Lung cancer is the leading cause of cancer-related death in men and women in the United States accounting for approximately 28% of total cancer deaths in 2010 despite comprising only ~15% of new cancer cases1. Decades of research have contributed to our understanding that lung cancer is a multi-step process involving genetic and epigenetic alterations where resulting DNA damage transforms normal lung epithelial cells into lung cancer2,3. It is not known whether all lung epithelial cells or only a subset of these cells (such as pulmonary epithelial stem cells or their immediate progenitors) are susceptible to full malignant transformation. Additionally, while the tumor initiating cell may have only a handful of mutations, as the tumor expands cells may acquire additional mutations4. Smoking damages the entire respiratory epithelium and thus “field cancerization” or “field defects” (molecular changes) are observed in histologically normal lung epithelium, as well as a variety of histologic preneoplastic/premalignant lesions, which also harbor molecular abnormalities common to the adjacent tumor5. The culmination of these changes leads to lung cancers exhibiting all the “hallmarks of cancer” (including self-sufficiency of growth signals, insensitivity to growth-inhibitory (anti-growth) signals, evasion of programmed cell death (apoptosis), limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis)6,7. Lung cancer is a heterogeneous disease clinically, biologically, histologically and molecularly. Understanding the molecular causes of this heterogeneity is the focus of current research and these could reflect changes occurring in different classes of epithelial cells or different molecular changes occurring in the same target lung epithelial cells. Identifying the genes and pathways involved, determining how they relate to the biologic behavior of lung cancer and their utility as diagnostic and therapeutic targets are important basic and translational research issues. Thus, current information on the key molecular steps in lung cancer pathogenesis and their timing in preneoplasia, primary cancer, and metastatic disease and the clinical implications is the subject of this review.
Molecular epidemiology and etiology
The two main types of lung cancer, non-small cell lung cancer (NSCLC) (representing 80–85% of cases) and small cell lung cancer (SCLC) (representing 15–20%) are identified based on histological, clinical and neuroendocrine characteristics. NSCLC and SCLC also differ molecularly with many genetic alterations exhibiting subtype specificity. NSCLC can be further histologically subdivided into adenocarcinoma, squamous carcinoma, large cell carcinoma (including large cell neuroendocrine lung cancers), bronchoalveolar lung cancer, and mixed histologic types (e.g. adenosquamous carcinoma). Common molecular differences between these major NSCLC subtypes and between NSCLC and SCLC are outlined in Table 1. These differences, as well as advances in both conventional and targeted therapy, signify the importance of stratifying NSCLC tumors by subtype for prognostic and predictive purposes and molecular studies8.
Table 1.
Common genetic alterations found in lung cancer
| Gene | SCLC (%)a | NSCLC (%)a | References | ||
|---|---|---|---|---|---|
| All | Adeno- carcinoma |
Squamous cell | |||
| Oncogenic alterations | |||||
| Mutation | |||||
| BRAF | Rare | 1–3 | 1–5 | Rare | 404,405 |
| EGFR | Rare | ~20 | 10–40 | Rare | 96,404,406–408 |
| ErbB2 (HER2) | Rare | 2 | 4 | Rare | 404,409 |
| KRAS | Rare | 10–30 | 15–35 | <5 | 404,410–412 |
| MET | 13 | 21 | 14 | 12 | 12 |
| PIK3CA | Rare | 1–5 | <5 | <5 | 57,387,413,414 |
| Amplification | |||||
| EGFR | Rare | 20–30 | 15 | 30 | 12 |
| ErbB2 (HER2) | 5–30 | 2–23 | 6 | 2 | 12,409,415,416 |
| MDM2 | 6–24 | 14 | 22 | 392,417 | |
| MET | 7–21 | 20 | 21 | 418,419 | |
| MYC | 18–30 | 8–22 | 130,420–422 | ||
| NKX2-1 (TITF1) | Rare | 12–30 | 10–15 | 3–15 | 12,74,423 |
| PIK3CA | ~5 | 9–17 | 6 | 33–36 | 12,387 |
| Increase in protein expression | |||||
| CRK | 8–30 | 8–30 | 424 | ||
| BCL2 | 75–95 | 10–35 | 412,425,426 | ||
| CCND1 | 0 | 43 | 35–55 | 30–35 | 398,427 |
| CD44 | Rare | Common | 3 | 48 | 428 |
| c-KIT | 46–91 | Rare | 429–435 | ||
| EGFR | Rare | 50–90 | 40–65 | 60–85 | 100–103,412 |
| ErbB2 (HER2) | <10 | 20–35 | 16–38 | 6–16 | 409,412,432,436–438 |
| MYC | 10–45 | <10 | 133,439–441 | ||
| PDGFRA | 65 | 2–100 | 100 | 89 | 442–445 |
| Tumor suppressing alterations | |||||
| Mutation | |||||
| CDKN2A (p16) | <1 | 10–40 | 412 | ||
| LKB1 | Rare | 30–40 | 30–60 | 5–30 | 186,187,189,446,447 |
| p53 | 75–90 | 50–60 | 50–70 | 60–70 | 412,448–450 |
| PTEN | 15–20 | <10 | 412 | ||
| Rb | 80–100 | 20–40 | 412,451–453 | ||
| Deletion/LOHb | |||||
| CDKN2A (p16) | 37 | 75–80 | 72,397,454 | ||
| FHIT | 100 | 55–75 | 72,454,455 | ||
| p53 | 86–93 | 74–86 | 72,454 | ||
| Rb | 93 | 62 | 72,454 | ||
| Loss of protein expression | |||||
| CAV1 | 95 | 24 | 456 | ||
| CDKN2A (p14ARF) | 65 | 40–50 | 393,394,397,457 | ||
| CDKN2A (p16) | 3–37 | 30–79 | ~55 | 60–75 | 72,454 |
| FHIT | 80–95 | 40–70 | 72,412,454 | ||
| PTEN | 25–74 | 77 | 70 | 391,457 | |
| Rb | 90 | 15–60 | 23–57 | 6–14 | 72 |
| TUSC2 (FUS1) | 100 | 82 | 79 | 87 | 458 |
| Tumor-acquired DNA methylation | |||||
| APC | 15–26 | 24–96 | 172,173,459 | ||
| CAV1 | 93 | 9 | 456 | ||
| CDH1 | 60 40 | 20–35 | 173,459–461 | ||
| CDH13 | 15–20 | 45 | 172,173 | ||
| CDKN2A (p14ARF) | ndb | 6–8 | 173 | ||
| CDKN2A (p16) | 5, 0 | 15–41 | 21–36 | 24–33 | 296,462,463 |
| DAPK1 | nd | 16–45 | 173,298,459 | ||
| FHIT | 64 | 37 | 172,173 | ||
| GSTP1 | 16 | 7–15 | 173,464 | ||
| MGMT | 16 | 10–30 | 173,459 | ||
| PTEN | 26 | 24 | 30 | 391 | |
| RARβ | 45–70 | 40–43 | 172,173,465 | ||
| RASSF1A | 72–85 | 15–45 | 31 | 43 | 169,173,175,297,459,466 |
| SEMA3B | nd | 41–50 | 46 | 47 | 169,170 |
| TIMP3 | nd | 19–26 | 173 | ||
| Telomeres | |||||
| Telomerase activity | 75–100 | 50–80 | 65–85 | 80–90 | 261–263,412,467 |
| Chromosomal aberrations | |||||
| EML4-ALK fusion | 2–13 | 136 | |||
| Large-scale loss | 1p, 3p, 4p, 4q, 5q, 8p, 10q, 13q, 17p | 3p, 5q, 8p, 9p, 13q, 17p, 18q, 19p, 19q, 21q, 22q | 2q, 3p, 4q, 8p, 9p, 9q, 10p, 10q, 13q, 15q, 18, 20 | 3p, 4q, 9p, 10p, 10q, 18, 20 | 63,64,90,454,468–471 |
| Focal deletions | 2q22.1, 3p14.2, 3q25.1, 5q11.2, 7q11.22, 7q34, 9p23, 9p21.3, 10q23.31, 11q11, 13q12.11, 13q14.2, 13q32.2, 18q23, 21p11.2 | 74,84,86 | |||
| Large-scale gain | 3q, 5p, 8q, 18q | 1q, 3q, 5p, 6p, 7p, 7q, 8q, 20p, 20q | 5p, 7p, 7q, 8q, 11q, 19, 20q | 2q, 3q, 5p, 7, 8q, 11q, 13q, 19, 20q | 63,64,90,454,468–471 |
| Focal amplifications | 1p36.32, 1p34.3, 1q32.2, 1q21.2, 2p24.3, 2q11.2, 2q31.1, 3q26.31, 5p15.33, 5p15.31, 5p14.3, 5q31.3, 6p21.1, 7p11.2, 8p12, 8q21.13, 8q24.21, 10q24.1, 10q26.3, 11q13.3, 12p12.1, 12q13.2, 12q14.1, 12q15, 14q13.3, 14q32.13, 16q22.2, 17q12, 18q12.1, 19q12, 19q13.33, 20q13.32, 22q11.21 | 74,84,86 | |||
nd, not determined
LOH, loss of heterozygosity
Approximately 85% of lung cancers are caused by carcinogens present in tobacco smoke, while worldwide, 15–25% of lung cancer cases occur in life time “never smokers” (less than 100 cigarettes in a lifetime). These etiologic differences are associated with distinct differences in tumor acquired molecular changes and are discussed later in this review9,10. While the general public associates lung cancer with smoking, due to the number of lung cancer cases overall, lung cancer occurring in life time never smokers is also a huge public health problem. Likewise, over 50% of newly diagnosed lung cancers in the USA occur in “former smokers” who changed their lifestyle – but the damage caused by past smoking still led to the development of lung cancer. Thus, it will be important to identify the non-smoking related etiologies of lung cancer arising in “never smokers” as well as methods to identify which former smokers are most likely to develop clinically evident lung cancer.
Genetic susceptibility to lung cancer
There has been intense study of inherited predisposition to lung cancer including study of polymorphisms associated with lung cancer risk (reviewed11,12) and familial linkage studies. In 2008, three independent genome-wide association studies (GWASs) identified single nucleotide polymorphism (SNP) variations at 15q24-q25.1 were associated with an increased risk of both nicotine dependence and developing lung cancer13–15. This locus includes genes encoding nicotinic acetylcholine receptor (nAChR) subunits (CHRNA5, CHRNA3, and CHRNB4). More recently, two meta-analyses have provided further evidence that variation at 15q25.1, 5p15.33, and 6p21.33 influences lung cancer risk16,17. It has not yet been elucidated whether there is a mechanistic association with these nAChR polymorphisms and nicotine addiction, carcinogenic derivatives of nicotine exposure, or the effect of nicotine acting on nAChRs known to be expressed in lung epithelial cells18–26. In addition, a genome-wide linkage study of pedigrees containing multiple generations of lung cancer from the Genetic Epidemiology of Lung Cancer Consortium (GELCC) mapped a familial susceptibility locus to 6q23-2527,28. A member of the regulator of G-protein signaling (RGS) family, RGS17, was identified as a potential causal gene within this locus where common variants were associated with familial, but not sporadic lung cancer29; however, it is likely that more than one genetic locus in the 6q region is influencing susceptibility.
Lung cancer in never-smokers
Never smoking lung cancers represent a distinct epidemiological, clinical and molecular disease from smoking lung cancers. If considered independently, never smoking lung cancers comprise the seventh most common cause of cancer death30. Never smoking lung cancer occurs more frequently in women and East Asians, has a peak incidence at a younger age, targets the distal airways, are usually adenocarcinomas, and frequently have acquired EGFR mutations making them very responsive to EGFR targeted therapies9,31–36. Table 2 outlines the molecular differences between smoking and never smoking lung cancers.
Table 2.
Molecular differences between smoking and never smoking lung cancers
| Gene | Never smoking | Smoking |
|---|---|---|
| TP53 mutations – overall | Less common | More common |
| TP53 mutations – G:C to T:A mutations | Less common | More common |
| KRAS mutations | Less common (0–7%) | More common (30–43%) |
| EGFR mutations | More common (45%) | Less common (7%) |
| STK11 mutations | Less common | More common |
| EML4-ALK fusions | More common | Less common |
| HER2 mutations | More common | Less common |
| Methylation index | Low | High |
| p16 methylation | Less common | More common |
| APC methylation | Less common | More common |
| Loss of hMSH2 expression | Common (40%) | Rare (10%) |
Human papilloma virus (HPV)-mediated lung cancer
Human papilloma virus (HPV), an established human carcinogen (for both uterine cervical and head and neck cancer), has been proposed to play a role in lung cancer pathogenesis; however, published data remains controversial. The presence of HPV oncoproteins E6 and E7 lead to inactivation of tumor suppressors p53 and Rb, respectively37,38. A meta-analysis of 53 publications comprising 4,508 cases found a mean incidence of HPV positive lung cancer of 25%, detected in all subtypes of lung cancer39. Geographically, European and American studies had a lower incidence of 15–17% while Asian lung cancer cases reported a mean incidence of 38%. In an effort to overcome sample and detection limitations of earlier studies, a recent case-control study of ~400 lung cancer patients of European descent, representing the largest study to date, found no evidence of an association of HPV and lung cancer40. While HPV will likely be primarily found in lung cancer arising in Asian populations, the detection of oncogenic variants of HPV in some tumors and the wealth of knowledge of the role of HPV oncoproteins suggest that a subset of lung cancer will have HPV infection as a major etiologic feature. It will be important to characterize other molecular alterations in these lung cancers, and how they respond to various therapies, given the differences in response of head and neck cancer associated with HPV to EGFR targeted therapy.
Molecular changes in lung carcinogenesis: Therapeutic implications from both oncogenic changes and the cellular adaptations necessary to tolerate these changes
Characterization of the molecular changes in lung cancer and associated preneoplastic cells is becoming increasingly well-defined, aided immeasurably by the continued advancement of both clinical and genomic tools. Improved detection and sampling of clinical samples using fluorescent bronchoscopy, endobronchial ultrasounds and laser capture microdissection techniques for instance, enables precise analysis of abnormal epithelial cells. Introduction of high-resolution and high-throughput genomic tools (described in more detail later in this review) has facilitated the identification and characterization of key molecular changes – often involving oncogenes and tumor suppressor genes (TSGs) – and importantly, the associated “tumor cell acquired vulnerabilities” that accompany these oncogenotype changes (Figure 1). The key new concept that applies to many cancers, including lung cancer, is that with the genetic and epigenetic changes that occur during carcinogenesis the cancer becomes both dependent (“addicted”) to the continued presence/function of these changes and also must make other cellular adaptations including mutations to minimize the “oncogene stress” induced by these changes. While mutated oncogenic proteins themselves are therapeutic targets (see discussion of mutant EGFR below), the other cellular adaptations which are present in tumor but not normal cells also become cancer specific therapeutic targets. The cancer needs both the oncogenic changes as well as the cellular adaptations to tolerate the oncogenic changes – that is the oncogenic changes are “synthetically lethal” with the adaptation changes. Thus, both of these are potential therapeutic targets that can be discovered by genome wide functional approaches such as siRNA library screening (see below). Together, these advances promote our understanding of the development and progression of lung cancer, which is of fundamental importance for improving the prevention, early detection, and treatment of this disease. Ultimately these findings need to be translated to the clinic by using molecular alterations as: biomarkers for early detection and risk assessment; targets for prevention; signatures for personalizing prognosis and therapy selection for each patient; and as therapeutic targets to selectively kill or inhibit the growth of lung cancer.
Figure 1. Oncogene addiction and synthetic lethality in targeting acquired tumor cell vulnerability.
A) Oncogene addiction. A tumor cell contains many abnormalities in oncogenes and tumor suppressor genes (TSGs) however while some gene mutations may be critical for tumor cell survival (“driver” mutations) other gene mutations are not (“passenger” mutations). Inactivation of a critical “driver” gene in a tumor cell will result in cell death or differentiation into a normal phenotype. Inactivation of non-critical “passenger” mutations however, will not affect the tumor cell. B) Synthetic lethality arises when inactivation of two of more genes (A + B) leads to cell death whereas inactivation of either gene alone does not affect viability of the cell as the remaining gene acts in a compensatory manner. C) Synthetic lethality to target tumor cells. If a tumor cell has a non-drugable oncogene or inactivation of a TSG (Gene A), the cell will be vulnerable to inactivation of Gene B whereas a normal cell will not thus creating a second therapeutic target in addition to targeting the “driver” mutation. Adapted from94,375,376.
