Abstract
Genome-wide microRNA (miRNA) profiling of 82 glioblastomas demonstrated that miR-181d was inversely associated with patient overall survival after correcting for age, Karnofsky performance status, extent of resection, and temozolomide (TMZ) treatment. This association was validated using the Cancer Genome Atlas (TCGA) dataset (n= 424) and an independent cohort (n= 35). In these independent cohorts, an association of miR-181d with survival was evident in patients who underwent TMZ treatment but was not observed in patients without TMZ therapy. Bioinformatic analysis of potential genes regulated by miR-181d revealed methyl-guanine-methyl-transferase (MGMT) as a downstream target. Indeed, transfection of miR-181d downregulated MGMT mRNA and protein expression. Furthermore, luciferase reporter assays and coprecipitation studies showed a direct interaction between miR-181d and MGMT 3′UTR. The suppressive effect of miR-181d on MGMT expression was rescued by the introduction of an MGMT cDNA. Finally, MGMT expression inversely correlated with miR-181d expression in independent glioblastoma cohorts. Together, these results suggest that miR-181d is a predictive biomarker for TMZ response and that its role is mediated, in part, by posttranscriptional regulation of MGMT.
Keywords: biomarker, glioblastoma, MGMT, miR-181d, temozolomide
Glioblastoma is the most common form of primary brain cancer1 and is well known for its aggressive clinical course. A major advance in glioblastoma therapy entailed the discovery that the DNA alkylating agent temozolomide (TMZ) confers survival benefit when combined with radiation therapy.2,3 Despite data suggesting that only approximately 10% of the patients undergoing this therapy achieve 5-year survival,2 all patients with glioblastoma undergo the therapy because it is currently impossible to predict the subset of patients who will derive significant therapeutic benefit. Thus, a major challenge in neuro-oncology involves developing biomarkers that will facilitate such prediction.
One important determinant of TMZ response in patients with glioblastoma involves the DNA repair protein methyl-guanine-methyl transferase (MGMT), a protein responsible for repairing TMZ-induced DNA damage.4 In several clinical series, high MGMT expression level was associated with TMZ therapy failure.5 Here, we identify miR-181d, a microRNA that is strongly associated with survival in patients with glioblastoma who underwent TMZ therapy. We demonstrate that the predictive value of miR-181d is, in part, related to downregulation of MGMT.
Methods
Patients and Samples
This study was approved by the institutional review boards of all participating hospitals, and written informed consent was obtained from all patients. All patients underwent surgical resection and had histologically confirmed glioblastomas. The characteristics of the patients are listed in Supplementary material, Table S1. Of importance, all patients underwent radiation therapy. However, only a subset of patients was additionally treated with TMZ. All tissue samples were snap-frozen within 5 min of tumor removal. Samples harboring <80% tumor tissues were excluded. The extent of resection was graded as gross total resection (GTR) or non-GTR using MRIs obtained within 72 h of surgical resection by 2 independent radiologists.
DNA, RNA, and miRNA Profiling
Isolation of DNA (QIAamp DNA Mini Kit; Qiagen) and miRNA (mirVana miRNA Isolation kit; Ambion) from tumor specimens was performed in accordance with the manufacturer's instructions. miRNA profiling was performed using the Human v2.0 miRNA Expression BeadChip (Illumina), and the microarray dataset was deposited in GEO (accession number GSE25632) in accordance with MIAME guidelines. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using an ABI 7900 real-time PCR system (Applied Biosystems) with the comparative Ct method (for details, see Supplemental Methods). Pyrophosphate sequencing was performed as previously described6 (for details, see Supplemental Methods).
