuscript preparation, and Diana Meister and Sushma Jasti for manuscript review.
Conflict of interest statement. None declared.
References
- 1.Legler JM, Ries LA, Smith MA, et al. Cancer surveillance series [corrected]: brain and other central nervous system cancers: recent trends in incidence and mortality. J Natl Cancer Inst. 1999;91:1382–1390. doi: 10.1093/jnci/91.16.1382. doi:10.1093/jnci/91.16.1382. [DOI] [PubMed] [Google Scholar]
- 2.Garden AS, Maor MH, Yung WK, et al. Outcome and patterns of failure following limited-volume irradiation for malignant astrocytomas. Radiother Oncol. 1991;20:99–110. doi: 10.1016/0167-8140(91)90143-5. doi:10.1016/0167-8140(91)90143-5. [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Bexell D, Scheding S, Bengzon J. Toward brain tumor gene therapy using multipotent mesenchymal stromal cell vectors. Mol Ther. 2010;18:1067–1075. doi: 10.1038/mt.2010.58. doi:10.1038/mt.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. doi:10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Clement V, Dutoit V, Marino D, Dietrich PY, Radovanovic I. Limits of CD133 as a marker of glioma self-renewing cells. Int J Cancer. 2009;125:244–248. doi: 10.1002/ijc.24352. doi:10.1002/ijc.24352. [DOI] [PubMed] [Google Scholar]
- 7.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. doi:10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 8.Bassing CH, Suh H, Ferguson DO, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. doi:10.1016/S0092-8674(03)00566-X. [DOI] [PubMed] [Google Scholar]
- 9.Celeste A, Difilippantonio S, Difilippantonio MJ, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. doi:10.1016/S0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. doi:10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ismail IH, Hendzel MJ. The gamma-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ Mol Mutagen. 2008;49:73–82. doi: 10.1002/em.20358. doi:10.1002/em.20358. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Tseng M, Perdreau SA, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67:2685–2692. doi: 10.1158/0008-5472.CAN-06-3497. doi:10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 13.Solovjeva LV, Svetlova MP, Chagin VO, Tomilin NV. Inhibition of transcription at radiation-induced nuclear foci of phosphorylated histone H2AX in mammalian cells. Chromosome Res. 2007;15:787–797. doi: 10.1007/s10577-007-1162-x. doi:10.1007/s10577-007-1162-x. [DOI] [PubMed] [Google Scholar]
- 14.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. doi:10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 15.Ku JH, Kwak C, Lee HS, Park HK, Lee E, Lee SE. Expression of survivin, a novel inhibitor of apoptosis, in superficial transitional cell carcinoma of the bladder. J Urol. 2004;171:631–635. doi: 10.1097/01.ju.0000106082.44786.3b. doi:10.1097/01.ju.0000106082.44786.3b. [DOI] [PubMed] [Google Scholar]
- 16.LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247–3259. doi: 10.1038/sj.onc.1202569. doi:10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- 17.Cheung WL, Albadine R, Chan T, Sharma R, Netto GJ. Phosphorylated H2AX in noninvasive low grade urothelial carcinoma of the bladder: correlation with tumor recurrence. J Urol. 2009;181:1387–1392. doi: 10.1016/j.juro.2008.10.146. doi:10.1016/j.juro.2008.10.146. [DOI] [PubMed] [Google Scholar]
- 18.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. doi:10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 19.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. doi:10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 20.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. doi:10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 21.Sivaparvathi M, Sawaya R, Wang SW, et al. Overexpression and localization of cathepsin B during the progression of human gliomas. Clin Exp Metastasis. 1995;13:49–56. doi: 10.1007/BF00144018. doi:10.1007/BF00144018. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, Sawaya R, Mohanam S, et al. Expression and localization of urokinase-type plasminogen activator receptor in human gliomas. Cancer Res. 1994;54:5016–5020. [PubMed] [Google Scholar]