Technologic revolution has allowed genome wide analyses of molecular changes occurring in lung cancer
Chronic exposure to tobacco smoke carcinogens propels genetic and epigenetic damage which can result in lung epithelial cells steadily acquiring growth and/or survival advantages. Malignant transformation is characterized by genetic instability which can exist at the chromosomal level (with large-scale loss or gain of genomic material, translocations, and microsatellite instability), at the nucleotide level (with single or several nucleotide base changes), or in the transcriptome (with altered gene expression). Abnormalities are typically targeted to proto-oncogenes, TSGs, DNA repair genes and other genes that can promote outgrowth of affected cells. Activation of telomerase (the telomere-lengthening enzyme required for cell immortality) and disruption or escape from apoptotic pathways are other common events in cancer cells. Over the past 5–10 years there has been a revolution in technologies that can be applied to determining all of the genetic and epigenetic changes in lung cancer as well as other cancers. These include genome-wide mRNA expression profiles, genome-wide DNA copy number variation changes, genome-wide DNA methylation changes, miRNA changes and mass spectroscopy proteomics analyses. The recent application of “next generation” (“NexGen”) sequencing technologies has led to the first genome-wide mutational analyses of lung cancers compared to normal germline DNA41–43. These have demonstrated a huge number of mutations occurring in lung cancers arising in smokers, many changes that do not alter the coding sequences, and many changes that are idiotypic to the particular tumor (see below in “Genomics” section). Within the next several years there will be similar data on perhaps 1,000 lung cancers which will provide an unprecedented amount of information. The key issues will be to determine which of these mutations are “actionable” – that is provide a guide for targeting therapy, which are “passenger” and which are “driver” mutations, how frequent the mutations are, how the mutations are related to other molecular changes (e.g. in the epigenome and miRNAs), and which mutations provide information to identify important subgroups (“molecular portraits”) of lung cancer that provide prognostic (survival information independent of therapy) and/or predictive (survival information dependent on the administration of specific therapies) utility. Of course this will require large scale multidisciplinary and international collaboration to unite clinically annotated with molecularly annotated lung cancer specimens. Examples of this are the USA NCI “The Cancer Genome Anatomy” Program (TCGA), the NCI Lung Cancer Mutation Consortium (LCMC), as well as international lung cancer sequencing consortiums. A key component of this is to be able to perform mutation testing of clinically available materials (such as formalin fixed paraffin embedded [FFPE] specimens) in a timely fashion using clinical laboratory practices (CLIA certified laboratory methods). Recently, the NCI’s LCMC performed such a study on >800 lung adenocarcinoma tumor specimens examining mutations in established lung cancer driver genes (EGFR, KRAS, BRAF, HER2, AKT1, NRAS, PIK3CA, MEK1, EML4-ALK, MET amplification). Mutations in at least one of these genes were found in ~60% of tumor specimens and >90% were “exclusive” – only one mutation was found in a particular tumor44. Table 1 describes the current state of our knowledge of the common genetic alterations found in lung cancer. A key element will be to make this information accessible and understandable to patients and physicians not expert in cancer genomics. An example of how patients and their physicians can interface with this data is the “My Cancer Genome” website established by the Vanderbilt Cancer Center (http://www.vicc.org/mycancergenome/).
Genetic instability: Chromosomal aberration and loss of heterozygosity
Like many solid tumors, genomic instability is a hallmark of lung cancer3. Mapping high-level amplifications and deletions in copy-number throughout the cancer genome has led to the identification of many oncogenes and TSGs45–62. Many genetic alterations have been associated with lung cancer, with the more frequently observed changes including aneuploidy, specific allelic loss at 3p, 4q, 9p, and 17p and gain at 1q, 3q, 5p, and 17q63–65. Additionally, genetic alterations in several genes have been implicated in lung cancer development, including activation of MYC, RAS, EGFR, NKX2-1, ERBB2, SOX2, BCL2, FGFR2, and CRKL as well as inactivation of RB1, CDKN2A, STK11 and FHIT3,63,65–80.
Identification of the genetic alterations that occur in tumors has long been an important approach to understanding tumorigenesis. Early techniques to analyze the cancer genome involved cytogenetic karyotyping, loss of heterozygosity (LOH) and microsatellite analyses, followed later by comparative genomic hybridization (CGH) using metaphase spreads or fluorescence in situ hybridization (FISH). These techniques identified multiple numeric and structural chromosomal alterations in the cancer genome; however, the shift of CGH into a microarray-based format improved upon previous techniques by providing high-resolution detection of copy-number gain and loss56,79,81–92. Thus, due to low resolution of earlier cytogenetic and CGH techniques, which made it difficult to identify focal aberrations and the causal genes critical for tumorigenesis, aberrant loci/genes in lung carcinogenesis continue to be defined75–80.
Oncogenes and growth stimulatory pathways and targeted therapeutics
Oncogene activation occurs in probably all lung cancers (typically by gene amplification, over-expression, point mutation, or DNA rearrangements) and can result in persistent upregulation of mitogenic growth signals which induce cell growth as well as “oncogene addiction” whereby the cell becomes dependent upon this aberrant oncogenic signaling for survival (Table 1)48,50–52,56,58,60,62,74,93,94. In lung cancer, commonly activated oncogenes include EGFR, ERBB2, MYC, KRAS, MET, CCND1, CDK4, MET, EML4-ALK fusion, and BCL2. These “driver” oncogenes or oncogene “addictions” represent acquired conditional (on the oncogene) vulnerabilities in lung cancer cells, and present as significant therapeutic targets by offering specificity of killing tumor but not normal cells. Oncogenic signaling pathways commonly found in lung cancer and potential targeted therapies are summarized in Figures 2–5 and Table 3, (also see article in this issue by Gettinger at al.).
Figure 2. EGFR mutations found in lung cancer.
Activating mutations, which are found with increased frequency in certain subsets of lung cancer patients, occur as three different types of somatic mutations – deletions, insertions, and missense point mutations – and are located in exons 19–21 which code for the tyrosine kinase domain of EGFR50,51. Mutant EGFRs (either by exon 19 deletion or exon 21 L858R mutation) show an increased amount and duration of EGFR activation compared with wildtype receptors50, and have preferential activation of the PI3K/AKT and STAT3/STAT5 pathways rather than the RAS/RAF/MEK/MAPK pathway98. EGFR mutant tumors are initially highly sensitive to EGFR tyrosine kinase inhibitors (TKIs)50–52 however, despite an initial response, patients treated with EGFR TKIs eventually develop resistance to TKIs which is linked (in approximately 50% tumors) to the acquiring of a second mutation at T790M in exon 20107,108,377–380. Interestingly, the presence of the T790M mutation in a primary lung cancer that had not been treated with EGFR-TKIs however, suggests that this resistance mutation may develop with tumor progression and not necessarily as a response to treatment381. Adapted from104,382.
Figure 5. The p53 and RB pathways.
Regulation of p53 can occur through the MDM2 oncogene which reduces p53 levels through degradation by ubiquitination. MDM2 can in turn be inhibited by the tumor suppressor p14ARF, an isoform of CDKN2A. As such, the genes that encode MDM2 and p14ARF are commonly altered in lung cancer through amplification and loss of expression, respectively392–394. The CDKN2A/RB1 pathway controls G1 to S phase cell cycle progression. RB acts as a tumor suppressor by acting with E2F proteins to repress transcription of genes necessary for the G1-S phase transition. RB is inhibited by hyperphosphorylation by CDK-CCND1 complexes (complexes between CDK4 or CDK6 and CCND1), and in turn, formation of CDK-CCND1 complexes can be inhibited by the p16 isoform of CDNK2A395. Nearly all constituents of the CDKN2A/RB pathway have been shown to be altered in lung cancer through mutations (CDK4 and CDKN2A), deletions (RB1 and CDKN2A), amplifications (CDK4 and CCDN1), methylation silencing (CDKN2A and RB1), and phosphorylation (RB)396–401.
Table 3.
Targeted therapies against oncogenic pathways in lung cancer
| Gene | Drug |
|---|---|
| AKT | MK-2206, Nelfinavir, Perifosine |
| ALK | Crizotinib, GSK1838705A, nVP-TAE684 |
| Aurora kinase | AZD 1152, MLN 8237, MK0457, MK5108 |
| BCL2 | ABT-737, Gossypol, Navitoclax (ABT-263), Oblimersen, Obatoclax |
| CDK | Purvalanol |
| COX-2 | Celecoxib |
| EGFR | AEE 788, AV-412, BMS-599626, BMS-690514, Canertinib, Cetuximab, CUDC-101EKB-569, Erlotinib, Gefitinib, Icotinib, Lapatinib, Matuzumab, Neratinib, Nimotuzumab, Panitumumab, Pelitinib, Vandetanib, XL647, Zalutumumab |
| ErbB2 | CI-1033, HKI-272, Lapatinib, Trastuzumab |
| FGFR | BIBF 1120, Brivanib Alaninate, E-7080, FP-1039, PD-173074, Regorafenib, TSU-68TKI-258, XL999 |
| FLT-3 | MK-0457, Sorafenib, Sunitinib, XL999 |
| FUS1 | fus1 liposome complex |
| HDACs | Belinostat, CUDC-101, Entinostat, Panobinostat, Pivanex, Romidepsin, SB939, Vorinostat |
| HER2 | AEE 788, Afatinib, AV-412, BMS-599626, BMS-690514, CUDC-101, EKB-569, Lapatinib, Neratinib, Pertuzumab, Trastuzumab, XL647, |
| Hh (SMO) | BMS-33923, Cyclopamine, GDC-0449, IPI-926, LDE 225 |
| HIF1 | Oncothyreon |
| HSP70 | 17-AAG |
| HSP90 | Alvespimycin, Retaspimycin, Tanespimycin |
| IAPs | HGS01029 |
| IGF-1R | AMG 479, BIIB022, BMS-754807, Cixutumumab, Figitumumab, MK-0646, OSI906 |
| c-KIT | AMG-706, Axitinib, Cediranib, Dasatinib, Imatinib, Motesanib, Pazopanib, Regorafenib, Sorafenib, Sunitinib, Vatalanib |
| MDM2 | JNJ-26854165, RO5045337 |
| MEK | AS 703026, AZD6244 (selumetinib), AZD8330, GDC-0973, GSK1120212, PD325901, RDEA119, Sorafenib |
| c-MET | AMG 102, AMG 208, ARQ197, Crizotinib, Foretinib, GSK1363089, PF-04217903, PHA-665752, SCH900105, SGX523, SU11274, XL184 |
| mTOR | AZD 8055, BEZ235, Everolimus, OSI 027, PX-866, Ridaforolimus, Sirolimus/Rapamycin, Temsirolimus |
| Notch (γ-secretase) | MK0752, MRK-003, PF03084014, RO 4929097 |
| p53 | p53 peptide vaccine, PRIMA-1 |
| PARP | AG014699, Iniparib, Olaparib, Veliparib |
| PDGFR | AMG-706, Axitinib, BIBF 1120, Cediranib, Dasatinib, E7080, Imatinib, IMC-3G3, Linifanib, Motesanib, Pazopanib, Ramucirumab, Regorafenib, Sorafenib, Sunitinib, TKI-258, TSU-68, Vatalanib, XL999 |
| PI3K | BEZ235, BGT226, GDC-0941, LY294002, PX-866, XL147, XL765 |
| PPARγ | BSI-201, CS 7017, Olaparib |
| Proteasome | Bortezomib, Carfilzomib, CEP-18770, MLN9708, Salinosporamide A |
| RAF | AZ628, GSK2118436, ISIS 5132, Regorafenib, Sorafenib, XL281 |
| RAS | lonafarnib, ISIS 2503 (H-Ras), Tipifarnib |
| SRC/BCR-ABL | AZD0530, Dasatinib, Imatinib, KX2-391, XL999 |
| Telomerase | Imetelstat, Sodium metaarsenite |
| TGF-β | Trabedersen |
| TRAIL | Apomab, Conatumumab, Dulanermin, Lexatumumab, Mapatumumab, rhApo2L |
| VEGF | Aflibercept, Bevacizumab |
| VEGFR | Adnectin, AEE 788, Axitinib, BIBF 1120, BMS-690514, Brivanib Alaninate, Cediranib, E7080, Foretinib, Linifanib, Motesanib, Neovasat, Pazopanib, Ramucirumab, Regorafenib, Sorafenib, Sunitinib, Tivozanib, TKI-258, TSU-68, Vandetanib, Vatalanib, XL184, XL647, XL999 |
Epidermal growth factor receptor signaling in lung cancer
The ErbB family of tyrosine kinase receptors includes four members – EGFR, ErbB-2 (HER2), ErbB-3, and ErbB-4 – with ability to form homo- and heterodimers and bind different ligands leading to receptor activation (Figure 2)95. EGFR exhibits over-expression or aberrant activation in 50–90% of NSCLCs; therefore, much effort has been focused on the development of targeted inhibitors for this molecule96. Initial research used monoclonal antibodies that target the extracellular domain but this was supplanted by the development of small molecules that inhibit intracellular EGFR tyrosine kinase activity: EGFR tyrosine kinase inhibitors (TKIs). In 2004, a significant advancement was made in the treatment of NSCLC following the observation that somatic mutations in the kinase domain of EGFR strongly correlated with sensitivity to EGFR TKIs50,51. Exquisite sensitivity and marked tumor response has since been shown with EGFR TKIs (such as erlotinib and gefitinib) and antibodies (such as cetuximab) in EGFR mutant tumors50–52,97,98 – an example of oncogene addiction in lung cancer where tumors initiated through EGFR mutation-activation of EGF signaling rely on continued EGF signaling for survival. Mutant EGFRs (either by exon 19 deletion or exon 21 L858R mutation) show an increased amount and duration of EGFR activation compared with wildtype receptors50, and have preferential activation of the PI3K/AKT and STAT3/STAT5 pathways rather than the RAS/RAF/MEK/MAPK pathway98. EGFR mutations are particularly prevalent in certain patient subgroups: adenocarcinoma histology, women, never smokers, and East Asian ethnicity52,99–103. Resistance to TKI therapy has been associated with EGFR exon 20 insertions or a secondary T790M mutation, KRAS mutation, or amplification of the MET proto-oncogene104–109 where MET activates the PI3K pathway through phosphorylation of ERBB3, independent of EGFR and ERBB2109. Importantly, the authors found inhibition of MET signaling can restore sensitivity to TKIs109. In lung adenocarcinomas, activated mutant EGFR has been shown to induce levels of IL-6 leading to activation of STAT3110. IL-6 also plays an important role by activation of JAK family tyrosine kinases111, which in turn activate multiple pathways through signaling molecules such as STAT3, MAPK, and PI3K112.
The RAS/RAF/MEK/MAPK pathway signaling in lung cancer
Activation of the RAS/RAF/MEK/MAPK pathway occurs frequently in lung cancer (Figure 3), most commonly via activating mutations in KRAS which occur in ~20% of lung cancers, particularly adenocarcinomas113,114. In lung cancer, 90% of mutations are located in KRAS (80% in codon 12, and the remainder in codons 13 and 61) with HRAS and NRAS mutations only occasionally documented115. Mutation results in constitutive activation of downstream signaling pathways, such as PI3K and MAPK, rendering KRAS mutant tumors independent of EGFR signaling and therefore resistant to EGFR TKIs as well as chemotherapy97,106,116. KRAS mutations are mutually exclusive with EGFR and ERBB2 mutations and are primarily observed in lung adenocarcinomas of smokers97,117. The prevalence and importance of KRAS in lung tumorigenesis make it an attractive therapeutic target. Two unsuccessful approaches were farnesyltransferase inhibitors, to inhibit posttranslational processing and membrane localization of RAS proteins, and antisense oligonucleotides against RAS113. More recently, efforts have been centered on downstream effectors of RAS signaling: RAF kinase and mitogen-activated protein kinase (MAPK) kinase (MEK)113,118. BRAF is the direct effector of RAS and while commonly mutated in melanoma (~70%) mutations are rare in lung cancer (~3%), predominantly in adenocarcinoma, and mutually exclusive to EGFR and KRAS mutations119–122. Strategies to inhibit RAF kinase include degradation of RAF1 mRNA through antisense oligodeoxyribonucleotides, and inhibition of kinase activity with multikinase inhibitor such as sorafenib. Several MEK inhibitors have commenced Phase II testing in lung cancer patients and are listed in Table 3. Attempts to directly inhibit or perturb mutant KRAS continue with the advent of whole-genome approaches. Synthetic lethal siRNA screens have identified small interfering RNAs (siRNAs) that specifically kill human lung cancer cells with KRAS mutations in vitro123–125. Additionally, combination of anti-KRAS strategies (such as depletion with short-hairpin RNAs (shRNAs)) with other targeted drugs has shown potential therapeutic utility126–128.
Figure 3. The RAS/RAF/MEK/MAPK pathway.
The RAS proto-oncogene family (KRAS, HRAS, NRAS and RRAS) encode four highly homologous 21kDa membrane-bound proteins involved in signal transduction. Proteins encoded by the RAS genes exist in two states: an active state, in which GTP is bound to the molecule and an inactive state, where the GTP has been cleaved to GDP383. Activating point mutations can confer oncogenic potential through a loss of intrinsic GTPase activity resulting in an inability to cleave GTP to GDP. This can initiate unchecked cell proliferation through the RAS/RAF/MEK/MAPK pathway, downstream of the EGFR signaling pathway384. Ras signaling also activates the PI3K/AKT pathway (leading to cell growth, proliferation, and survival), RalGDS and RASSF1. Adapted from12,385.
MYC
One of the major downstream effectors of the RAS/RAF/MEK/MAPK pathway is the MYC proto-oncogene (Figure 3). In normal conditions this transcription factor functions to keep tight control of cellular proliferation; however, aberrant expression through amplification or over-expression is commonly found in lung cancer129,130. MYC proto-oncogene members (MYC, MYCN and MYCL) are targets of RAS signaling and key regulators of numerous downstream pathways such as cell proliferation131 where enforced Myc expression drives cell cycle in an autonomous fashion. It can also sensitize cells to apoptosis through activation of the mitochondrial apoptosis pathway – thus, Myc driven tumorigenesis often requires co-expression of anti-apoptotic BCL2 proteins132. Activation of MYC members often occurs through gene amplification. MYC is most frequently activated in NSCLC133, while the other two members, MYCN and MYCL along with MYC, are usually activated in SCLC64,134.