Statistical Analysis
Arrays were scanned with the Illumina BeadArray Reader and analyzed using Illumina BeadStudio. Clinical correlations were evaluated by χ2 test or 1-way analysis of variance. The coefficient of variance (CV) was calculated for each miRNA, and those with CV >0.8 were further analyzed. Normalization of z-scores across samples was performed, followed by using univariate Cox proportional hazard regression analysis to estimate the significance of the association for each miRNA expression with overall survival. The P value for each miRNA was then re-estimated by randomly assigning the survival time and status (repeated 10 000 times) to obtain the P value distribution. Permutation P value was then calculated for each miRNA. After correction using the Benjamini-Hochberg false discovery rate, 9 candidate miRNAs were selected for further analysis. These miRNAs were evaluated by Pearson correlation and Spearman correlation analyses. miRNAs exhibiting statistically significant association in all 3 analyses were then incorporated into a Cox multivariate analysis that included Karnofsky performance status (KPS), TMZ therapy, and extent of resection. Information with regard to the Cancer Genome Atlas (TCGA) dataset and validation analysis can be found in Supplemental Methods. All statistical analyses were performed using Matlab 2009b, JMP (SAS Institute), or GraphPad Prism (GraphPad Software).
Cells, Transfection, Immunoblotting, Luciferase Assay, and Co-Precipitation Studies
A1207, T98G, LN340, and LN18 GBM cells were grown in DMEM supplemented with 10% FBS and 1% Pen-Strep (Invitrogen) in a 5% CO2 incubator. The cells were transfected with either miR-181d mimic or control miRNA (Qiagen) using Hiperfect (Qiagen). Cells were collected after 72 h. For RNA extraction, total RNA isolation was performed using the QiagenRNeasy kit (Qiagen). cDNAs were then generated using the iScript cDNA synthesis kit (Biorad). MGMT and ACTB cDNA levels were assessed using qRT-PCR (Supplemental Methods). Immunoblotting was performed as previously described.7 Primary antibodies used included anti-MGMT (Abcam, ab7045, 1:500) and α-tubulin (Sigma Aldrich, T9026, 1:3000). Luciferase assays and coprecipitation studies were performed as previously described8,9 (details in Supplemental Methods). For the MGMT rescue experiments, A1207, T98G, LN340, or LN18, glioblastoma cells were cotransfected with combinations of control vectors, miR-181d expression vectors (OriGene Technologies), or MGMT cDNA expression vectors (OriGene Technologies). MGMT expression was determined 72 h after transfection. Cell viability was determined either by trypan blue staining performed 72 h after transfection or by clonogenic assays.
Results
miR-181d Level Is Inversely Correlated with Overall Survival Among Patients with Glioblastoma
To identify miRNAs with potential prognostic or predictive value, we profiled the miRNAs isolated from 82 clinically annotated glioblastoma samples collected at Tiantan Hospital (termed the “discovery” set). All patients underwent radiation therapy, with a subset additionally treated with TMZ. The clinical characteristics of this cohort are shown in Supplementary material, Table S1. The miRNA profiling data were first analyzed using a univariate Cox proportional hazards model with a correction for multiple comparisons. Candidates were further tested for their associations with overall survival using the Pearson's and Spearman's correlation methods10 (Table 1a). Three miRNAs (miR-936, miR-1238, and miR-181d) were consistently associated with overall survival among patients with glioblastoma by all 3 methods used.
Table 1.