- 23.Jo M, Eastman BM, Webb DL, Stoletov K, Klemke R, Gonias SL. Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res. 2010;70:8948–8958. doi: 10.1158/0008-5472.CAN-10-1936. doi:10.1158/0008-5472.CAN-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strojnik T, Rosland GV, Sakariassen PO, Kavalar R, Lah T. Neural stem cell markers, nestin and musashi proteins, in the progression of human glioma: correlation of nestin with prognosis of patient survival. Surg Neurol. 2007;68:133–143. doi: 10.1016/j.surneu.2006.10.050. doi:10.1016/j.surneu.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 25.Malla R, Gopinath S, Alapati K, et al. Downregulation of uPAR and cathepsin B induces apoptosis via regulation of Bcl-2 and Bax and inhibition of the PI3K/Akt pathway in gliomas. PLoS One. 2010;5:e13731. doi: 10.1371/journal.pone.0013731. doi:10.1371/journal.pone.0013731. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. doi:10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 27.Gondi CS, Kandhukuri N, Kondraganti S, et al. Down-regulation of uPAR and cathepsin B retards cofilin dephosphorylation. Int J Oncol. 2006;28:633–639. [PMC free article] [PubMed] [Google Scholar]
- 28.Malla RR, Gopinath S, Gondi CS, et al. Cathepsin B and uPAR knockdown inhibits tumor-induced angiogenesis by modulating VEGF expression in glioma. Cancer Gene Ther. 2011;18:419–434. doi: 10.1038/cgt.2011.9. doi:10.1038/cgt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopinath S, Malla RR, Gondi CS, et al. Co-depletion of cathepsin B and uPAR induces G0/G1 arrest in glioma via FOXO3a mediated p27 upregulation. PLoS One. 2010;5:e11668. doi: 10.1371/journal.pone.0011668. doi:10.1371/journal.pone.0011668. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Altaner C. Glioblastoma and stem cells. Neoplasma. 2008;55:369–374. [PubMed] [Google Scholar]
- 31.Naidu MD, Mason JM, Pica RV, Fung H, Pena LA. Radiation resistance in glioma cells determined by DNA damage repair activity of Ape1/Ref-1. J Radiat Res (Tokyo). 2010;51:393–404. doi: 10.1269/jrr.09077. doi:10.1269/jrr.09077. [DOI] [PubMed] [Google Scholar]
- 32.Gupta R, Nalla AK, Gogineni VR, et al. uPAR/cathepsin B overexpression reverse angiogenesis by rescuing FAK phosphorylation in uPAR/cathepsin B down regulated meningioma. PLoS One. 2011;6:e17123. doi: 10.1371/journal.pone.0017123. doi:10.1371/journal.pone.0017123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Leskov KS, Criswell T, Antonio S, et al. When X-ray-inducible proteins meet DNA double strand break repair. Semin Radiat Oncol. 2001;11:352–372. doi: 10.1053/srao.2001.26912. doi:10.1053/srao.2001.26912. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Halicka HD, Darzynkiewicz Z. Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage (DNA double-strand breaks) Curr Protoc Cytom. 2004;7:27. doi: 10.1002/0471142956.cy0727s30. [DOI] [PubMed] [Google Scholar]
- 35.Steger DJ, Workman JL. Transcriptional analysis of purified histone acetyltransferase complexes. Methods. 1999;19:410–416. doi: 10.1006/meth.1999.0877. doi:10.1006/meth.1999.0877. [DOI] [PubMed] [Google Scholar]
- 36.Chakravarti A, Zhai GG, Zhang M, et al. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene. 2004;23:7494–7506. doi: 10.1038/sj.onc.1208049. doi:10.1038/sj.onc.1208049. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci USA. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. doi:10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriyama T, Kataoka H, Hamasuna R, et al. Simultaneous up-regulation of urokinase-type plasminogen activator (uPA) and uPA receptor by hepatocyte growth factor/scatter factor in human glioma cells. Clin Exp Metastasis. 1999;17:873–879. doi: 10.1023/a:1006729611241. doi:10.1023/A:1006729611241. [DOI] [PubMed] [Google Scholar]