EML4-ALK fusion proteins
In 2007, a novel fusion gene with transforming ability was reported in a small subset of NSCLC patients135. Formed by the inversion of two closely located genes on chromosome 2p, fusion of PTK echinoderm microtubule-associated protein like-4 (EML4) with anaplastic lymphoma kinase (ALK), a transmembrane tyrosine kinase, yields the EML4-ALK fusion protein. The fusion results in constitutive oligomerization leading to persistent mitogenic signaling and malignant transformation and a recent meta-analysis of 13 studies encompassing 2,835 tumors reported the EML4-ALK fusion protein is present in 4% of NSCLCs136. EML4-ALK fusions are found exclusive of EGFR and KRAS mutations, and occur predominantly in adenocarcinomas and never or light smokers. Tumors with EML4-ALK fusions exhibit dramatic clinical responses to ALK targeted therapy137–141 and the ALK inhibitor crizotinib (PF-02341066) has now entered a Phase III clinical trial.
The PI3K/AKT/mTOR pathway
Phosphoinositide 3-kinases (PI3Ks) are lipid kinases that regulate cellular processes such as proliferation, survival, adhesion and motility142. The PI3K/AKT/mTOR pathway is a downstream signaling pathway of several receptor tyrosine kinases, such as EGFR, and can also be activated via binding of PI3K to activated RAS143. In lung tumorigenesis, activation of the PI3K/AKT/mTOR pathway occurs early in pathogenesis, generally through mutations in PI3K or PTEN as well as EGFR or KRAS, amplification of PIK3CA, PTEN loss, or activation of AKT144 and results in cell survival through inhibition of apoptosis (Figure 4). The pathway has two negative regulators: the tumor suppressor gene, PTEN, and TUSC1/TUSC2 complex which act upstream and downstream of AKT, respectively. The serine/threonine kinase mTOR, a downstream effector of AKT, is an important intracellular signaling enzyme in the regulation of cell growth, motility, and survival in tumor cells145. Targeted therapies to the PI3K/AKT/mTOR pathway (such as LY294002 and rapamycin) have shown significant efficacy in both NSCLC and SCLC cells with activated AKT signaling146–148.
Figure 4. The PI3K/AKT/mTOR pathway.
Downstream targets of AKT are involved in cell growth, angiogenesis, cell metabolism, protein synthesis, and suppression of apoptosis directly or via the activation of mTOR. Activation of the PI3K/AKT pathway can occur through the binding of the SH2-domains of p85, the regulatory subunit of PI3K, to phosphotyrosine residues of activated RTKs such as EGFR143. Alternatively, activation can occur via binding of PI3K to activated RAS. Mutation and more commonly, amplification of PIK3CA, which encodes the catalytic subunit of phosphatidylinositol 3-kinase (PI3K), occurs most commonly in squamous cell carcinomas56,90,386,387. AKT, a serine/threonine kinase that acts downstream from PI3K can also have mutations that lead to pathway activation. One of the primary effectors of AKT is mTOR, a serine/threonine kinase involved in regulating proliferation, cell cycle progression, mRNA translation, cytoskeletal organization, and survival388. The tumor suppressor PTEN, which negatively regulates the PI3K/AKT pathway via phosphatase activity on phosphatidylinositol 3,4,5-trisphosphate (PIP3), a product of PI3K389 is commonly suppressed in lung cancer by inactivating mutations or loss of expression390,391.
SOX2 and NKX2-1 (TITF1) – lung cancer lineage dependent oncogenes
Genome-wide screens for DNA copy number changes in primary NSCLCs has led to the identification of recurrent, histologic subtype-specific focal amplification at 14q13.3 (adenocarcinoma) and 3q26.33 (squamous cell carcinoma) 74,75,80,93,149. Functional analysis identified NKX2-1 (also termed TITF1) and SOX2 as the respective targets of these amplifications. NKX2-1 encodes a lineage-specific transcription factor essential for branching morphogenesis in lung development and the formation of type II pneumocytes – the cells lining lung alveoli150,151. Initial studies reported on the oncogenic role of NKX2-1 in lung adenocarcinoma74,93,149,152; however, recent in vivo data suggests it also has a tumor suppressive role153. SOX2 amplification was identified specifically in squamous cell carcinomas and is required for normal esophageal squamous development75,80. Amplification of tissue-specific transcription factors in cancer has been previously observed in prostate cancer (AR)154, melanoma (MITF)155, and breast cancer (ESR1)156. These findings have led to the development of a “lineage-dependency” concept in tumors157 where the survival and progression of a tumor is dependent upon continued signaling through a specific lineage pathways (i.e. abnormal expression of pathways involved in normal cell development) rather than continued signaling through the pathway of oncogenic transformation as seen with oncogene addiction94.
Tumor suppressor genes (TSGs) and growth inhibitory pathways
Loss of TSG function is an important step in lung carcinogenesis and usually results from inactivation of both alleles with LOH inactivating one allele through chromosomal deletion or translocation, and point mutation, epigenetic or transcriptional silencing inactivating the second allele158,159. Commonly inactivated TSGs in lung cancer include TP53, RB1, STK11, CDKN2A, FHIT, RASSF1A and PTEN.
The p53 pathway
TP53 (17p13) encodes a phosphoprotein which prevents accumulation of genetic damage in daughter cells. In response to cellular stress, p53 induces the expression of downstream genes such as cyclin-dependent kinase (CDK) inhibitors which regulate cell cycle checkpoint signals, causing the cell to undergo G1 arrest and allowing DNA repair or apoptosis159 (Figure 5). p53 inactivating mutations are the most common alterations in lung cancer where 17p13 frequently demonstrates hemizygous deletion and mutational inactivation in the remaining allele160–162. Some point mutations in TP53 confer a gain-of-function phenotype leading to increased aggressiveness of lung cancer163. Due to the prevalence of p53 inactivating mutations in human cancers large scale efforts have been focused on therapeutic strategies to restore normal p53 function. These include re-introduction of wildtype p53 using gene therapy, pharmacological rescue of mutant p53 with small molecule agents and peptides, blocking of MDM2 expression, inhibiting MDM2 ubiquitin ligase activity, and targeting the p53-MDM2 interaction with small molecule inhibitors. In vivo restoration of p53 expression in a subpopulation of tumor cells has been achieved with p53 gene therapy of lung cancer patients164.
The CDKN2A/RB pathway
The CDKN2A-RB1 pathway controls G1 to S phase cell cycle progression (Figure 5). Hypophosphorylated retinoblastoma (RB) protein, encoded by RB1, halts the G1/S phase transition by binding to the transcription factor E2F1 and was the first tumor suppresser gene identified in lung cancer165,166. Absent or mutant RB protein is found in approximately 90% of SCLCs compared to only 10–15% of NSCLCs while abnormalities in p16 (encoded by CDKN2A) and an upstream regulator of RB phosphorylation are predominantly found in NSCLCs167.
Chromosome 3p TSGs
Loss of one copy of chromosome 3p is one of the most frequent and early events in human cancer, found in 96% of lung tumors and 78% of lung preneoplastic lesions168. Mapping of this loss identified several genes with functional tumor suppressing capacity including FHIT (3p14.2), RASSF1A, TUSC2 (also called FUS1), and semaphorin family members SEMA3B and SEMA3F (all at 3p21.3), and RARβ (3p24). In addition to LOH or allele loss, some of these 3p genes (FHIT, RASSF1A, SEMA3B and RARβ) often exhibit decreased expression in lung cancer cells by means of epigenetic mechanisms such as promoter hypermethylation169–173. Furthermore, FHIT, RASSF1A, TUSC2, and SEMA3B will reduce growth when re-introduced into lung cancer cells. FHIT, located in the most common fragile site in the human genome (FRA3B), has been shown to induce apoptosis in lung cancer174. RASSF1A can induce apoptosis, as well as stabilize microtubules, and affect cell cycle regulation175. The tumor suppressing effect of TUSC2 is thought to occur via through inhibition of protein tyrosine kinases such as EGFR, PDGFR, c-Abl, c-Kit, and AKT176 as well as inhibition of MDM2-mediated degradation of p53177. The candidate TSG SEMA3B encodes a secreted protein which can decrease cell proliferation and induce apoptosis when re-expressed in lung, breast and ovarian cancer cells169,170,178,179 in part, by inhibiting the AKT pathway180. Another family member, SEMA3F may inhibit vascularization and tumorigenesis by acting on VEGF and ERK1/2 activation181,182 and RARβ exerts its tumor suppressing function by binding retinoic acid, thereby limiting cell growth and differentiation.
STK11 (LKB1)
The serine/threonine kinase STK11 (also called LKB1) functions as a TSG by regulating cell polarity, motility, differentiation, metastasis and cell metabolism183. Germline inactivating mutations of STK11 cause Peutz-Jeghers syndrome184, but somatic inactivation through point mutation and frequent deletion on 19p13 occurs in ~30% of lung cancers – ranking it the third most commonly mutated gene in lung adenocarcinoma after p53 and RAS119,185,186. STK11 mutations often correlate with KRAS activation and result in the promotion of cell growth187. Its tumor suppressing effect is thought to function, in part, through inhibition of the mTOR pathway via AMP-activated protein kinase188 (Figure 3). STK11 inactivation appears to be particularly prevalent in NSCLC while rare in SCLCs, and inactivating mutations are more common in tumors from males and smokers, and poorly differentiated adenocarcinomas78,185–187,189. Mutation in both KRAS and STK11 appears to confer increased sensitivity to MEK inhibition in NSCLC cell lines compared to either mutation alone190.
Lung cancer stem cells: Detection, signaling pathways and therapeutic targeting
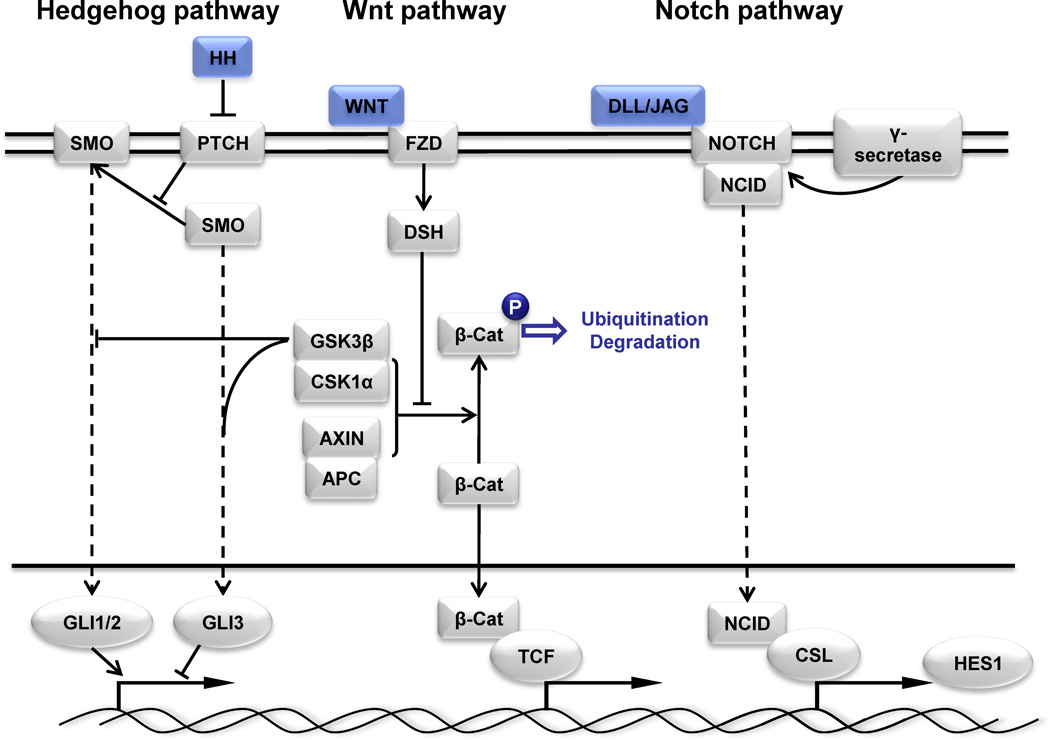
The cancer stem cell (CSC) model hypothesizes there is a population of rare, stem-like tumor cells capable of self-renewing and undergoing asymmetric division thereby giving rise to differentiated progeny that comprise the bulk of the tumor191–193. While the first evidence for CSCs (also termed tumor initiating cells) was reported in acute myeloid leukemia194, support for their existence in solid tumors, including lung cancer, is becoming increasingly common137,139,195–199. Several cell surface biomarkers have been reported for the detection and isolation of putative lung CSCs (Table 4). Interestingly, it is becoming apparent that in addition to significant variability of the utility of CSC biomarkers between different solid tumor types, no single biomarker can reliably detect CSCs in tumors from the same tissue – possible reflecting tumor heterogeneity. Regulation of CSCs in lung cancer is likely by the Hedgehog (Hh), Wnt and Notch stem cell signaling pathways200 (Figure 6). Important in normal lung development, specifically progenitor cell development and pulmonary organogenesis, these pathways are now also being studied in regards to their role in tumor development. Increased signaling of the HH pathway results in activation of the transcription regulating GLI oncogenes (GLI1, GLI2, and GLI3)201–203 and persistent activation is found in both SCLC and NSCLC204,205. The Wnt pathway has critical roles in organogenesis, cancer initiation and progression, and maintenance of stem cell pluripotency. In NSCLC, studies have found dysregulation of Wnt pathway members such as Wnt1, Wnt2 and Wnt7a, as well as upregulation of Wnt pathway agonists (Dvl proteins, LEF1, and Ruvb11) and underexpression or silencing of antagonists (WIF-1, sFRP1, CTNNBIP1, and WISP2)206–212. Notch signaling is important in cell fate determination but can also promote and maintain survival in many human cancers213–216. These signaling pathways are thought to be involved in the regulation of stem/progenitor cell self-renewal and maintenance and while normally a tightly regulated process; genes that comprise these pathways are often mutated in human cancers217–219, leading to abnormal activation of downstream effectors.
Table 4.
Published putative markers for the isolation of lung cancer stem cells
| Sample type | Tumor type | Marker/Property for CSC Isolation | Reference |
|---|---|---|---|
| Cell lines | NSCLC | Hoechst exclusion | 198,472 |
| Chemoresistance | 473 | ||
| ALDH activity | 196,474,475 | ||
| CD133+ | 195 | ||
| SCLC | ALDH activity | 474 | |
| CD133+ | 476 | ||
| uPAR | 477 | ||
| Tumor tissue | NSCLC | Hoechst exclusion | 198 |
| ALDH activity | 474 | ||
| CD133+ | 137,195,199 | ||
| SCLC | CD133+ | 199 |
Figure 6. Stem cell self-renewal pathways and therapeutic strategies to block these pathways in cancer.
Notch, Wnt, and Hedgehog (Hh) are stem cell self-renewal pathways that are often deregulated and aberrantly activated in lung cancer, thus representing key therapeutic targets. The hedgehog pathway signals through Hh ligands binding to the Patched (PTCH) receptor and inhibiting its repression of Smoothened (SMO), allowing SMO activation which results in nuclear translocation of GLI transcription factors. Wnt signaling functions through Wnt ligands binding to the Frizzled (FZD) receptor and signaling through disheveled (DSH) leading to the stabilization of β-catenin. In the absence of Hh or Wnt ligands, GSK3 phosphorylates GLI1/2 and β-catenin, respectively, resulting in ubiquitination and degradation. Notch signaling functions through Notch ligands (DLL and JAG) binding to the Notch receptor which results in the cleavage of Notch intracellular binding domain (NICD) by γ-secretase enabling it to translocate to the nucleus, bind to CLS transcription factors and activate transcription. Some components of the pathways were omitted (dashed lines) for simplicity. Adapted from402,403.
Clinical Implications
CSCs are thought to have higher resistance to cytotoxic therapies and radiotherapy than the bulk tumor cells. Thus, while conventional treatment strategies may initially “de-bulk” the primary tumor through elimination of differentiated tumor cells, the small population of CSCs eventually regenerate the tumor, giving rise to recurrence. In lung cancer, evidence of this increased resistance has been shown in primary tumors199 and lung cancer mouse xenografts137. Approaches to specifically treating the CSC population include selective targeting using CSC detection molecules, sensitization of CSCs to conventional therapies and differentiation therapies, and inhibition of signaling pathways important to CSCs, such as Hh, Wnt and Notch signaling pathways, and telomerase an important enzyme in normal stem cell function that is activated in most lung cancers (see below). In lung, progress towards the latter approach has been shown in lung cancer cells204,220. Inhibition of the Hh pathway has been demonstrated with cyclopamine, a naturally occurring inhibitor of SMO which has led to the development of synthetic oral inhibitors which show clinical activity in basal cell carcinoma221. Inhibition of the Notch signaling pathway shows potential with γ-secretase inhibitors. Several inhibitors have shown efficacy in NSCLC222,223 and a Phase II trial using a γ-secretase inhibitor as second line therapy has commenced. Lastly, analysis of CSC biomarkers as diagnostic and prognostic biomarkers has recently shown clinical utility196,224–226.
Angiogenesis and the tumor microenvironment
Angiogenesis is one of the hallmarks of cancer, essential for a microscopic tumor to expand into a macroscopic, clinically relevant tumor. Thus, angiogenic growth factors are required early in pathogenesis. A number of angiogenic proteins have been characterized including vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), interleukin-8, and angiopoietins 1 and 2. VEGF is an important inducer of angiogenesis and is known to stimulate proliferation and migration, inhibit apoptosis, promote survival and regulate endothelial cell permeability227. VEGF signaling is stimulated by tumor hypoxia, growth factors and cytokines, and oncogenic activation228. VEGF is highly expressed in both NSCLC and SCLC229 and its expression is associated with poor prognosis in NSCLC230–232, therefore inhibition of VEGF signaling in tumor cells is an important therapeutic target.