miRNAs associated with survival in Tiantan cohort of 82 glioblastomas
| (a) | |||||||
|---|---|---|---|---|---|---|---|
| Variable | Univariate cox regression |
Pearson correlation |
Spearman correlation |
||||
| Type | P value | HR | Coefficient | P-value | Coefficient | P-value | |
| miR-181d | Protective | .004 | 0.654 | 0.241 | .0293 | 0.244 | .027 |
| miR-936 | Risky | .001 | 1.468 | –0.262 | .0175 | –0.261 | .017 |
| miR-1238 | Risky | .002 | 1.464 | –0.245 | .0262 | –0.233 | .035 |
| miR-346 | Risky | .001 | 1.528 | –0.263 | .0175 | >.05 | |
| miR-551a | Risky | .002 | 1.464 | –0.232 | .0356 | >.05 | |
| miR-297 | Risky | .001 | 1.496 | –0.226 | .0410 | >.05 | |
| miR-299-3p | Risky | .005 | 1.401 | –0.242 | .0287 | >.05 | |
| miR-541* | Risky | .001 | 1.474 | –0.248 | .0246 | >.05 | |
| miR-661 | Risky | .001 | 1.484 | –0.260 | .0185 | >.05 | |
| (b) | |||||||
| Variable |
Univariate Cox Regression |
Multivariate Cox Regression |
|||||
| HR | 95% CI | P-value | HR | 95% CI | P-Value | ||
| Sex | 1.313 | 0.750–2.298 | .34 | ||||
| Age | 1.006 | 0.985–1.027 | .59 | ||||
| KPS score | 0.378 | 0.222–0.643 | <.001 | 0.769 | 0.371–1.595 | .481 | |
| Gross total resection | 0.427 | 0.251–0.726 | .002 | 0.382 | 0.182–0.804 | .011 | |
| Temozolomide | 0.386 | 0.213–0.699 | .002 | 0.416 | 0.219–0.793 | .008 | |
| miR-181d | 0.654 | 0.491–0.872 | .004 | 0.808 | 0.659–0.969 | .026 | |
| miR-936 | 1.468 | 1.173–1.836 | .001 | 1.601 | 0.991–2.588 | .055 | |
| miR-1238 | 1.464 | 1.153–1.857 | .002 | 0.926 | 0.559–1.533 | .765 | |
Abbreviation: HR, hazard ratio.
Under the “Type” column, “Protective” denotes instances where the miRNA expression level is inversely correlated with overall survival and “Risky” denotes instances where the miRNA expression level is correlated with overall survival. Coefficients of correlation by Pearson's or Spearman's methods are as shown. P-values reaching statistical significance are shown in italics.
Extent of resection (GTR versus subtotal resection) and patient characteristics (KPS, age, sex) were also analyzed by univariate Cox regression analysis (Table 1b). This analysis revealed that KPS, gross total resection, and TMZ treatment were associated with overall survival. We then performed a stepwise multivariate Cox proportional hazards analysis incorporating KPS, GTR, TMZ therapy, and the candidate miRNAs. This analysis found that GTR, TMZ therapy, and miR-181d remained significantly associated with survival. In contrast to previous reports,11 age and KPS were not significantly associated with survival in our regression analysis. We believe that this is attributable to patient selection bias, because older patients or patients with poor KPS rarely undergo resection in China and would not be included in this cohort.
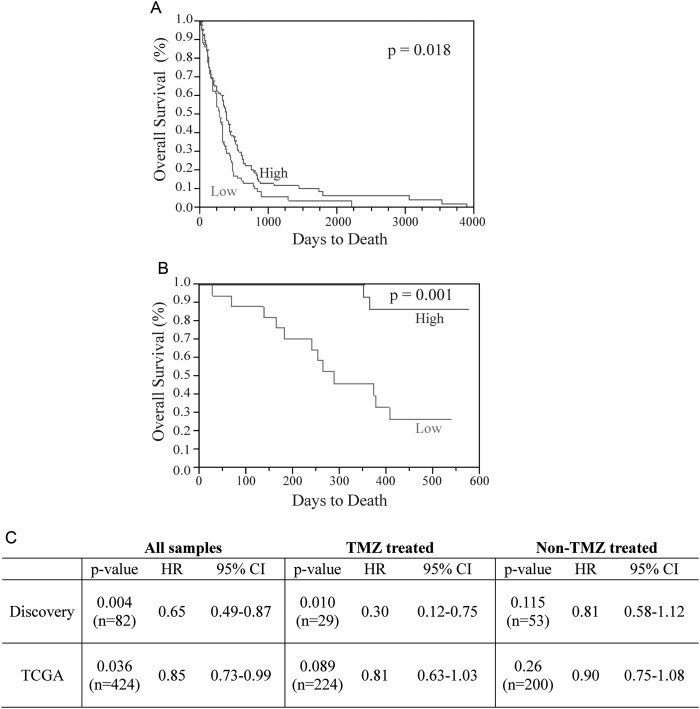
We then used the TCGA glioblastoma dataset (n= 424) to validate the association between miR-181d and overall survival. We found that TCGA glioblastomas with higher than the median level of miR-181d expression were associated with improved survival relative to those with lower than the median level of miR-181d (P= .018) (Fig. 1A). To further confirm this result, we performed qRT-PCR for miR-181d in an independent cohort of 35 patients with glioblastoma treated at Harbin Medical University (termed the “validation set,” Supplementary material, Table S1). Again, miR-181d expression was inversely correlated with overall survival in this validation set (P = .001) (Fig. 1B).