- 39.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–7273. doi: 10.1038/sj.onc.1207946. doi:10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 40.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. doi:10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badiga AV, Chetty C, Kesanakurti D, et al. MMP-2 siRNA Inhibits Radiation-Enhanced Invasiveness in Glioma Cells. PLoS One. 2011;6:e20614. doi: 10.1371/journal.pone.0020614. doi:10.1371/journal.pone.0020614. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ganji PC, Nalla AK, Gupta R, et al. siRNA-Mediated Downregulation of MMP-9 and uPAR in Combination with Radiation Induces G2/M Cell-Cycle Arrest in Medulloblastoma. Mol Cancer Res. 2011;9:51–66. doi: 10.1158/1541-7786.MCR-10-0399. doi:10.1158/1541-7786.MCR-10-0399. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Firat E, Gaedicke S, Tsurumi C, Esser N, Weyerbrock A, Niedermann G. Delayed cell death associated with mitotic catastrophe in gamma-irradiated stem-like glioma cells. Radiat Oncol. 2011;6:71. doi: 10.1186/1748-717X-6-71. doi:10.1186/1748-717X-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gondi CS, Lakka SS, Dinh D, Olivero W, Gujrati M, Rao JS. Downregulation of uPA, uPAR and MMP-9 using small, interfering, hairpin RNA (siRNA) inhibits glioma cell invasion, angiogenesis and tumor growth. Neuron Glia Biology. 2004;1:165–176. doi: 10.1017/s1740925x04000237. doi:10.1017/S1740925X04000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. RNAi-mediated inhibition of cathepsin B and uPAR leads to decreased cell invasion, angiogenesis and tumor growth in gliomas. Oncogene. 2004;23:8486–8496. doi: 10.1038/sj.onc.1207879. doi:10.1038/sj.onc.1207879. [DOI] [PubMed] [Google Scholar]
- 46.Kim IA, Bae SS, Fernandes A, et al. Selective inhibition of Ras, phosphoinositide 3 kinase, and Akt isoforms increases the radiosensitivity of human carcinoma cell lines. Cancer Res. 2005;65:7902–7910. doi: 10.1158/0008-5472.CAN-05-0513. [DOI] [PubMed] [Google Scholar]
- 47.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. doi:10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 48.Wu RS, Panusz HT, Hatch CL, Bonner WM. Histones and their modifications. CRC Crit Rev Biochem. 1986;20:201–263. doi: 10.3109/10409238609083735. doi:10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- 49.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. doi:10.1016/S0959-437X(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 50.Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. doi:10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 51.Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids. 2010 doi: 10.4061/2010/920161. vol. 2010. pii: 920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen W, Zhu F, Zhang J, et al. MST1 promotes apoptosis through phosphorylation of histone H2AX. J Biol Chem. 2010;285:39108–39116. doi: 10.1074/jbc.M110.151753. doi:10.1074/jbc.M110.151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Parry JA, Chin A, Duensing S, Duensing A. Soluble histone H2AX is induced by DNA replication stress and sensitizes cells to undergo apoptosis. Mol Cancer. 2008;7:61. doi: 10.1186/1476-4598-7-61. doi:10.1186/1476-4598-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu C, Zhu F, Cho YY, et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. doi:10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaughlin N, Annabi B, Bouzeghrane M, et al. The Survivin-mediated radioresistant phenotype of glioblastomas is regulated by RhoA and inhibited by the green tea polyphenol (-)-epigallocatechin-3-gallate. Brain Res. 2006;1071:1–9. doi: 10.1016/j.brainres.2005.10.009. doi:10.1016/j.brainres.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Shirai K, Suzuki Y, Oka K, et al. Nuclear survivin expression predicts poorer prognosis in glioblastoma. J Neurooncol. 2009;91:353–358. doi: 10.1007/s11060-008-9720-4. doi:10.1007/s11060-008-9720-4. [DOI] [PubMed] [Google Scholar]
- 57.Gondi CS, Kandhukuri N, Dinh DH, Gujrati M, Rao JS. Down-regulation of uPAR and uPA activates caspase-mediated apoptosis and inhibits the PI3K/AKT pathway. Int J Oncol. 2007;31:19–27. [PMC free article] [PubMed] [Google Scholar]
- 58.Svensson C, Part K, Kunnis-Beres K, Kaldmae M, Fernaeus SZ, Land T. Pro-survival effects of JNK and p38 MAPK pathways in LPS-induced activation of BV-2 cells. Biochem Biophys Res Commun. 2011;406:488–492. doi: 10.1016/j.bbrc.2011.02.083. doi:10.1016/j.bbrc.2011.02.083. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi S, Ebihara A, Kajiho H, Kontani K, Nishina H, Katada T. RASSF7 negatively regulates pro-apoptotic JNK signaling by inhibiting the activity of phosphorylated-MKK7. Cell Death Differ. 2011;18:645–655. doi: 10.1038/cdd.2010.137. doi:10.1038/cdd.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.