Clinical Implications
Two main approaches to anti-VEGF therapy are blocking VEGF from binding to its extracellular receptors using VEGF-specific antibodies and recombinant fusion proteins, or using small molecule TKIs that bind to the intracellular region of VEGFR233. The humanized monoclonal antibody bevacizumab blocks the binding of VEGF-A to its receptors VEGFR1 and VEGFR1 and is now approved for use in some solid cancers, including lung234. Interestingly, VEGF expression does not always correlate with response to bevacizumab235. One possible reason could be single nucleotide polymorphisms (SNPs) in VEGF. Numerous SNPs have been reported in VEGF with some being associated with lower plasma levels of VEGF236, better outcome in NSCLC237, or recently, response to bevacizumab238.
The tumor microenvironment describes the complex and dynamic milieu of stromal cells, endothelial cells, innate cells and lymphoblasts that surround tumor cells. Cells that comprise the tumor microenvironment interact both with each other and with tumor cells, and as a consequence, they can affect tumor growth, invasion and metastasis239. This supports the “seed and soil” hypothesis proposed by Stephen Paget in 1889240 who observed that the patterns of organ metastasis were a result of favorable conditions between metastatic tumor cells (the “seed”) and the organ microenvironment (the “soil). Modulation of critical tumor microenvironment biomarkers could improve current treatment of lung cancers. For example, hypoxia is associated with an increased risk of metastasis and increased resistance to radiotherapy and possible chemotherapy. Inhibition of HIF1α, a master transcription factor activated in response to hypoxia, or VEGFR, a target of HIF1α, can increase sensitivity to radiotherapy241,242.
Metastasis and epithelial to mesenchymal transition (EMT)
Many of the molecular changes discussed above promote metastatic capability of a tumor cell, enabling it to detach from the primary tumor, invade tissue and enter circulation and lastly colonize and grow in a secondary site. Recently, the cell-biological program epithelial to mesenchymal transition (EMT), involved in embryogenesis and normal development in the differentiation of multiple tissues and organs, has been the focus of tumor progression and metastasis due, in part, to evidence of EMT in many in vitro cancer cell models243. EMT describes the loss of cell polarity into a motile, mesenchymal phenotype typically characterized by loss of E-cadherin expression244. Conversion of epithelial cells to a mesenchymal state promotes motility and invasiveness allowing the tumor cells to detach from the primary tumor and relocate to a secondary site. The cells will then undergo a mesenchymal to epithelial transition (MET) to revert to an epithelial state to enable proliferative growth245. While initial reports demonstrated the role of EMT in invasion and metastasis, EMT has since been associated with early events in carcinogenesis246, the acquirement of stem cell-like properties246–248, and resistance to cell death, senescence and conventional chemotherapies245. In lung cancer, mesenchymal markers and EMT inducers (e.g. Vimentin, Twist and Snail) have been shown to be strong prognostic markers249–251. EMT has also been linked to resistance to EGFR TKIs252,253 and COX-2 and LKB1 have been implicated promoting EMT in lung cancer254–256. The miR-200 family of miRNAs is an important negative regulator of EMT257–260 and is discussed later in this review.
Activation of telomerase in lung cancer pathogenesis
Activation of telomerase, the telomere-lengthening enzyme, in premalignant cells prevents loss of telomere ends beyond critical points and is essential for cell immortality. Although silenced in normal cells, telomerase is activated in >80% of NSCLCs and almost uniformly in SCLCs (Table 1)261–263.
Clinical Implications
The prevalence of activated telomerase in cancer cells has made it an attractive target for therapeutic inhibition. Inhibition of telomerase in such cells leads to telomere shortening and ultimately either cellular senescence or apoptosis264,265. Approaches to telomerase inhibition include using antisense oligonucleotides that bind to human telomerase RNA265 (such as Imetelstat, which has started Phase II trials266) and immunotherapy whereby a patient’s own immune system is stimulated with a vaccine to recognize tumor cells containing a major histocompatibility complex presenting hTERT peptide on the cell surface267,268.
Epigenetic changes in lung carcinogenesis
Methylation and histone modification
Epigenetic events can lead to changes in gene expression without any changes in DNA sequence and therefore, importantly, are potentially reversible269. Aberrant promoter hypermethylation is an epigenetic change that occurs early in lung tumorigenesis resulting in silencing of gene transcription and therefore a common method for inactivation of TSGs in lung cancer (Table 1)270. They include genes involved in tissue invasion, DNA repair, detoxification of tobacco carcinogens, and differentiation. The prevalence of promoter methylation has been reported to differ between smokers and never-smokers. Promoter methylation of p16, MGMT, RASSF1, MTHFR, and FHIT was significantly higher among smokers than never-smokers whilst RASSF2, TNFRSF10C, BHLHB5, and BOLL was more common in never-smokers271–275. Recent advances in whole-genome microarray profiling have allowed researchers to globally study DNA methylation patterns in lung cancer – the lung cancer epigenome or methylome – and indicate the role of methylation in lung tumorigenesis may have been underestimated276–285. Initial genome-wide studies analyzed the effect on gene expression following treatment of lung cancer cell lines with demethylating agents (such as 5-azacytidine); however, development of methylation-specific microarrays enables epigenomic analysis of tumor specimens276–281.
Clinical Implications
Aberrant methylation occurs early in lung cancer pathogenesis and can be detected in circulating DNA; thus, many studies have investigated the utility of methylation status in lung cancer for risk assessment, early detection, disease progression and prognosis (reviewed286,287). Table 5 summarizes published candidate early detection, prognostic and predictive methylation biomarkers where hypermethylation of p16, APC, FHIT, RASSF1A, DAPK and CDH1 being repeatedly reported as potential prognostic markers288–302.
Table 5.
DNA methylation as a biomarker in lung cancer
| Early detection | Prognostic marker | Predictive marker |
|---|---|---|
| APC | APC | SFN (14-3-3 sigma) |
| CDH13 | CDH1 | |
| DAPK1 | CDH13 | |
| DNMT1 | CXCL12 | |
| FHIT | DAPK1 | |
| GATA5 | DLEC1 | |
| GSTP1 | EPB41L3 (DAL-1) | |
| MAGEA1 | ESR1 | |
| MAGEB2 | FHIT | |
| MGMT | IGFBP-3 | |
| p16 | MGMT | |
| PAX5-b | MLH1 | |
| RARβ2 | MSH2 | |
| RASSF1A | p16 | |
| RASSF5 | PYCARD (ASC) | |
| RUNX3 | PTEN | |
| TCF21 | RASSF1A | |
| RRAD | ||
| RUNX3 | ||
| SPARC | ||
| TIMP3 | ||
| TMS1 | ||
| TSLC1 | ||
| WIF1 |
DNA is methylated by DNA methyltransferases (DNMTs) which are responsible for both de novo and maintenance of pre-existing methylation in a cell303. Histone modification is another mechanism for epigenetic control of gene transcription where histone deacetylation results in condensing of chromatin resulting in transcriptionally inactive DNA. Inhibitors of DNMTs or histone deacetylases (HDACs) resulting in pharmacologic restoration of expression of epigenetically silenced genes is an exciting targeted therapeutic approach and show promise in lung cancer304,305 (Table 3).
MicroRNA-mediated regulation of lung cancer
MicroRNAs (miRNAs) are a class of non-protein encoding small RNAs capable of regulating gene expression by either direct cleavage of a targeted mRNA or inhibiting translation by interacting with the 3’ untranslated region (UTR) of a target mRNA. miRNAs commonly have multiple target genes therefore a single miRNA can often affect multiple cellular processes. Furthermore, a mRNA may be targeted by more than one miRNA resulting in a complex network of molecular pathways to elucidate. Aberrant expression of miRNAs has been found to play an important role in the pathogenesis of cancer as either oncogenes or TSGs306–316. Microarray-based analyses of miRNA expression have identified many lung cancer-associated miRNAs313,314,317–328, and a review of experimentally validated miRNAs has been published previously329. One of the most widely-studied lung cancer-associated miRNAs is the let-7 miRNA family. Functioning as a tumor suppressor, it has been shown to regulate N-RAS, K-RAS, MYC and HMGA2330–332 via binding to the let-7 binding sites in their respective 3’ UTRs330,333. It is frequently under-expressed in lung tumors, particularly NSCLC, compared to normal lung, and decreased expression has also been associated with poor prognosis313,318. Induction of let-7 miRNA expression has been found to inhibit in vitro growth313,331,334,335 and reduce tumor development in a murine model of lung cancer335,336. Other miRNAs that exhibit tumor suppressing effects in lung cancer include miR-29a/b/c, miR-34a/b/c, miR-16, and miR-126318–321,337,338, and recently, miR-128b was reported to be a direct regulator of EGFR with frequent LOH occurring in NSCLC cell lines322. Oncogenic miRNAs found to be over-expressed in lung cancer include the miR-17-92 cluster of seven miRNAs (that target PTEN, E2F1-3 and BIM), miR-21 (suggested to be positively regulated by the EGFR signaling pathway, specifically EGFR mutations), miR-93, miR-98, miR-197, miR-221/222, and miR-155314,323,327,328. Additionally, hsa-miR-146b, miR-155 and miR-21 and have been reported to be strong predictors of poor prognosis in lung cancer318,326,339,340. Recent evidence shows a strong link between miRNAs and invasion and metastasis with several miRNAs found to regulate key regulators of EMT, a process central to cancer metastasis258–260,341. These include miR-10b (through inhibition of HOXD10), miR-126, and the miRNA-200 family (which inhibit EMT inducers ZEB1 and ZEB2)257–259,320,341.
Clinical Implications
There is currently a strong research focus on miRNAs as potential diagnostic and prognostic biomarkers, and therapeutic targets. Restoration of aberrantly expressed miRNAs can be achieved in vitro and in vivo using miRNA mimics (for under-expressed miRNAs) or miRNA inhibitors (termed antisense oligonucleotides or antagomirs) (for over-expressed miRNAs)342–346. miRNA profiles for histologic347,348 and prognostic318,326,337,338,340 classification of lung tumors and detection of miRNAs in peripheral blood and sputum349–351 illustrate the potential of miRNAs as diagnostic and early detection biomarkers in lung cancer. Additionally, concurrent inhibition or over-expression of miRNAs with conventional therapies has resulted in an increased response to EGFR TKIs and radiotherapy327,352. These studies illustrate the immense potential of miRNAs in therapeutics development; however, limitations in pharmacokinetics, delivery and toxicity need to be addressed353,354.
The search for new biomarkers: Tools and model systems
Genomics: Tools for identification, prediction and prognosis
Genetic and epigenetic mechanisms underlying lung cancer development and progression continue to emerge, spearheaded by the development of technologies allowing genome-wide analysis of DNA copy-number, mutations, gene expression, SNPs and methylation.
Transcriptome Profiling
Profiling the lung cancer transcriptome has imparted biologically- and clinically-relevant information such as novel dysregulated genes and pathways and gene signatures that can predict patient prognosis, response to treatment, and histology reviewed in355–357. In an effort to overcome limitations of sample size and heterogeneity in previous studies, a multi-site, blinded validation study of 442 lung adenocarcinomas comprehensively examined whether the mRNA profile of primary tumors robustly predicts patient outcome either alone or in combination with clinicopathological factors358. This study developed several models (or signatures) which for the most part predicted outcome better than current clinical methods. A recent critical review of published prognostic signatures in lung cancer, however, found little evidence of any published signature being ready for clinical application due, for the most part, to problems with study design and analysis359. The role of expression of the 48 nuclear receptors (and later their co-regulators) has been studied in lung cancer and found to provide as good or better prognostic information than other mRNA expression signatures360. Since the nuclear receptors are also targets for therapeutic manipulation (via hormone agonists and antagonists) the expression of nuclear receptor patterns in individual lung cancers may also provide insight for targeted therapy. Despite complexities of mRNA profiling, the success of prognostic signatures in breast cancer, as seen with Oncotype DX361, impels further research efforts.
Genome-wide copy number profiling
High resolution mapping of copy number alterations in the lung cancer genome has been able to identify single genes as targets of genomic gain or loss through improved definition of known aberrant regions or by identification of focal alterations undetectable with earlier technology74–76,79,80,83,84,86. A large-scale analysis of 371 primary lung adenocarcinomas identified 57 significant recurrent copy-number alterations, of which 31 were focal events and many were new lung cancer loci74; for example, amplification at 14q13.3 was reported as the most common event targeting the transcription factor NKX2-1, discussed earlier. Similar studies in NSCLC and squamous cell carcinoma cohorts have identified other novel ‘drivers’ of lung carcinogenesis75,76,79,80.
Genome wide sequencing of lung cancers
Large-scale sequencing and SNP analyses have also led to the identification of novel somatic mutations in the lung cancer genome13–15,119. In a screen of 188 lung adenocarcinomas Ding et al119 identified somatic mutations in putative oncogenes (ERBB4, KDR, FGFR4, EPHA3) and TSGs (NF1, RB1, ATM, and APC). A major breakthrough has come with the development of “next generation” (also termed second-generation) DNA sequencing technologies which enable sequencing of expressed genes (‘transcriptomes’), known exons (‘exomes’) and complete genomes of tumors362. Data analysis can detect point mutations, insertions/deletions, copy number alterations, translocations and non-human sequences. Comparison of a primary lung NSCLC of adenocarcinoma histology with adjacent normal tissue identified many somatic mutations at an estimated rate of ~18 per megabase, including >50,000 single nucleotide variants41. Sequencing of a SCLC cell line revealed over 22,000 somatic substitutions42 while another study which sequenced a SCLC cell line and a neuroendocrine lung cancer cell line found a higher rate of somatic and germline rearrangements in the SCLC cell line43. Sequencing of the coding exons of ~1,500 genes across 441 tumors, including 134 lung, found lung adenocarcinomas and squamous cell carcinomas displayed high protein-altering mutation rates363, perhaps indicative of the inherent heterogeneity found in lung tumors compared with tumors from other tissues. One hurdle in second-generation sequencing is storage and analysis of the immense amount of data that is produced and separating biologically meaningful data from noise. However, the potential insight we will have into cancer genomes and its applicability to diagnostic sampling brings us even closer to the goal of ‘personalized medicine’.
Genome-wide functional (siRNA, shRNA library) screening
“Synthetic lethal” screens using RNAi (siRNAs and shRNA libraries) technology have allowed unbiased, genome-wide approaches to identification of genes whose perturbation can selectively kill lung cancer cells (Figure 1). The ability to identify “synthetic lethality” associated with oncogenic changes in tumor cells has particular utility in identifying new therapeutic targets or molecules to treat traditionally hard to target tumors, such as those with oncogenic KRAS. siRNA and shRNA screens have identified genes whose perturbation can selectively sensitize NSCLC cell lines to sub-lethal doses of chemotherapeutic agents364, sensitize KRAS mutant cells to targeted drugs126–128, suppress tumorigenicity in cells with specific gene dysregulation such as oncogenic KRAS123–125,365, or aberrant EGFR366,367, or identify novel genes critical for tumorigenic processes such as metastasis368.
Public databases and bioinformatics
Although the challenges in gathering reliable and clinically- and pathologically-annotated data are not trivial, high throughput technologies and publicly stored genome-wide databases related to lung cancer are resources with the potential to drive a global collaborative effort in identifying new targets for lung cancer diagnostics and therapeutics. Currently, and within the near future, all lung cancer investigators will have access to all of the genome-wide studies performed on lung cancers with the attached clinical annotation. This will allow independent confirmation on the role of the different molecular changes for prognosis, prediction, and targeting of therapy. With these tools researchers have enhanced ability to correlate patient subsets with augmented sensitivity to conventional or targeted therapeutics, distinguish driver versus passenger mutations, and better focus the design on novel therapeutic targets.
In vitro and in vivo model systems
While genome-wide approaches have the capacity of identifying novel genes or interactions in relation to lung cancer, the functional relevance of these findings need to be elucidated using preclinical model systems, namely in vitro models (such as tumor cell lines or immortalized human bronchial epithelial cells) and in vivo xenograft and transgenic mouse models of lung carcinogenesis. Experimental disease models play a crucial role in developing our understanding of lung carcinogenesis. Lung cancer cell lines and xenografts provide one set of important models. However, due to the genetic complexity of lung cancers they will usually have hundreds if not thousands of genetic/epigenetic changes. By contrast, two much simpler and equally valuable models, particularly to study the progression of lung carcinogenesis, are immortalized human bronchial epithelial cells (HBECs) and genetically engineered mouse models (GEMMs). These systems provide methods to reduce the inherent complexity and heterogeneity of the lung cancer genome and allow characterization of single or sequential genetic alterations in relation to the development, maintenance, and progression of lung cancer.
HBECs are derived from primary human airway epithelial cells and immortalized with either viral oncoproteins (such as SV40 early region) and hTERT369 or overexpression of Cdk4 and hTERT260,370. Stepwise transformation of these cells can be studied by the introduction of defined genetic manipulations commonly found in lung cancer371,372.
GEMMs allow the study of lung cancer pathogenesis with defined changes in the setting of the whole organism. They were critical in developing our understanding of oncogene dependence94, as observed in conditional KrasD12-induced lung adenocarcinomas, where switching off the driving oncogene was sufficient to induce tumor regression even in the presence of other non-driving oncogenic alterations373. Ensuing research has characterized several conditional lung tumor inducing combinations of oncogenic activations in mice (summarized in Table 6) which have been used to test new targeted therapies, improve effectiveness of conventional chemotherapies, identify biomarkers and imaging strategies for early detection, and study disease relapse and metastasis374.