Fig. 1.
miR-181d inversely correlates with overall survival. (A) miR-181d inversely correlated with overall survival in the TCGA database. Blue: higher than median expression. Red: lower than median expression. (B) miR-181d correlated with overall survival in an independent cohort of 35 patients as assessed by qRT-PCR. Blue: higher than median expression. Red: lower than median expression. (C) Correlation of miR-181d with survival after stratification of TMZ treatment status.
Having validated miR-181d in 3 independent cohorts, we next investigated whether miR-181d constituted a prognostic or predictive biomarker by the univariate Cox analysis. The association of miR-181d with overall survival was evident in the TCGA and discovery set patients who underwent TMZ treatment (P = .010 and .089, respectively). Such correlation was not observed in patients without TMZ therapy. These results suggest miR-181d as a predictive biomarker for TMZ response (Fig. 1C). This analysis could not be performed in the validation set because of poorly documented TMZ treatment status.
MGMT Is a Direct Target of miR-181d
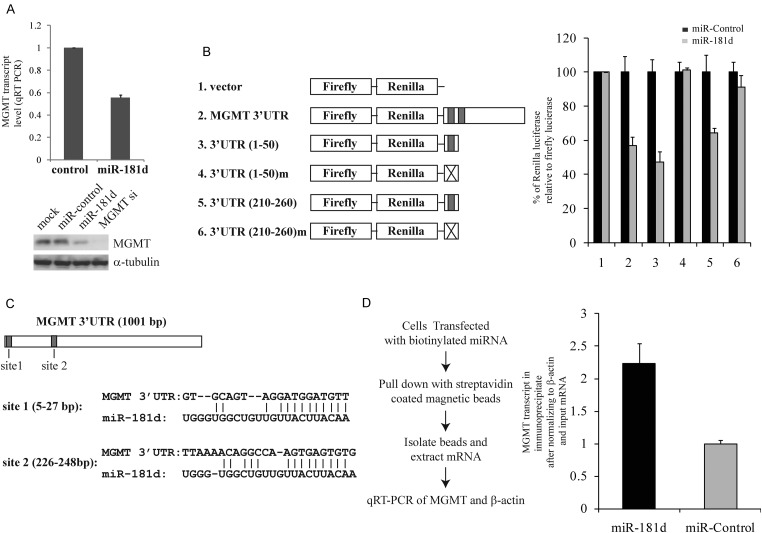
Bioinformatic analysis of potential miR-181d targets revealed MGMT as a candidate (http://www.microrna.org/microrna/home.do). Because of the critical importance of MGMT in TMZ response,4 we hypothesized that miR-181d downregulates MGMT, thereby rendering glioblastomas more susceptible to TMZ. Indeed, transfection of a miR-181d mimic into the A1207 glioblastoma cell line caused decreased MGMT mRNA and protein expressions (Fig. 2A). We then tested whether MGMT is a direct target of miR-181d using the pSiCheck-2 luciferase reporter constructs (Promega). The luciferase activity from the construct containing the MGMT 3′UTR was reduced by approximately 50% after transfection of miR-181d relative to a control miRNA (Fig. 2B, lanes 1 and 2). This effect was not observed in the construct without the MGMT 3′UTR. To further establish the MGMT 3′UTR as the direct target of miR-181d, we identified 2 predicted miR-181d binding (MRE) sites using TargetScan 4.212 and cloned the 50 nucleotides containing each putative MRE into pSiCheck-2 vectors (Fig. 2C). The luciferase activity from the constructs containing either putative MRE was reduced by approximately 50% after transfection of miR-181d. This suppressive effect was not observed with a control miRNA. Of importance, mutating the putative MREs abolished these effects (Fig. 2B, lanes 3–6). To demonstrate a direct physical interaction between miR-181d and MGMT mRNA, biotinylated miR-181d or control miRNA was transfected into A1207 cells, and mRNA-biotinylated miRNA complexes were pulled down using streptavidin beads.8 We found an approximately 2.5-fold enrichment of MGMT transcript in the miR-181d pull-down relative to the control miRNA (Fig. 2D). Together, these results provide evidence that miR-181d directly interacts with MGMT transcript to downregulate its expression.
Fig. 2.
miR-181d downregulates MGMT expression. (A) Transfection of miR-181d suppressed MGMT mRNA (top panel) and protein expression in the A1207 glioblastoma cell line (bottom panel). For qRT-PCR, MGMT mRNA was normalized either to β-actin mRNA (shown here) or U6 rRNA (with comparable results, data not shown). For the Western blotting, α-tubulin was used as the loading control. (B) miR-181d binding sites in the MGMT 3′UTR suppressed luciferase expression in the presence of miR-181d. (C) Predicted miR-181d binding sites in the MGMT 3′UTR. (D), miR-181d physically interacts with MGMT transcript. β-actin was used as an internal control.
MGMT Transcript Level Inversely Correlates with miR-181d Level
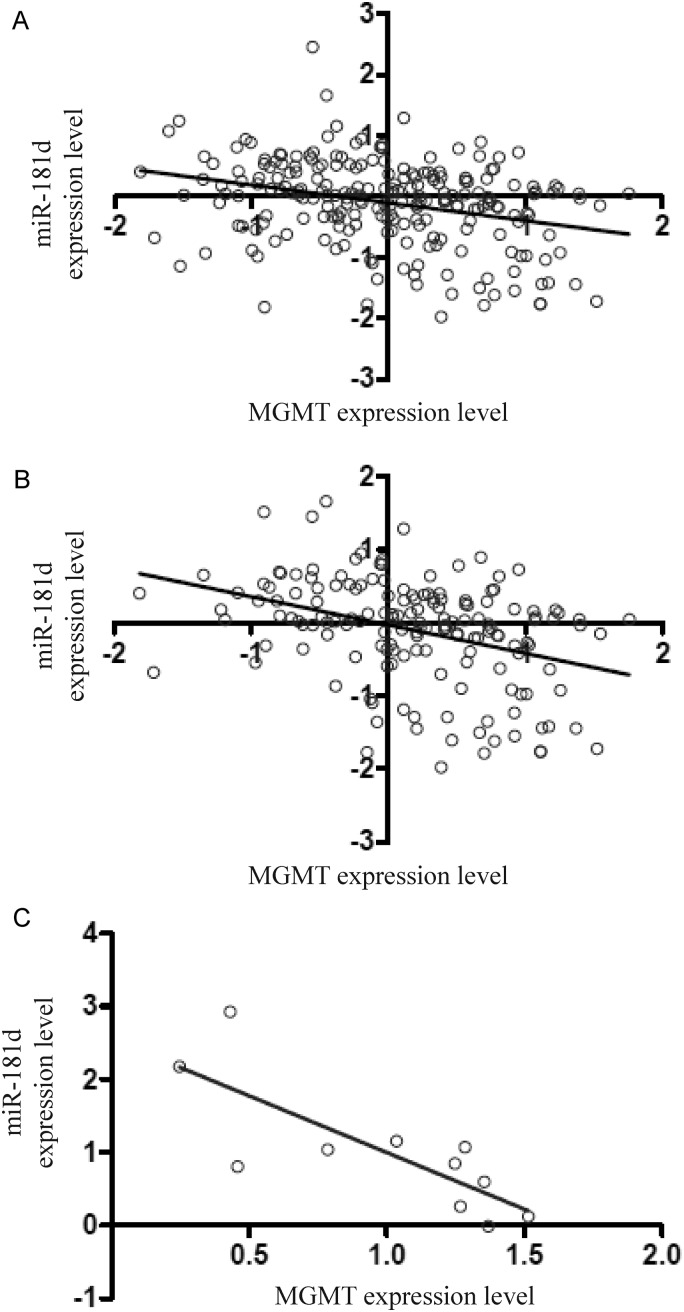
Given the results suggesting that miR-181d post-transcriptionally regulates MGMT, we anticipated an inverse correlation between miR-181d expression level and MGMT transcript level in glioblastoma specimens. We tested this hypothesis by analyzing 223 glioblastomas from the TCGA dataset (see Supplemental Methods).13 The analysis indeed revealed an inverse relationship between the miR-181d expression level and the MGMT transcript level, albeit not one reaching statistical significance (P = .09, R2 = 0.09) (Fig. 3A). We reasoned that the MGMT promoter methylation could affect MGMT transcript level5,14 in addition to miR-181d regulation. We