Table 6.
Conditional genetically engineered mouse models of lung cancer
| Genetic manipulationa | Histopathology of lungs | Metastasis | Reference |
|---|---|---|---|
| BRAFV600E | Adenocarcinoma | 479 | |
| EGFRL858R or Del | Adenocarcinoma with BAC features | 480,481 | |
| EGFRT790M or T790M+L858R or T790M+Del | Adenocarcinoma | 482,483 | |
| EGFRvIII | Adenocarcinoma | None | 484 |
| EML4/ALK | Adenocarcinoma with BAC features | 485 | |
| HER2YVMA | Adenosquamous | Yes | 486 |
| KrasD12 | Adenocarcinoma | None | 487 |
| KrasV12 | Adenocarcinoma | Lymph node, kidney | 488 |
| KrasD12 + HIF2α | Yes | 489 | |
| KrasD12 + Ink4a/Arf−/− | Adenocarcinoma | None | 446 |
| KrasD12 + p16Ink4a−/− | Adenocarcinoma | Yes | 446 |
| KrasD12 + p53L/L− | Adenocarcinoma | Yes | 446 |
| KrasD12 + p53F/F | Adenocarcinoma | Yes | 490 |
| KrasD12 + p53R270H/F | Adenocarcinoma | 490 | |
| KrasD12 + p53R172H/F | Adenocarcinoma | 490 | |
| KrasD12 + PTENΔ5/Δ5 | Adenocarcinoma | 491 | |
| KrasD12 + Lkb1L/L or L/− | Squamous cell, adenosquamous, large cell | Yes (lymph node, skeletal) | 446 |
| KrasD12 + Lkb1 L/+ or +/− | Adenocarcinoma | Yes (lymph node, skeletal) | 446 |
| PIK3CAH1047R | Adenocarcinoma with BAC features | 128 | |
| p16Ink4a−/− + p53L/L | None | 446 | |
| p53+/− | Adenocarcinoma | 492 | |
| p53F/F | Adenocarcinoma | 493 | |
| p53R172H/+ or R172H/− | Adenocarcinoma | 492 | |
| p53R270H/+ or R270H/− | Adenocarcinoma, squamous cell | 492 | |
| p53F/F + RbF/F | SCLC | Yes | 493 |
-, germline null allele; L or F, conditional knockout allele;
Summary
This review has outlined some of the significant molecular alterations known to be involved in the initiation and/or progression of lung cancer. Continued development of targeted therapies for the treatment of lung cancer is dependent upon increased understanding of involved molecules and pathways. Cancer genome analyses are identifying 100s to 1000s of candidate targets but these all require molecular and clinical validation. Furthermore, it is becoming increasingly apparent that targeting a single molecule will not be enough due to the non-linearity of pathways involved in carcinogenesis. Rather, targeting multiple molecules at once to combat the inter-connective and complex signaling pathways will improve efficacy. Recent next-generation sequencing efforts are revealing the lung cancer genome is mutated at a high rate, likely contributing to the known heterogeneity of these tumors and explaining the lack of identifying effective conventional and targeted therapies that have a universal effect in lung cancer. Systematic understanding of the molecular basis of lung cancer through comprehensive characterization of aberrations in the cancer genome and their functionality will provide the means to evaluate their use in diagnosis, prognosis and therapy. Integration of clinical and biological factors will ultimately lead to improved detection, diagnosis, treatment, and prognosis of lung cancer by achieving “personalized medicine”, the selection of the best treatment for each patient based on tumor associated biomarkers.
Acknowledgements
This research was supported by:
National Cancer Institute Lung Cancer Specialized Program of Research Excellence (SPORE) (P50CA70907), Department of Defense VITAL (W81XWH0410142) and PROSPECT (W81XWH0710306), NASA NSCOR (NNJ05HD36G), NASA (NNJ05HD36G) and by the Office of Science (BER) U.S. Department of Energy, Grant Number DE-AI02-05ER64068. JEL supported by NH&MRC Biomedical Fellowship (494511).
We thank the many current and past members of the Minna lab for their contributions to lung cancer translational research and our especially our long term collaborator Dr. Adi Gazdar. Also we apologize to other investigators for omission of any references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010 Sep-Oct;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331–348. doi: 10.1146/annurev.pathol.1.110304.100103. [DOI] [PubMed] [Google Scholar]
- 3.Sekido Y, Fong KM, Minna JD. Progress in understanding the molecular pathogenesis of human lung cancer. Biochim Biophys Acta. 1998 Aug 19;1378(1):F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976 Oct 1;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 5.Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer cell international. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Gazdar AF. Should we continue to use the term non-small-cell lung cancer? Ann Oncol. 2010 Oct;21(Suppl 7):vii225–vii229. doi: 10.1093/annonc/mdq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007 Oct;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 10.Scagliotti GV, Longo M, Novello S. Nonsmall cell lung cancer in never smokers. Curr Opin Oncol. 2009 Mar;21(2):99–104. doi: 10.1097/CCO.0b013e328321049e. [DOI] [PubMed] [Google Scholar]
- 11.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008 Jul 1;123(1):1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 12.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008 Sep 25;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008 May;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008 Apr 3;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008 Apr 3;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafnar T, Sulem P, Besenbacher S, et al. Genome-wide significant association between a sequence variant at 15q15.2 and lung cancer risk. Cancer Res. 2011 Feb 15;71(4):1356–1361. doi: 10.1158/0008-5472.CAN-10-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009 Aug 15;69(16):6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Marchand L, Derby KS, Murphy SE, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008 Nov 15;68(22):9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24–25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008 Nov 5;100(21):1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiraishi K, Kohno T, Kunitoh H, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009 Jan;30(1):65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- 21.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007 Jan 1;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008 Jul;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens VL, Bierut LJ, Talbot JT, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008 Dec;17(12):3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. The American journal of psychiatry. 2008 Sep;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paliwal A, Vaissiere T, Krais A, et al. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res. 2010 Apr 1;70(7):2779–2788. doi: 10.1158/0008-5472.CAN-09-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Liu P, Wen W, et al. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24–25.1. Cancer Res. 2009 Oct 1;69(19):7844–7850. doi: 10.1158/0008-5472.CAN-09-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet. 2004 Sep;75(3):460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amos CI, Pinney SM, Li Y, et al. A susceptibility locus on chromosome 6q greatly increases lung cancer risk among light and never smokers. Cancer Res. 2010 Mar 15;70(6):2359–2367. doi: 10.1158/0008-5472.CAN-09-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You M, Wang D, Liu P, et al. Fine mapping of chromosome 6q23–25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin Cancer Res. 2009 Apr 15;15(8):2666–2674. doi: 10.1158/1078-0432.CCR-08-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005 Mar-Apr;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 31.Tan YK, Wee TC, Koh WP, et al. Survival among Chinese women with lung cancer in Singapore: a comparison by stage, histology and smoking status. Lung Cancer. 2003 Jun;40(3):237–246. doi: 10.1016/s0169-5002(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 32.Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest. 2004 Aug;126(2):347–351. doi: 10.1378/chest.126.2.347. [DOI] [PubMed] [Google Scholar]
- 33.Toh CK, Gao F, Lim WT, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol. 2006 May 20;24(15):2245–2251. doi: 10.1200/JCO.2005.04.8033. [DOI] [PubMed] [Google Scholar]
- 34.Janjigian YY, McDonnell K, Kris MG, et al. Pack-years of cigarette smoking as a prognostic factor in patients with stage IIIB/IV nonsmall cell lung cancer. Cancer. 2010 Feb 1;116(3):670–675. doi: 10.1002/cncr.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007 Feb 10;25(5):561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 36.Rudin CM, Avila-Tang E, Harris CC, et al. Lung cancer in never smokers: molecular profiles and therapeutic implications. Clin Cancer Res. 2009 Sep 15;15(18):5646–5661. doi: 10.1158/1078-0432.CCR-09-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990 Apr 6;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 38.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989 Feb 17;243(4893):934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 39.Klein F, Kotb WF, Petersen I. Incidence of human papilloma virus in lung cancer. Lung Cancer. 2008 Nov 17; doi: 10.1016/j.lungcan.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Koshiol J, Rotunno M, Gillison ML, et al. Assessment of human papillomavirus in lung tumor tissue. J Natl Cancer Inst. 2011 Mar 16;103(6):501–507. doi: 10.1093/jnci/djr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010 May 27;465(7297):473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 42.Pleasance ED, Stephens PJ, O' Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010 Jan 14;463(7278):184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008 Jun;40(6):722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kris MG, Johnson BE, Kwiatkowski DJ, et al. Identification of driver mutations in tumor specimens from 1000 patients with lung adenocarcinoma: The NCI’s lung cancer mutation consortium (LCMC) J Clin Oncol. 2011;29(suppl) abstr CRA7506. [Google Scholar]
- 45.Fleming TP, Saxena A, Clark WC, et al. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors. Cancer Res. 1992 Aug 15;52(16):4550–4553. [PubMed] [Google Scholar]
- 46.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003 Jan 31;299(5607):708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 47.Lin CR, Chen WS, Kruiger W, et al. Expression cloning of human EGF receptor complementary DNA: gene amplification and three related messenger RNA products in A431 cells. Science. 1984 May 25;224(4651):843–848. doi: 10.1126/science.6326261. [DOI] [PubMed] [Google Scholar]
- 48.Merlino GT, Xu YH, Ishii S, et al. Amplification and enhanced expression of the epidermal growth factor receptor gene in A431 human carcinoma cells. Science. 1984 Apr 27;224(4647):417–419. doi: 10.1126/science.6200934. [DOI] [PubMed] [Google Scholar]
- 49.Ullrich A, Coussens L, Hayflick JS, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984 May-Jun;309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 50.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 51.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004 Jun 4;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 52.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004 Sep 7;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semba K, Kamata N, Toyoshima K, Yamamoto T. A v-erbB-related protooncogene, c-erbB-2, is distinct from the c-erbB-1/epidermal growth factor-receptor gene and is amplified in a human salivary gland adenocarcinoma. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6497–6501. doi: 10.1073/pnas.82.19.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004 Sep 30;431(7008):525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 55.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999 Jan;21(1):99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 56.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002 Jul 1;62(13):3636–3640. [PubMed] [Google Scholar]
- 57.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004 Apr 23;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 58.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986 Oct 16–22;323(6089):643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 59.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994 Apr 15;264(5157):436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 60.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994 Apr 21;368(6473):753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997 Mar 28;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 62.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997 Apr;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 63.Fong KM, Kida Y, Zimmerman PV, Ikenaga M, Smith PJ. Loss of heterozygosity frequently affects chromosome 17q in non-small cell lung cancer. Cancer Res. 1995 Oct 1;55(19):4268–4272. [PubMed] [Google Scholar]
- 64.Fong KM, Sekido Y, Minna JD. Molecular pathogenesis of lung cancer. J Thorac Cardiovasc Surg. 1999 Dec;118(6):1136–1152. doi: 10.1016/S0022-5223(99)70121-2. [DOI] [PubMed] [Google Scholar]
- 65.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 66.Zimmerman PV, Hawson GA, Bint MH, Parsons PG. Ploidy as a prognostic determinant in surgically treated lung cancer. Lancet. 1987;2(8558):530–533. doi: 10.1016/s0140-6736(87)92923-0. [DOI] [PubMed] [Google Scholar]
- 67.Fong KM, Zimmerman PV, Smith PJ. Correlation of loss of heterozygosity at 11p with tumour progression and survival in non-small cell lung cancer. Genes, Chromosomes & Cancer. 1994;10(3):183–189. doi: 10.1002/gcc.2870100306. [DOI] [PubMed] [Google Scholar]
- 68.Fong KM, Zimmerman PV, Smith PJ. Microsatellite instability and other molecular abnormalities in non-small cell lung cancer. Cancer Res. 1995 Jan 1;55(1):28–30. [PubMed] [Google Scholar]
- 69.Fong KM, Zimmerman PV, Smith PJ. Tumor progression and loss of heterozygosity at 5q and 18q in non-small cell lung cancer. Cancer Res. 1995 Jan 15;55(2):220–223. [PubMed] [Google Scholar]
- 70.Fong KM, Kida Y, Zimmerman PV, Smith PJ. MYCL genotypes and loss of heterozygosity in non-small-cell lung cancer. Br J Cancer. 1996 Dec;74(12):1975–1978. doi: 10.1038/bjc.1996.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fong KM, Zimmerman PV, Smith PJ. KRAS codon 12 mutations in Australian non-small cell lung cancer. Aust N Z J Med. 1998 Apr;28(2):184–189. doi: 10.1111/j.1445-5994.1998.tb02967.x. [DOI] [PubMed] [Google Scholar]
- 72.Virmani AK, Fong KM, Kodagoda D, et al. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer. 1998 Apr;21(4):308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 73.Geradts J, Fong KM, Zimmerman PV, Minna JD. Loss of Fhit expression in non-small-cell lung cancer: correlation with molecular genetic abnormalities and clinicopathological features. Br J Cancer. 2000 Mar;82(6):1191–1197. doi: 10.1054/bjoc.1999.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007 Dec 6;450(7171):893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009 Nov;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010 Dec 15;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanchez-Cespedes M, Ahrendt SA, Piantadosi S, et al. Chromosomal alterations in lung adenocarcinoma from smokers and nonsmokers. Cancer Res. 2001 Feb 15;61(4):1309–1313. [PubMed] [Google Scholar]
- 78.Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011 May 2; doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim YH, Kwei KA, Girard L, et al. Genomic and functional analysis identifies CRKL as an oncogene amplified in lung cancer. Oncogene. 2010 Mar 11;29(10):1421–1430. doi: 10.1038/onc.2009.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan P, Kadara H, Behrens C, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010;5(2):e9112. doi: 10.1371/journal.pone.0009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang F, Yin Z, Caraway NP, Li R, Katz RL. Genomic profiles in stage I primary non small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia. 2004 Sep-Oct;6(5):623–635. doi: 10.1593/neo.04142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim TM, Yim SH, Lee JS, et al. Genome-wide screening of genomic alterations and their clinicopathologic implications in non-small cell lung cancers. Clin Cancer Res. 2005 Dec 1;11(23):8235–8242. doi: 10.1158/1078-0432.CCR-05-1157. [DOI] [PubMed] [Google Scholar]
- 83.Shibata T, Uryu S, Kokubu A, et al. Genetic classification of lung adenocarcinoma based on array-based comparative genomic hybridization analysis: its association with clinicopathologic features. Clin Cancer Res. 2005 Sep 1;11(17):6177–6185. doi: 10.1158/1078-0432.CCR-05-0293. [DOI] [PubMed] [Google Scholar]
- 84.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005 Jul 5;102(27):9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wikman H, Nymark P, Vayrynen A, et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3–q14.1 in lung cancer. Genes Chromosomes Cancer. 2005 Feb;42(2):193–199. doi: 10.1002/gcc.20122. [DOI] [PubMed] [Google Scholar]
- 86.Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005 Jul 1;65(13):5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 87.Choi JS, Zheng LT, Ha E, et al. Comparative genomic hybridization array analysis and real-time PCR reveals genomic copy number alteration for lung adenocarcinomas. Lung. 2006 Nov-Dec;184(6):355–362. doi: 10.1007/s00408-006-0009-0. [DOI] [PubMed] [Google Scholar]
- 88.Choi YW, Choi JS, Zheng LT, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung Cancer. 2007 Jan;55(1):43–51. doi: 10.1016/j.lungcan.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 89.Zhu H, Lam DCL, Han KC, et al. High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Letters. 2007 Jan 8;255(1–2):303–314. doi: 10.1016/j.canlet.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 90.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006 Mar 15;118(6):1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 91.Dehan E, Ben-Dor A, Liao W, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007 May;56(2):175–184. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 92.Girard N, Ostrovnaya I, Lau C, et al. Genomic and mutational profiling to assess clonal relationships between multiple non-small cell lung cancers. Clin Cancer Res. 2009 Aug 15;15(16):5184–5190. doi: 10.1158/1078-0432.CCR-09-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008 Jun 5;27(25):3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002 Jul 5;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 95.Normanno N, Bianco C, Strizzi L, et al. The ErbB receptors and their ligands in cancer: an overview. Curr Drug Targets. 2005 May;6(3):243–257. doi: 10.2174/1389450053765879. [DOI] [PubMed] [Google Scholar]
- 96.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003 Oct 15;21(20):3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 97.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005 Sep 1;23(25):5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 98.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004 Aug 20;305(5687):1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 99.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005 Mar 2;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 100.Rusch V, Baselga J, Cordon-Cardo C, et al. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res. 1993 May 15;53(10) Suppl:2379–2385. [PubMed] [Google Scholar]
- 101.Franklin WA, Veve R, Hirsch FR, Helfrich BA, Bunn PA., Jr Epidermal growth factor receptor family in lung cancer and premalignancy. Semin Oncol. 2002 Feb;29(1) Suppl 4:3–14. doi: 10.1053/sonc.2002.31520. [DOI] [PubMed] [Google Scholar]
- 102.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(2) Suppl:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 103.Fujino S, Enokibori T, Tezuka N, et al. A comparison of epidermal growth factor receptor levels and other prognostic parameters in non-small cell lung cancer. Eur J Cancer. 1996 Nov;32A(12):2070–2074. doi: 10.1016/s0959-8049(96)00243-2. [DOI] [PubMed] [Google Scholar]
- 104.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009 Aug;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomas RK, Greulich H, Yuza Y, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- 106.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005 Jan;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005 Mar;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007 May 18;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 110.Gao SP, Mark KG, Leslie K, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007 Dec;117(12):3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ishihara K, Hirano T. Molecular basis of the cell specificity of cytokine action. Biochim Biophys Acta. 2002 Nov 11;1592(3):281–296. doi: 10.1016/s0167-4889(02)00321-x. [DOI] [PubMed] [Google Scholar]