therefore selected glioblastomas with unmethylated MGMT promoter from the TCGA database using the previously established criteria.13 In the 159 specimens with unmethylated MGMT promoter, the MGMT transcript level was inversely correlated with the miR-181d level (P = .004, R2 = 0.13) (Fig. 3B). To validate this observation, we analyzed the MGMT promoter methylation using an independent set of 20 glioblastomas. Of the 20 tumors, 9 showed heavy MGMT promoter methylation. All methylated tumors exhibited low or undetectable levels of MGMT mRNA. In the remaining 11 specimens, the MGMT transcript level was inversely correlated with miR-181d expression (P = .04, R2 = 0.62) (Fig. 3C).
Fig. 3.
miR-181d expression level inversely correlates with MGMT mRNA level. (A) Correlation between miR-181d and MGMT transcript level in 223 TCGA patients with characterized MGMT promoter, MGMT mRNA level, and miR181-d level (P = .09, R2 = 0.09). (B) Correlation of miR-181d and MGMT transcript level in 159 patients with unmethylated MGMT promoter (P = .004, R2 = 0.13). Relative abundance of MGMT and miR-181d is plotted on the x- and y-axes. (C) Inverse correlation between miR-181d and MGMT transcript level in an independent set of 11 glioblastomas (P = .04, R2 = 0.62). Expression levels of miR-181d and MGMT were assessed by qRT-PCR. Relative abundance of each is plotted on the x- and y-axes.