- 112.Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for Translational Therapeutics. Cancer. 2007 Nov 1;110(9):1911–1928. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 113.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003 Jan;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 114.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008 Jul;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992 May 1;52(9) Suppl:2665s–2669s. [PubMed] [Google Scholar]
- 116.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008 Oct;9(10):962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 117.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008 Sep 15;14(18):5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adjei AA. K-ras as a target for lung cancer therapy. J Thorac Oncol. 2008 Jun;3(6) Suppl 2:S160–S163. doi: 10.1097/JTO.0b013e318174dbf9. [DOI] [PubMed] [Google Scholar]
- 119.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008 Oct 23;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 121.Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002 Dec 1;62(23):7001–7003. [PubMed] [Google Scholar]
- 122.Sasaki H, Kawano O, Endo K, et al. Uncommon V599E BRAF mutations in Japanese patients with lung cancer. J Surg Res. 2006 Jun 15;133(2):203–206. doi: 10.1016/j.jss.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 123.Scholl C, Frohling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009 May 29;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 124.Guo W, Wu S, Liu J, Fang B. Identification of a small molecule with synthetic lethality for K-ras and protein kinase C iota. Cancer Res. 2008 Sep 15;68(18):7403–7408. doi: 10.1158/0008-5472.CAN-08-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009 May 29;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009 Jun 2;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sunaga N, Shames DS, Girard L, et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011 Feb;10(2):336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008 Dec;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krystal G, Birrer M, Way J, et al. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Richardson GE, Johnson BE. The biology of lung cancer. Semin Oncol. 1993 Apr;20(2):105–127. [PubMed] [Google Scholar]
- 131.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005 Aug;6(8):635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 132.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008 Dec;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 133.Nau MM, Brooks BJ, Jr, Carney DN, et al. Human small-cell lung cancers show amplification and expression of the N-myc gene. Proc Natl Acad Sci U S A. 1986 Feb;83(4):1092–1096. doi: 10.1073/pnas.83.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Broers JL, Viallet J, Jensen SM, et al. Expression of c-myc in progenitor cells of the bronchopulmonary epithelium and in a large number of non-small cell lung cancers. Am J Respir Cell Mol Biol. 1993 Jul;9(1):33–43. doi: 10.1165/ajrcmb/9.1.33. [DOI] [PubMed] [Google Scholar]
- 135.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007 Aug 2;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 136.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009 Dec;4(12):1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 137.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.De Francesco F, Tirino V, Desiderio V, et al. Human CD34/CD90 ASCs are capable of growing as sphere clusters, producing high levels of VEGF and forming capillaries. PLoS One. 2009;4(8):e6537. doi: 10.1371/journal.pone.0006537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tirino V, Camerlingo R, Franco R, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009 Sep;36(3):446–453. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- 140.De Rosa A, De Francesco F, Tirino V, et al. A new method for the cryopreserving ASCs: an attractive and suitable large-scale and long-term cell banking technology. Tissue Eng Part C Methods. 2009 Mar 2; doi: 10.1089/ten.TEC.2008.0674. [DOI] [PubMed] [Google Scholar]
- 141.Costantino E, Maddalena F, Calise S, et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009 Jun 28;279(1):39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 142.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006 Aug;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 143.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002 Jul;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 144.West KA, Linnoila IR, Belinsky SA, Harris CC, Dennis PA. Tobacco carcinogen-induced cellular transformation increases activation of the phosphatidylinositol 3'-kinase/Akt pathway in vitro and in vivo. Cancer Res. 2004 Jan 15;64(2):446–451. doi: 10.1158/0008-5472.can-03-3241. [DOI] [PubMed] [Google Scholar]
- 145.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005 Sep;8(3):179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 146.Maulik G, Madhiwala P, Brooks S, et al. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. J Cell Mol Med. 2002 Oct-Dec;6(4):539–553. doi: 10.1111/j.1582-4934.2002.tb00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001 May 15;61(10):3986–3997. [PubMed] [Google Scholar]
- 148.Tsurutani J, West KA, Sayyah J, Gills JJ, Dennis PA. Inhibition of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway but not the MEK/ERK pathway attenuates laminin-mediated small cell lung cancer cellular survival and resistance to imatinib mesylate or chemotherapy. Cancer Res. 2005 Sep 15;65(18):8423–8432. doi: 10.1158/0008-5472.CAN-05-0058. [DOI] [PubMed] [Google Scholar]
- 149.Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007 Oct 16;104(42):16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bingle CD. Thyroid transcription factor-1. Int J Biochem Cell Biol. 1997 Dec;29(12):1471–1473. doi: 10.1016/s1357-2725(97)00007-1. [DOI] [PubMed] [Google Scholar]
- 151.Ikeda K, Clark JC, Shaw-White JR, Stahlman MT, Boutell CJ, Whitsett JA. Gene structure and expression of human thyroid transcription factor-1 in respiratory epithelial cells. J Biol Chem. 1995 Apr 7;270(14):8108–8114. doi: 10.1074/jbc.270.14.8108. [DOI] [PubMed] [Google Scholar]
- 152.Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007 Jul 1;67(13):6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 153.Winslow MM, Dayton TL, Verhaak RG, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011 May 5;473(7345):101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Visakorpi T, Hyytinen E, Koivisto P, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995 Apr;9(4):401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 155.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005 Jul 7;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 156.Holst F, Stahl PR, Ruiz C, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007 May;39(5):655–660. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 157.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006 Aug;6(8):593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 158.Knudson AG., Jr The ninth Gordon Hamilton-Fairley memorial lecture. Hereditary cancers: clues to mechanisms of carcinogenesis. Br J Cancer. 1989 May;59(5):661–666. doi: 10.1038/bjc.1989.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Breuer RH, Postmus PE, Smit EF. Molecular pathology of non-small-cell lung cancer. Respiration. 2005 May-Jun;72(3):313–330. doi: 10.1159/000085376. [DOI] [PubMed] [Google Scholar]
- 160.Takahashi T, Nau MM, Chiba I, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989 Oct 27;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 161.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 162.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994 Sep 15;54(18):4855–4878. [PubMed] [Google Scholar]
- 163.van Oijen MG, Slootweg PJ. Gain-of-function mutations in the tumor suppressor gene p53. Clin Cancer Res. 2000 Jun;6(6):2138–2145. [PubMed] [Google Scholar]
- 164.Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007 Feb 8;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 165.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988 Jul 15;241(4863):353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yokota J, Mori N, Akiyama T, Shimosato Y, Sugimura T, Terada M. Multiple genetic alterations in small-cell lung carcinoma. Princess Takamatsu Symp. 1989;20:43–48. [PubMed] [Google Scholar]
- 167.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994 Nov;9(11):3375–3378. [PubMed] [Google Scholar]
- 168.Wistuba II, Behrens C, Virmani AK, et al. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000 Apr 1;60(7):1949–1960. [PubMed] [Google Scholar]
- 169.Ito M, Ito G, Kondo M, et al. Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer Lett. 2005 Jul 8;225(1):131–139. doi: 10.1016/j.canlet.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 170.Kuroki T, Trapasso F, Yendamuri S, et al. Allelic loss on chromosome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-small cell lung cancer. Cancer Res. 2003 Jun 15;63(12):3352–3355. [PubMed] [Google Scholar]
- 171.Feng Q, Hawes SE, Stern JE, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008 Mar;17(3):645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001 Apr;28(2) Suppl 4:3–13. [PubMed] [Google Scholar]
- 173.Zochbauer-Muller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist. 2002;7(5):451–457. doi: 10.1634/theoncologist.7-5-451. [DOI] [PubMed] [Google Scholar]
- 174.Siprashvili Z, Sozzi G, Barnes LD, et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005 May 1;65(9):3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 176.Ji L, Roth JA. Tumor suppressor FUS1 signaling pathway. J Thorac Oncol. 2008 Apr;3(4):327–330. doi: 10.1097/JTO.0b013e31816bce65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Deng WG, Kawashima H, Wu G, et al. Synergistic tumor suppression by coexpression of FUS1 and p53 is associated with down-regulation of murine double minute-2 and activation of the apoptotic protease-activating factor 1-dependent apoptotic pathway in human non-small cell lung cancer cells. Cancer Res. 2007 Jan 15;67(2):709–717. doi: 10.1158/0008-5472.CAN-06-3463. [DOI] [PubMed] [Google Scholar]
- 178.Tomizawa Y, Sekido Y, Kondo M, et al. Inhibition of lung cancer cell growth and induction of apoptosis after reexpression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13954–13959. doi: 10.1073/pnas.231490898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ochi K, Mori T, Toyama Y, Nakamura Y, Arakawa H. Identification of semaphorin3B as a direct target of p53. Neoplasia. 2002 Jan-Feb;4(1):82–87. doi: 10.1038/sj.neo.7900211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Castro-Rivera E, Ran S, Brekken RA, Minna JD. Semaphorin 3B inhibits the phosphatidylinositol 3-kinase/Akt pathway through neuropilin-1 in lung and breast cancer cells. Cancer Res. 2008 Oct 15;68(20):8295–8303. doi: 10.1158/0008-5472.CAN-07-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brambilla E, Constantin B, Drabkin H, Roche J. Semaphorin SEMA3F localization in malignant human lung and cell lines: A suggested role in cell adhesion and cell migration. Am J Pathol. 2000 Mar;156(3):939–950. doi: 10.1016/S0002-9440(10)64962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kessler O, Shraga-Heled N, Lange T, et al. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res. 2004 Feb 1;64(3):1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 183.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 184.Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998 Jan 8;391(6663):184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 185.Sanchez-Cespedes M, Parrella P, Esteller M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002 Jul 1;62(13):3659–3662. [PubMed] [Google Scholar]
- 186.Carretero J, Medina PP, Pio R, Montuenga LM, Sanchez-Cespedes M. Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene. 2004 May 13;23(22):4037–4040. doi: 10.1038/sj.onc.1207502. [DOI] [PubMed] [Google Scholar]
- 187.Matsumoto S, Iwakawa R, Takahashi K, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007 Aug 30;26(40):5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shaw RJ, Bardeesy N, Manning BD, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004 Jul;6(1):91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 189.Onozato R, Kosaka T, Achiwa H, et al. LKB1 gene mutations in Japanese lung cancer patients. Cancer Sci. 2007 Nov;98(11):1747–1751. doi: 10.1111/j.1349-7006.2007.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mahoney CL, Choudhury B, Davies H, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer. 2009 Jan 27;100(2):370–375. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov 1;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 192.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005 Sep;15(9):494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 193.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006 Oct 1;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 194.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994 Feb 17;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 195.Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3(7):e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009 Mar;7(3):330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kitamura H, Okudela K, Yazawa T, Sato H, Shimoyamada H. Cancer stem cell: implications in cancer biology and therapy with special reference to lung cancer. Lung Cancer. 2009 Dec;66(3):275–281. doi: 10.1016/j.lungcan.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 198.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007 May 15;67(10):4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 199.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008 Mar;15(3):504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 200.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006 Feb 15;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895-1886. [DOI] [PubMed] [Google Scholar]
- 201.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006 Dec;5(12):1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 202.Riobo NA, Lu K, Emerson CP., Jr Hedgehog signal transduction: signal integration and cross talk in development and cancer. Cell Cycle. 2006 Aug;5(15):1612–1615. doi: 10.4161/cc.5.15.3130. [DOI] [PubMed] [Google Scholar]
- 203.Lauth M, Toftgard R. Non-canonical activation of GLI transcription factors: implications for targeted anti-cancer therapy. Cell Cycle. 2007 Oct 15;6(20):2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- 204.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003 Mar 20;422(6929):313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 205.Yuan Z, Goetz JA, Singh S, et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007 Feb 15;26(7):1046–1055. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 206.He B, You L, Uematsu K, et al. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia. 2004 Jan-Feb;6(1):7–14. doi: 10.1016/s1476-5586(04)80048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.You L, He B, Xu Z, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004 Aug 12;23(36):6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 208.Winn RA, Van Scoyk M, Hammond M, et al. Antitumorigenic effect of Wnt 7a and Fzd 9 in non-small cell lung cancer cells is mediated through ERK-5-dependent activation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2006 Sep 15;281(37):26943–26950. doi: 10.1074/jbc.M604145200. [DOI] [PubMed] [Google Scholar]
- 209.Fukui T, Kondo M, Ito G, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005 Sep 15;24(41):6323–6327. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- 210.Mazieres J, He B, You L, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004 Jul 15;64(14):4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 211.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003 Oct 16;22(46):7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 212.Wissmann C, Wild PJ, Kaiser S, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003 Oct;201(2):204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 213.Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003 Apr 3;22(13):1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 214.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004 Oct;14(5):341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 215.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004 Nov;14(5):779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 216.Hainaud P, Contreres JO, Villemain A, et al. The role of the vascular endothelial growth factor-Delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions. Cancer Res. 2006 Sep 1;66(17):8501–8510. doi: 10.1158/0008-5472.CAN-05-4226. [DOI] [PubMed] [Google Scholar]
- 217.Daniel VC, Peacock CD, Watkins DN. Developmental signalling pathways in lung cancer. Respirology. 2006 May;11(3):234–240. doi: 10.1111/j.1440-1843.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 218.Olsen CL, Hsu PP, Glienke J, Rubanyi GM, Brooks AR. Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer. 2004 Aug 4;4:43. doi: 10.1186/1471-2407-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003 Sep 29;22(42):6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 220.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008 Aug 15;68(16):6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 221.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009 Sep 17;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 222.Luistro L, He W, Smith M, et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009 Oct 1;69(19):7672–7680. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wei P, Walls M, Qiu M, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010 Jun;9(6):1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 224.Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010 Dec 1;70(23):9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010 Aug;34(8):1193–1198. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sholl LM, Long KB, Hornick JL. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2010 Jan;18(1):55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- 227.Korpanty G, Smyth E, Sullivan LA, Brekken RA, Carney DN. Antiangiogenic therapy in lung cancer: focus on vascular endothelial growth factor pathway. Experimental biology and medicine. 2010 Jan;235(1):3–9. doi: 10.1258/ebm.2009.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocrine reviews. 2004 Aug;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 229.Stefanou D, Batistatou A, Arkoumani E, Ntzani E, Agnantis NJ. Expression of vascular endothelial growth factor (VEGF) and association with microvessel density in small-cell and non-small-cell lung carcinomas. Histol Histopathol. 2004 Jan;19(1):37–42. doi: 10.14670/HH-19.37. [DOI] [PubMed] [Google Scholar]
- 230.Kaya A, Ciledag A, Gulbay BE, et al. The prognostic significance of vascular endothelial growth factor levels in sera of non-small cell lung cancer patients. Respir Med. 2004 Jul;98(7):632–636. doi: 10.1016/j.rmed.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 231.Dudek AZ, Mahaseth H. Circulating angiogenic cytokines in patients with advanced non-small cell lung cancer: correlation with treatment response and survival. Cancer Invest. 2005;23(3):193–200. doi: 10.1081/cnv-200055949. [DOI] [PubMed] [Google Scholar]
- 232.Jantus-Lewintre E, Sanmartin E, Sirera R, et al. Combined VEGF-A and VEGFR-2 concentrations in plasma: Diagnostic and prognostic implications in patients with advanced NSCLC. Lung Cancer. 2011 Apr 7; doi: 10.1016/j.lungcan.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 233.Hasani A, Leighl NB. Targeting vascular endothelial growth factor in lung cancer. J Thorac Oncol. 2010 Dec;5(12) Suppl 6:S484–S486. doi: 10.1097/01.JTO.0000391377.17179.6b. [DOI] [PubMed] [Google Scholar]
- 234.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006 Dec 14;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 235.Yang JC, Haworth L, Sherry RM, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med. 2003 Jul 31;349(5):427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000 Aug;12(8):1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 237.Heist RS, Zhai R, Liu G, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008 Feb 20;26(6):856–862. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- 238.Schneider BP, Radovich M, Sledge GW, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat. 2008 Sep;111(1):157–163. doi: 10.1007/s10549-007-9755-9. [DOI] [PubMed] [Google Scholar]
- 239.Sautes-Fridman C, Cherfils-Vicini J, Damotte D, et al. Tumor microenvironment is multifaceted. Cancer Metastasis Rev. 2011 Mar;30(1):13–25. doi: 10.1007/s10555-011-9279-y. [DOI] [PubMed] [Google Scholar]
- 240.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 241.Schwartz DL, Powis G, Thitai-Kumar A, et al. The selective hypoxia inducible factor-1 inhibitor PX-478 provides in vivo radiosensitization through tumor stromal effects. Mol Cancer Ther. 2009 Apr;8(4):947–958. doi: 10.1158/1535-7163.MCT-08-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Williams KJ, Telfer BA, Shannon AM, Babur M, Stratford IJ, Wedge SR. Combining radiotherapy with AZD2171, a potent inhibitor of vascular endothelial growth factor signaling: pathophysiologic effects and therapeutic benefit. Mol Cancer Ther. 2007 Feb;6(2):599–606. doi: 10.1158/1535-7163.MCT-06-0508. [DOI] [PubMed] [Google Scholar]
- 243.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002 Jun;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 244.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006 Mar 27;172(7):973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009 Nov 25;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 246.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008 May 16;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chiou SH, Wang ML, Chou YT, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010 Dec 15;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 248.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009 Dec;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 249.Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS, Wu KJ. Prognostic significance of hypoxia-inducible factor-1alpha, TWIST1 and Snail expression in resectable non-small cell lung cancer. Thorax. 2009 Dec;64(12):1082–1089. doi: 10.1136/thx.2009.115691. [DOI] [PubMed] [Google Scholar]
- 250.Miura N, Yano T, Shoji F, et al. Clinicopathological significance of Sip1-associated epithelial mesenchymal transition in non-small cell lung cancer progression. Anticancer Res. 2009 Oct;29(10):4099–4106. [PubMed] [Google Scholar]
- 251.Soltermann A, Tischler V, Arbogast S, et al. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008 Nov 15;14(22):7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- 252.Yauch RL, Januario T, Eberhard DA, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005 Dec 15;11(24 Pt 1):8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 253.Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005 Oct 15;65(20):9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 254.Roy BC, Kohno T, Iwakawa R, et al. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010 Nov;70(2):136–145. doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 255.Dohadwala M, Yang SC, Luo J, et al. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006 May 15;66(10):5338–5345. doi: 10.1158/0008-5472.CAN-05-3635. [DOI] [PubMed] [Google Scholar]
- 256.Krysan K, Lee JM, Dohadwala M, et al. Inflammation, epithelial to mesenchymal transition, and epidermal growth factor receptor tyrosine kinase inhibitor resistance. J Thorac Oncol. 2008 Feb;3(2):107–110. doi: 10.1097/JTO.0b013e3181630ece. [DOI] [PubMed] [Google Scholar]
- 257.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008 Apr 1;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008 May;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 259.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009 Sep 15;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tellez CS, Juri DE, Do K, et al. EMT and Stem Cell-Like Properties Associated with miR-205 and miR-200 Epigenetic Silencing Are Early Manifestations during Carcinogen-Induced Transformation of Human Lung Epithelial Cells. Cancer Res. 2011 Apr 15;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Albanell J, Lonardo F, Rusch V, et al. High telomerase activity in primary lung cancers: association with increased cell proliferation rates and advanced pathologic stage. J Natl Cancer Inst. 1997 Nov 5;89(21):1609–1615. doi: 10.1093/jnci/89.21.1609. [DOI] [PubMed] [Google Scholar]
- 262.Hiyama K, Hiyama E, Ishioka S, et al. Telomerase activity in small-cell and non-small-cell lung cancers. J Natl Cancer Inst. 1995 Jun 21;87(12):895–902. doi: 10.1093/jnci/87.12.895. [DOI] [PubMed] [Google Scholar]
- 263.Frias C, Garcia-Aranda C, De Juan C, et al. Telomere shortening is associated with poor prognosis and telomerase activity correlates with DNA repair impairment in non-small cell lung cancer. Lung Cancer. 2008 Jun;60(3):416–425. doi: 10.1016/j.lungcan.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 264.Shay JW, Wright WE. Telomerase activity in human cancer. Curr Opin Oncol. 1996 Jan;8(1):66–71. doi: 10.1097/00001622-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 265.Ouellette MM, Wright WE, Shay JW. Targeting Telomerase-Expressing Cancer Cells. J Cell Mol Med. 2011 Feb 18; doi: 10.1111/j.1582-4934.2011.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Molckovsky A, Siu LL. First-in-class, first-in-human phase I results of targeted agents: highlights of the 2008 American society of clinical oncology meeting. Journal of hematology & oncology. 2008;1:20. doi: 10.1186/1756-8722-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Vonderheide RH, Hahn WC, Schultze JL, Nadler LM. The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity. 1999 Jun;10(6):673–679. doi: 10.1016/s1074-7613(00)80066-7. [DOI] [PubMed] [Google Scholar]
- 268.Vonderheide RH. Telomerase as a universal tumor-associated antigen for cancer immunotherapy. Oncogene. 2002 Jan 21;21(4):674–679. doi: 10.1038/sj.onc.1205074. [DOI] [PubMed] [Google Scholar]
- 269.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002 Jan 1;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 270.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001 Apr;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 271.Kim H, Kwon YM, Kim JS, et al. Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J Clin Oncol. 2004 Jun 15;22(12):2363–2370. doi: 10.1200/JCO.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 272.Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia. 2006 Jan;8(1):46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Vaissiere T, Hung RJ, Zaridze D, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009 Jan 1;69(1):243–252. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010 Apr 1;126(7):1630–1639. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 275.Kaira K, Sunaga N, Tomizawa Y, et al. Epigenetic inactivation of the RAS-effector gene RASSF2 in lung cancers. Int J Oncol. 2007 Jul;31(1):169–173. [PubMed] [Google Scholar]
- 276.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006 Mar;16(3):383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Christensen BC, Marsit CJ, Houseman EA, et al. Differentiation of lung adenocarcinoma, pleural mesothelioma, and nonmalignant pulmonary tissues using DNA methylation profiles. Cancer Res. 2009 Aug 1;69(15):6315–6321. doi: 10.1158/0008-5472.CAN-09-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008 Jan 8;105(1):252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Dai Z, Lakshmanan RR, Zhu WG, et al. Global methylation profiling of lung cancer identifies novel methylated genes. Neoplasia. 2001 Jul-Aug;3(4):314–323. doi: 10.1038/sj.neo.7900162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Brena RM, Morrison C, Liyanarachchi S, et al. Aberrant DNA methylation of OLIG1, a novel prognostic factor in non-small cell lung cancer. PLoS Med. 2007 Mar 27;4(3):e108. doi: 10.1371/journal.pmed.0040108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kim EH, Park AK, Dong SM, Ahn JH, Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene. 2010 Jun 7; doi: 10.1038/onc.2010.223. [DOI] [PubMed] [Google Scholar]
- 282.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006 Dec;3(12):e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhong S, Fields CR, Su N, Pan YX, Robertson KD. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2007 Apr 19;26(18):2621–2634. doi: 10.1038/sj.onc.1210041. [DOI] [PubMed] [Google Scholar]
- 284.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009 Jun;19(3):181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002 Jun;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 286.Suzuki M, Yoshino I. Aberrant methylation in non-small cell lung cancer. Surgery today. 2010 Jul;40(7):602–607. doi: 10.1007/s00595-009-4094-6. [DOI] [PubMed] [Google Scholar]
- 287.Heller G, Zielinski CC, Zochbauer-Muller S. Lung cancer: from single-gene methylation to methylome profiling. Cancer Metastasis Rev. 2010 Mar;29(1):95–107. doi: 10.1007/s10555-010-9203-x. [DOI] [PubMed] [Google Scholar]
- 288.Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007 Dec 15;110(12):2785–2792. doi: 10.1002/cncr.23113. [DOI] [PubMed] [Google Scholar]
- 289.Gu J, Berman D, Lu C, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006 Dec 15;12(24):7329–7338. doi: 10.1158/1078-0432.CCR-06-0894. [DOI] [PubMed] [Google Scholar]
- 290.Ota N, Kawakami K, Okuda T, et al. Prognostic significance of p16(INK4a) hypermethylation in non-small cell lung cancer is evident by quantitative DNA methylation analysis. Anticancer Res. 2006 Sep-Oct;26(5B):3729–3732. [PubMed] [Google Scholar]
- 291.Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH. Cohypermethylation of p16 and FHIT promoters as a prognostic factor of recurrence in surgically resected stage I non-small cell lung cancer. Cancer Res. 2006 Apr 15;66(8):4049–4054. doi: 10.1158/0008-5472.CAN-05-3813. [DOI] [PubMed] [Google Scholar]
- 292.Wang J, Lee JJ, Wang L, et al. Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin Cancer Res. 2004 Sep 15;10(18 Pt 1):6119–6125. doi: 10.1158/1078-0432.CCR-04-0652. [DOI] [PubMed] [Google Scholar]
- 293.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001 Dec 15;61(24):8659–8663. [PubMed] [Google Scholar]
- 294.Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002 Jan 15;62(2):371–375. [PubMed] [Google Scholar]
- 295.Brabender J, Usadel H, Danenberg KD, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001 Jun 14;20(27):3528–3532. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 296.Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001 Apr 15;61(8):3419–3424. [PubMed] [Google Scholar]
- 297.Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001 May 2;93(9):691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tang X, Khuri FR, Lee JJ, et al. Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst. 2000 Sep 20;92(18):1511–1516. doi: 10.1093/jnci/92.18.1511. [DOI] [PubMed] [Google Scholar]
- 299.Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004 Nov 15;22(22):4575–4583. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 300.Kim DH, Kim JS, Ji YI, et al. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003 Jul 1;63(13):3743–3746. [PubMed] [Google Scholar]
- 301.Tomizawa Y, Kohno T, Kondo H, et al. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin Cancer Res. 2002 Jul;8(7):2362–2368. [PubMed] [Google Scholar]
- 302.Toyooka S, Suzuki M, Maruyama R, et al. The relationship between aberrant methylation and survival in non-small-cell lung cancers. Br J Cancer. 2004 Aug 16;91(4):771–774. doi: 10.1038/sj.bjc.6602013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rhee I, Jair KW, Yen RW, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000 Apr 27;404(6781):1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 304.Ramalingam SS. Histone deacetylase, proteasome, and heat shock protein inhibitors for the treatment of lung cancer. J Thorac Oncol. 2010 Dec;5(12) Suppl 6:S458–S460. doi: 10.1097/01.JTO.0000391365.56258.16. [DOI] [PubMed] [Google Scholar]
- 305.Mukhopadhyay NK, Weisberg E, Gilchrist D, Bueno R, Sugarbaker DJ, Jaklitsch MT. Effectiveness of trichostatin A as a potential candidate for anticancer therapy in non-small-cell lung cancer. The Annals of thoracic surgery. 2006 Mar;81(3):1034–1042. doi: 10.1016/j.athoracsur.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 306.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004 Feb;39(2):167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 307.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003 Oct;1(12):882–891. [PubMed] [Google Scholar]
- 308.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005 Mar 8;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005 Jun 9;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004 May 1;64(9):3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 312.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006 Mar 24;124(6):1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 313.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004 Jun 1;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 314.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005 Nov 1;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 315.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005 Jun 9;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 316.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006 Apr;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 317.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008 Jan;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 318.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006 Mar;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 319.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Crawford M, Brawner E, Batte K, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008 Sep 5;373(4):607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 321.Nasser MW, Datta J, Nuovo G, et al. Downregulation of microRNA-1 (miR-1) in lung cancer: Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin induced apoptosis bymiR-1. J Biol Chem. 2008 Sep 25; doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 322.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008 Jun;19(6):1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 323.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic Value of Mature MicroRNA-21 and MicroRNA-205 Overexpression in Non-Small Cell Lung Cancer by Quantitative Real-Time RT-PCR. Clin Chem. 2008 Aug 21; doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 324.Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008 Jun 19;27(27):3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 325.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009 Jul 15;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 327.Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Du L, Schageman JJ, Subauste MC, et al. miR-93, miR-98, and miR-197 Regulate Expression of Tumor Suppressor Gene FUS1. Mol Cancer Res. 2009 Aug 11; doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Du L, Pertsemlidis A. microRNAs and lung cancer: tumors and 22-mers. Cancer Metastasis Rev. 2010 Mar;29(1):109–122. doi: 10.1007/s10555-010-9204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005 Mar 11;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 331.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007 May 1;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 2011 Jan;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 333.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007 Mar 16;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007 Aug 15;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 335.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008 Mar 15;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 336.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Wang Z, Chen Z, Gao Y, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011 Mar 1;11(5):490–496. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 338.Gallardo E, Navarro A, Vinolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009 Nov;30(11):1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 339.Donnem T, Eklo K, Berg T, et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. Journal of translational medicine. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011 Apr 1;17(7):1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010 Apr;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Mercatelli N, Coppola V, Bonci D, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008 Apr 15;68(8):2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 344.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009 May;37(8):2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009 Mar;9(5):1374–1384. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007 Apr 26;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 347.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009 Apr 20;27(12):2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 348.Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC medical genomics. 2008;1:61. doi: 10.1186/1755-8794-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Chen X, Hu Z, Wang W, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel non-invasive biomarkers for non-small cell lung cancer diagnosis. Int J Cancer. 2011 May 9; doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 350.Wei J, Gao W, Zhu CJ, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chinese journal of cancer. 2011 Jun;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010 Dec 15;127(12):2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007 Dec 1;67(23):11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Rupaimoole R, Han HD, Lopez-Berestein G, Sood AK. MicroRNA therapeutics: principles, expectations, and challenges. Chinese journal of cancer. 2011 Jun;30(6):368–370. doi: 10.5732/cjc.011.10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Nana-Sinkam SP, Croce CM. MicroRNA dysregulation in cancer: opportunities for the development of microRNA-based drugs. IDrugs : the investigational drugs journal. 2010 Dec;13(12):843–846. [PubMed] [Google Scholar]
- 355.Anguiano A, Nevins JR, Potti A. Toward the individualization of lung cancer therapy. Cancer. 2008 Oct 1;113(7) Suppl:1760–1767. doi: 10.1002/cncr.23644. [DOI] [PubMed] [Google Scholar]
- 356.Xie Y, Minna JD. Predicting the future for people with lung cancer. Nat Med. 2008 Aug;14(8):812–813. doi: 10.1038/nm0808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Sriram KB, Larsen JE, Yang IA, Bowman RV, Fong KM. Genomic medicine in non-small cell lung cancer: paving the path to personalized care. Respirology. 2011 Feb;16(2):257–263. doi: 10.1111/j.1440-1843.2010.01892.x. [DOI] [PubMed] [Google Scholar]
- 358.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008 Aug;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010 Apr 7;102(7):464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Jeong Y, Xie Y, Xiao G, et al. Nuclear receptor expression defines a set of prognostic biomarkers for lung cancer. PLoS Med. 2010;7(12):e1000378. doi: 10.1371/journal.pmed.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004 Dec 30;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 362.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010 Oct;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 363.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010 Aug 12;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 364.Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007 Apr 12;446(7137):815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 365.Vicent S, Chen R, Sayles LC, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest. 2010 Nov 1;120(11):3940–3952. doi: 10.1172/JCI44165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol Biol Cell. 2009 Mar;20(6):1833–1844. doi: 10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Yamanaka S, Gu Z, Sato M, et al. siRNA targeting against EGFR, a promising candidate for a novel therapeutic application to lung adenocarcinoma. Pathobiology. 2008;75(1):2–8. doi: 10.1159/000113789. [DOI] [PubMed] [Google Scholar]
- 368.Lara R, Mauri FA, Taylor H, et al. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene. 2011 Mar 21; doi: 10.1038/onc.2011.61. [DOI] [PubMed] [Google Scholar]
- 369.Lundberg AS, Randell SH, Stewart SA, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002 Jul 4;21(29):4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 370.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004 Dec 15;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 371.Sato M, Vaughan MB, Girard L, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006 Feb 15;66(4):2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 372.Sasai K, Sukezane T, Yanagita E, et al. Oncogene-mediated human lung epithelial cell transformation produces adenocarcinoma phenotypes in vivo. Cancer Res. 2011 Apr 1;71(7):2541–2549. doi: 10.1158/0008-5472.CAN-10-2221. [DOI] [PubMed] [Google Scholar]