miR-181d–Induced TMZ Sensitivity Can Be Restored by Expression of MGMT cDNA
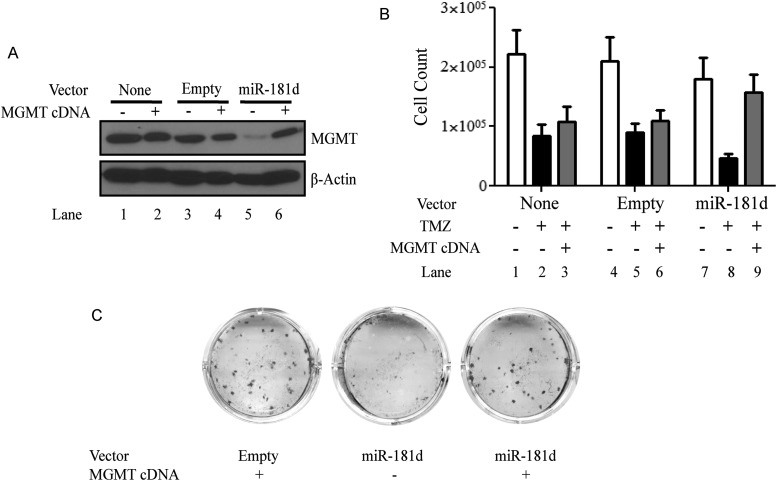
Our data suggested that miR-181d downregulates MGMT expression by direct binding of its 3′UTR. It follows that MGMT expression from a cDNA lacking the 3′UTR should be resistant to the effect of miR-181d. Furthermore, miR-181d–induced TMZ sensitivity should be restored by the introduction of an MGMT cDNA. Indeed, suppression of MGMT expression by miR-181d (Fig. 4A, lane 5) was restored by introduction of a construct expressing MGMT cDNA (Fig. 4A, lane 6). In addition, the TMZ sensitivity observed when miR-181d was introduced into the A1207 glioblastoma line (Fig. 4B, lane 8) was abolished by the expression of MGMT by a cDNA construct (Fig. 4B, lane 9). This result was confirmed using 3 independent glioblastoma lines: T98G, LN340, and LN18 (Fig. 4).
Fig. 4.
miR-181d regulates MGMT through its 3′UTR. (A) Suppressive effect of miR-181d on MGMT expression in A1207 glioblastoma cells was rescued by the transfection of MGMT cDNA. β-actin was used as a loading control. (B) miR-181d–induced temozolomide sensitivity in the A1207 glioblastoma line was rescued by the transfection of MGMT cDNA. Cell survival was determined by direct cell count after trypan blue staining. The result represents the average of 10 independent fields. The experiment was repeated 3 times. (C) miR-181d–induced temozolomide sensitivity in the LN18 glioblastoma line was rescued by the transfection of MGMT cDNA. Cell survival was determined by clonogenic assay. The experiment was repeated three times. Similar results were observed in A1207, LN340, and T98G cell lines.
Discussion
The critical role of MGMT as a cellular determinant of TMZ response is well established in model organisms spanning the entire evolutionary ladder.15,16 Here, we offer the first evidence that MGMT is posttranscriptionally regulated by miR-181d. Our transfection experiments, luciferase reporter assays, and coprecipitation studies suggested that miR-181d directly interacts with the MGMT 3′UTR to regulate MGMT at a posttranscriptional level. In support of this hypothesis, the expression level of miR-181d was inversely correlated with the MGMT transcript level in glioblastoma specimens. Furthermore, the suppressive effect of miR-181d on MGMT expression (and related TMZ sensitivity) can be reversed by expression of MGMT cDNA. Finally, high expression of miR-181d (and, thus, lower MGMT) was associated with improved overall survival in 3 independent cohorts totaling >500 patients with glioblastoma. Of importance, the results were robustly reproducible using high-quality glioblastoma specimens (>80% viable tumors) collected from different sites and using distinct profiling platforms and statistical methods.
From a biomarker perspective, our findings suggest 2 novel discovery strategies. First, it is known that each miRNA can modulate the expression of hundreds of gene transcripts.17 The resultant phenotype is likely to be the aggregate effect of many altered transcripts. In this context, we believe that miR-181d regulated other gene transcripts (apart from MGMT) that contributed to the prognostic or predictive value of miR-181d, and it would be of interest to identify such transcripts. Second, the modest correlation between miR-181d and MGMT (Fig. 3) suggests the existence of other MGMT-regulating miRNAs. These miRNAs, such as miR-181d, may serve as predictive biomarkers for TMZ response.
Previous studies have suggested that MGMT is both a predictive biomarker for TMZ therapy4 and a prognostic marker for patients with glioblastoma.5,18 The predictive value of MGMT can be related largely to its DNA repair capacity.4 It is difficult to explain why MGMT expression level was correlated with survival among patients who did not undergo TMZ therapy.5,18 It seems likely that MGMT may participate in the repair of endogenously occurring, toxic DNA damage typically found in glioblastoma cells.7 If one adopts this paradigm, one would anticipate that miR-181d, through its regulation of MGMT, has both predictive and prognostic value. Our study showed that the association of miR-181d with survival was most evident in patients treated with TMZ, suggesting that miR-181d is a predominantly predictive biomarker. We attribute this observation to the fact that miRNAs typically act to fine tune the expression level of their targets19 rather than function as an on-off switch. We speculate that the residual MGMT expression (Fig. 2A) may be sufficient for the repair of endogenous DNA damage but insufficient to repair the excess damage caused by TMZ therapy.
A potential clinical application of our finding relates to the use of MGMT promoter methylation as a glioblastoma biomarker. MGMT promoter methylation is inversely correlated with MGMT expression14,20 and is associated with TMZ response.5 A major limitation of MGMT promoter methylation–based diagnosis, however, is that it fails to account for other mechanisms regulating MGMT expression, such as posttranscriptional regulation. Indeed, a subset of patients with unmethylated MGMT promoter glioblastomas responds favorably to TMZ.2,3 We propose that MGMT-regulating miRNAs, such as miR-181d, may be useful for patient selection in this context. This thesis awaits prospective validation in future clinical trials.
Supplementary Material
Conflict of interest statement. None declared.
Funding
This work was supported by National Key Project of Science and Technology Supporting Programs (grant 2007BAI05B08), National Natural Science Foundation (grant 30772238 and 30730035), National Basic Research Program of China (grant 2010CB529406), and National High Technology Research and Development Program of China (grant 2012AA02A508) (to T.J.) and the Doris Duke Charitable Foundation, Sontag Foundation, Burroughs Wellcome Fund, Forbeck Foundation, and Kimmel Foundation (to C.C.C.).
Supplementary Material
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. doi:10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. doi:10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. doi: 10.1200/JCO.2007.11.5964. doi:10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. doi:10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. doi: 10.1038/sj.bjc.6605127. doi:10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitta M, Kozono D, Kennedy R, et al. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS One. 2010;5(5):e10767. doi: 10.1371/journal.pone.0010767. doi:10.1371/journal.pone.0010767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskwa P, Buffa FM, Pan Y, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41(2):210–220. doi: 10.1016/j.molcel.2010.12.005. doi:10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykxhoorn DM, Wu Y, Xie H, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4(9):e7181. doi: 10.1371/journal.pone.0007181. doi:10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel WW. Biostatistics: A Foundation for Analysis in the Health Sciences. New York: Wiley; 1987. [Google Scholar]
- 11.Scott CB, Scarantino C, Urtasun R, et al. Validation and predictive power of Radiation Therapy Oncology Group (RTOG) recursive partitioning analysis classes for malignant glioma patients: a report using RTOG 90-06. Int J Radiat Oncol Biol Phys. 1998;40(1):51–55. doi: 10.1016/s0360-3016(97)00485-9. doi:10.1016/S0360-3016(97)00485-9. [DOI] [PubMed] [Google Scholar]
- 12.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. doi:10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59(4):793–797. [PubMed] [Google Scholar]
- 15.Demple B, Sedgwick B, Robins P, Totty N, Waterfield MD, Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci USA. 1985;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. doi:10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tano K, Shiota S, Collier J, Foote RS, Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci USA. 1990;87(2):686–690. doi: 10.1073/pnas.87.2.686. doi:10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson DG, Hogan DJ, McCullough HL, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7(11):e1000238. doi: 10.1371/journal.pbio.1000238. doi:10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neurooncology. 2009;12(2):116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Bhakat KK, Mitra S. CpG methylation-dependent repression of the human O6-methylguanine-DNA methyltransferase gene linked to chromatin structure alteration. Carcinogenesis. 2003;24(8):1337–1345. doi: 10.1093/carcin/bgg086. doi:10.1093/carcin/bgg086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.