- 373.Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001 Dec 15;15(24):3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Politi K, Pao W. How genetically engineered mouse tumor models provide insights into human cancers. J Clin Oncol. 2011 Jun 1;29(16):2273–2281. doi: 10.1200/JCO.2010.30.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011 Feb;21(1):34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 376.Rehman FL, Lord CJ, Ashworth A. Synthetic lethal approaches to breast cancer therapy. Nature reviews. Clinical oncology. 2010 Dec;7(12):718–724. doi: 10.1038/nrclinonc.2010.172. [DOI] [PubMed] [Google Scholar]
- 377.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005 Feb 24;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 378.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006 Nov 1;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 379.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006 Oct 1;12(19):5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 380.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005 May 24;102(21):7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004 Dec 15;64(24):8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 382.Mitsudomi T, Kosaka T, Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int J Clin Oncol. 2006 Jun;11(3):190–198. doi: 10.1007/s10147-006-0583-4. [DOI] [PubMed] [Google Scholar]
- 383.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005 Jan 17;92(1):131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ' it ain't over 'til it's over'. Trends Cell Biol. 2000 Apr;10(4):147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 385.Harris TJ, McCormick F. The molecular pathology of cancer. Nature reviews. Clinical oncology. 2010 May;7(5):251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 386.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer. 2006 Nov;54(2):209–215. doi: 10.1016/j.lungcan.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 387.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008 Sep 1;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007 Jul;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 389.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998 May 29;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 390.Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002 May;8(5):1178–1184. [PubMed] [Google Scholar]
- 391.Marsit CJ, Zheng S, Aldape K, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005 Jul;36(7):768–776. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 392.Higashiyama M, Doi O, Kodama K, et al. MDM2 gene amplification and expression in non-small-cell lung cancer: immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br J Cancer. 1997;75(9):1302–1308. doi: 10.1038/bjc.1997.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Gazzeri S, Della Valle V, Chaussade L, Brambilla C, Larsen CJ, Brambilla E. The human p19ARF protein encoded by the beta transcript of the p16INK4a gene is frequently lost in small cell lung cancer. Cancer Res. 1998 Sep 1;58(17):3926–3931. [PubMed] [Google Scholar]
- 394.Vonlanthen S, Heighway J, Tschan MP, et al. Expression of p16INK4a/p16alpha and p19ARF/p16beta is frequently altered in non-small cell lung cancer and correlates with p53 overexpression. Oncogene. 1998 Nov 26;17(21):2779–2785. doi: 10.1038/sj.onc.1202501. [DOI] [PubMed] [Google Scholar]
- 395.Ohtani N, Yamakoshi K, Takahashi A, Hara E. The p16INK4a-RB pathway: molecular link between cellular senescence and tumor suppression. J Med Invest. 2004 Aug;51(3–4):146–153. doi: 10.2152/jmi.51.146. [DOI] [PubMed] [Google Scholar]
- 396.Reissmann PT, Koga H, Takahashi R, et al. Inactivation of the retinoblastoma susceptibility gene in non-small-cell lung cancer. The Lung Cancer Study Group. Oncogene. 1993 Jul;8(7):1913–1919. [PubMed] [Google Scholar]
- 397.Merlo A, Gabrielson E, Askin F, Sidransky D. Frequent loss of chromosome 9 in human primary non-small cell lung cancer. Cancer Res. 1994 Feb 1;54(3):640–642. [PubMed] [Google Scholar]
- 398.Brambilla E, Moro D, Gazzeri S, Brambilla C. Alterations of expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. J Pathol. 1999 Aug;188(4):351–360. doi: 10.1002/(SICI)1096-9896(199908)188:4<351::AID-PATH385>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 399.Sato M, Takahashi K, Nagayama K, et al. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer. 2005 Dec;44(4):405–414. doi: 10.1002/gcc.20253. [DOI] [PubMed] [Google Scholar]
- 400.Esteller M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003;532:39–49. doi: 10.1007/978-1-4615-0081-0_5. [DOI] [PubMed] [Google Scholar]
- 401.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007 Jan 1;21(1):49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007 Oct;117(10):2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Larsen JE, Spinola M, Gazdar AF, Minna JD. An overview of the molecular biology of lung cancer. In: Pass HI, Carbone DP, Johnson DH, Minna JD, Scagliotti GV, Turrisi AT, editors. Principles and Practice of Lung Cancer: The Official Reference Text of the International Association for the Study of Lung Cancer (IASLC) 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2010. pp. 59–74. [Google Scholar]
- 404.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006 Jan 15;118(2):257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 405.Yousem SA, Nikiforova M, Nikiforov Y. The histopathology of BRAF-V600E-mutated lung adenocarcinoma. Am J Surg Pathol. 2008 Sep;32(9):1317–1321. doi: 10.1097/PAS.0b013e31816597ca. [DOI] [PubMed] [Google Scholar]
- 406.Nakamura H, Saji H, Ogata A, et al. Correlation between encoded protein overexpression and copy number of the HER2 gene with survival in non-small cell lung cancer. Int J Cancer. 2003 Jan 1;103(1):61–66. doi: 10.1002/ijc.10795. [DOI] [PubMed] [Google Scholar]
- 407.Hirashima N, Takahashi W, Yoshii S, Yamane T, Ooi A. Protein overexpression and gene amplification of c-erb B-2 in pulmonary carcinomas: a comparative immunohistochemical and fluorescence in situ hybridization study. Mod Pathol. 2001 Jun;14(6):556–562. doi: 10.1038/modpathol.3880350. [DOI] [PubMed] [Google Scholar]
- 408.Tatematsu A, Shimizu J, Murakami Y, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin Cancer Res. 2008 Oct 1;14(19):6092–6096. doi: 10.1158/1078-0432.CCR-08-0332. [DOI] [PubMed] [Google Scholar]
- 409.Swanton C, Futreal A, Eisen T. Her2-targeted therapies in non-small cell lung cancer. Clin Cancer Res. 2006 Jul 15;12(14 Pt 2):4377s–4383s. doi: 10.1158/1078-0432.CCR-06-0115. [DOI] [PubMed] [Google Scholar]
- 410.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987 Oct 8;317(15):929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 411.De Biasi F, Del Sal G, Hand PH. Evidence of enhancement of the ras oncogene protein product (p21) in a spectrum of human tumors. Int J Cancer. 1989 Mar 15;43(3):431–435. doi: 10.1002/ijc.2910430315. [DOI] [PubMed] [Google Scholar]
- 412.Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer Lett. 2008 Jul 12; doi: 10.1016/j.canlet.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Lee JW, Soung YH, Kim SY, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005 Feb 17;24(8):1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 414.Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005 Sep 1;65(17):7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 415.Micke P, Hengstler JG, Ros R, et al. c-erbB-2 expression in small-cell lung cancer is associated with poor prognosis. Int J Cancer. 2001 May 15;92(4):474–479. doi: 10.1002/ijc.1229. [DOI] [PubMed] [Google Scholar]
- 416.Potti A, Willardson J, Forseen C, et al. Predictive role of HER-2/neu overexpression and clinical features at initial presentation in patients with extensive stage small cell lung carcinoma. Lung Cancer. 2002 Jun;36(3):257–261. doi: 10.1016/s0169-5002(01)00488-3. [DOI] [PubMed] [Google Scholar]
- 417.Dworakowska D, Jassem E, Jassem J, et al. MDM2 gene amplification: a new independent factor of adverse prognosis in non-small cell lung cancer (NSCLC) Lung Cancer. 2004 Mar;43(3):285–295. doi: 10.1016/j.lungcan.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 418.Cappuzzo F, Janne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2008 Oct 3; doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol. 2008 Apr;3(4):331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 420.Johnson BE, Russell E, Simmons AM, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl. 1996;24:210–217. doi: 10.1002/jcb.240630516. [DOI] [PubMed] [Google Scholar]
- 421.Ibson JM, Waters JJ, Twentyman PR, Bleehen NM, Rabbitts PH. Oncogene amplification and chromosomal abnormalities in small cell lung cancer. J Cell Biochem. 1987 Apr;33(4):267–288. doi: 10.1002/jcb.240330405. [DOI] [PubMed] [Google Scholar]
- 422.Shiraishi M, Noguchi M, Shimosato Y, Sekiya T. Amplification of protooncogenes in surgical specimens of human lung carcinomas. Cancer Res. 1989 Dec 1;49(23):6474–6479. [PubMed] [Google Scholar]
- 423.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008 Jan 21; doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Miller CT, Chen G, Gharib TG, et al. Increased C-CRK proto-oncogene expression is associated with an aggressive phenotype in lung adenocarcinomas. Oncogene. 2003 Sep 11;22(39):7950–7957. doi: 10.1038/sj.onc.1206529. [DOI] [PubMed] [Google Scholar]
- 425.Pezzella F, Turley H, Kuzu I, et al. bcl-2 protein in non-small-cell lung carcinoma. N Engl J Med. 1993 Sep 2;329(10):690–694. doi: 10.1056/NEJM199309023291003. [DOI] [PubMed] [Google Scholar]
- 426.Kaiser U, Schilli M, Haag U, et al. Expression of bcl-2--protein in small cell lung cancer. Lung Cancer. 1996 Aug;15(1):31–40. doi: 10.1016/0169-5002(96)00568-5. [DOI] [PubMed] [Google Scholar]
- 427.Reissmann PT, Koga H, Figlin RA, Holmes EC, Slamon DJ. Amplification and overexpression of the cyclin D1 and epidermal growth factor receptor genes in non-small-cell lung cancer. Lung Cancer Study Group. J Cancer Res Clin Oncol. 1999;125(2):61–70. doi: 10.1007/s004320050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Eren B, Sar M, Oz B, Dincbas FH. MMP-2, TIMP-2 and CD44v6 expression in non-small-cell lung carcinomas. Ann Acad Med Singapore. 2008 Jan;37(1):32–39. [PubMed] [Google Scholar]
- 429.Junker K, Wiethege T, Muller KM. Pathology of small-cell lung cancer. J Cancer Res Clin Oncol. 2000 Jul;126(7):361–368. doi: 10.1007/PL00008483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Micke P, Basrai M, Faldum A, et al. Characterization of c-kit expression in small cell lung cancer: prognostic and therapeutic implications. Clin Cancer Res. 2003 Jan;9(1):188–194. [PubMed] [Google Scholar]
- 431.Cook RM, Miller YE, Bunn PA., Jr Small cell lung cancer: etiology, biology, clinical features, staging, and treatment. Curr Probl Cancer. 1993 Mar-Apr;17(2):69–141. doi: 10.1016/0147-0272(93)90010-y. [DOI] [PubMed] [Google Scholar]
- 432.Araki K, Ishii G, Yokose T, et al. Frequent overexpression of the c-kit protein in large cell neuroendocrine carcinoma of the lung. Lung Cancer. 2003 May;40(2):173–180. doi: 10.1016/s0169-5002(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 433.Rygaard K, Nakamura T, Spang-Thomsen M. Expression of the proto-oncogenes c-met and c-kit and their ligands, hepatocyte growth factor/scatter factor and stem cell factor, in SCLC cell lines and xenografts. Br J Cancer. 1993 Jan;67(1):37–46. doi: 10.1038/bjc.1993.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Plummer H, 3rd, Catlett J, Leftwich J, et al. c-myc expression correlates with suppression of c-kit protooncogene expression in small cell lung cancer cell lines. Cancer Res. 1993 Sep 15;53(18):4337–4342. [PubMed] [Google Scholar]
- 435.Hibi K, Takahashi T, Sekido Y, et al. Coexpression of the stem cell factor and the c-kit genes in small-cell lung cancer. Oncogene. 1991 Dec;6(12):2291–2296. [PubMed] [Google Scholar]
- 436.Weiner DB, Nordberg J, Robinson R, et al. Expression of the neu gene-encoded protein (P185neu) in human non-small cell carcinomas of the lung. Cancer Res. 1990 Jan 15;50(2):421–425. [PubMed] [Google Scholar]
- 437.Schneider PM, Hung MC, Chiocca SM, et al. Differential expression of the c-erbB-2 gene in human small cell and non-small cell lung cancer. Cancer Res. 1989 Sep 15;49(18):4968–4971. [PubMed] [Google Scholar]
- 438.Fernandes A, Hamburger AW, Gerwin BI. ErbB-2 kinase is required for constitutive stat 3 activation in malignant human lung epithelial cells. Int J Cancer. 1999 Nov 12;83(4):564–570. doi: 10.1002/(sici)1097-0215(19991112)83:4<564::aid-ijc20>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 439.Rygaard K, Vindelov LL, Spang-Thomsen M. Expression of myc family oncoproteins in small-cell lung-cancer cell lines and xenografts. Int J Cancer. 1993 Apr 22;54(1):144–152. doi: 10.1002/ijc.2910540123. [DOI] [PubMed] [Google Scholar]
- 440.Takahashi T, Obata Y, Sekido Y, et al. Expression and amplification of myc gene family in small cell lung cancer and its relation to biological characteristics. Cancer Res. 1989 May 15;49(10):2683–2688. [PubMed] [Google Scholar]
- 441.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 442.Zhang P, Gao WY, Turner S, Ducatman BS. Gleevec (STI-571) inhibits lung cancer cell growth (A549) and potentiates the cisplatin effect in vitro. Mol Cancer. 2003 Jan 3;2:1. doi: 10.1186/1476-4598-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007 Dec 14;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 444.Johnson FM, Krug LM, Tran HT, et al. Phase I studies of imatinib mesylate combined with cisplatin and irinotecan in patients with small cell lung carcinoma. Cancer. 2006 Jan 15;106(2):366–374. doi: 10.1002/cncr.21640. [DOI] [PubMed] [Google Scholar]
- 445.Rossi G, Cavazza A, Marchioni A, et al. Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large-cell neuroendocrine carcinoma of the lung. J Clin Oncol. 2005 Dec 1;23(34):8774–8785. doi: 10.1200/JCO.2005.02.8233. [DOI] [PubMed] [Google Scholar]
- 446.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007 Aug 16;448(7155):807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 447.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008 Jul 22;99(2):245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Carbone DP, Mitsudomi T, Chiba I, et al. p53 immunostaining positivity is associated with reduced survival and is imperfectly correlated with gene mutations in resected non-small cell lung cancer. A preliminary report of LCSG 871. Chest. 1994 Dec;106(6) Suppl:377S–381S. [PubMed] [Google Scholar]
- 449.Wistuba II, Berry J, Behrens C, et al. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res. 2000 Jul;6(7):2604–2610. [PMC free article] [PubMed] [Google Scholar]
- 450.Chiba I, Takahashi T, Nau MM, et al. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Lung Cancer Study Group. Oncogene. 1990 Oct;5(10):1603–1610. [PubMed] [Google Scholar]
- 451.Shimizu E, Zhao M, Shinohara A, et al. Differential expressions of cyclin A and the retinoblastoma gene product in histological subtypes of lung cancer cell lines. J Cancer Res Clin Oncol. 1997;123(10):533–538. doi: 10.1007/s004320050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J Clin Oncol. 1998 Mar;16(3):1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 453.Hensel CH, Hsieh CL, Gazdar AF, et al. Altered structure and expression of the human retinoblastoma susceptibility gene in small cell lung cancer. Cancer Res. 1990 May 15;50(10):3067–3072. [PubMed] [Google Scholar]
- 454.Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000 Sep 1;60(17):4894–4906. [PubMed] [Google Scholar]
- 455.Thiberville L, Payne P, Vielkinds J, et al. Evidence of cumulative gene losses with progression of premalignant epithelial lesions to carcinoma of the bronchus. Cancer Res. 1995 Nov 15;55(22):5133–5139. [PubMed] [Google Scholar]
- 456.Sunaga N, Miyajima K, Suzuki M, et al. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004 Jun 15;64(12):4277–4285. doi: 10.1158/0008-5472.CAN-03-3941. [DOI] [PubMed] [Google Scholar]
- 457.Mori S, Ito G, Usami N, et al. p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer. 2004 Apr 15;100(8):1673–1682. doi: 10.1002/cncr.20164. [DOI] [PubMed] [Google Scholar]
- 458.Prudkin L, Behrens C, Liu DD, et al. Loss and reduction of FUS1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin Cancer Res. 2008 Jan 1;14(1):41–47. doi: 10.1158/1078-0432.CCR-07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Safar AM, Spencer H, Jr, Su X, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res. 2005 Jun 15;11(12):4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 460.Toyooka S, Toyooka KO, Maruyama R, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001 Nov;1(1):61–67. [PubMed] [Google Scholar]
- 461.Shimamoto T, Ohyashiki JH, Hirano T, Kato H, Ohyashiki K. Hypermethylation of E-cadherin gene is frequent and independent of p16INK4A methylation in non-small cell lung cancer: potential prognostic implication. Oncol Rep. 2004 Aug;12(2):389–395. [PubMed] [Google Scholar]
- 462.Jarmalaite S, Kannio A, Anttila S, Lazutka JR, Husgafvel-Pursiainen K. Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer. 2003 Oct 10;106(6):913–918. doi: 10.1002/ijc.11322. [DOI] [PubMed] [Google Scholar]
- 463.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999 Jan 1;59(1):67–70. [PubMed] [Google Scholar]
- 464.Kim DS, Cha SI, Lee JH, et al. Aberrant DNA methylation profiles of non-small cell lung cancers in a Korean population. Lung Cancer. 2007 Oct;58(1):1–6. doi: 10.1016/j.lungcan.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 465.Suh YA, Lee HY, Virmani A, et al. Loss of retinoic acid receptor beta gene expression is linked to aberrant histone H3 acetylation in lung cancer cell lines. Cancer Res. 2002 Jul 15;62(14):3945–3949. [PubMed] [Google Scholar]
- 466.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000 Jul;25(3):315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 467.Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996 Apr;12(4):368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 468.Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002 Oct 7;21(45):6877–6883. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- 469.Luk C, Tsao MS, Bayani J, Shepherd F, Squire JA. Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001 Mar;125(2):87–99. doi: 10.1016/s0165-4608(00)00363-0. [DOI] [PubMed] [Google Scholar]
- 470.Petersen I, Bujard M, Petersen S, et al. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997 Jun 15;57(12):2331–2335. [PubMed] [Google Scholar]
- 471.Petersen I, Langreck H, Wolf G, et al. Small-cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer. 1997;75(1):79–86. doi: 10.1038/bjc.1997.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Sung JM, Cho HJ, Yi H, et al. Characterization of a stem cell population in lung cancer A549 cells. Biochem Biophys Res Commun. 2008 Jun 20;371(1):163–167. doi: 10.1016/j.bbrc.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 473.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3(8):e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 2010 Mar;29(1):61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chemico-biological interactions. 2009 Mar 16;178(1–3):48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Cui F, Wang J, Chen D, Chen YJ. CD133 is a temporary marker of cancer stem cells in small cell lung cancer, but not in non-small cell lung cancer. Oncol Rep. 2011 Mar;25(3):701–708. doi: 10.3892/or.2010.1115. [DOI] [PubMed] [Google Scholar]
- 477.Gutova M, Najbauer J, Gevorgyan A, et al. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS One. 2007;2(2):e243. doi: 10.1371/journal.pone.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Wen J, Fu J, Zhang W, Guo M. Genetic and epigenetic changes in lung carcinoma and their clinical implications. Mod Pathol. 2011 Mar 18; doi: 10.1038/modpathol.2011.46. [DOI] [PubMed] [Google Scholar]
- 479.Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007 May 15;67(10):4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 480.Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9(6):485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 481.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006 Jun 1;20(11):1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Regales L, Balak MN, Gong Y, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS One. 2007;2(8):e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009 Dec 24;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006 May 16;103(20):7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Chen Z, Sasaki T, Tan X, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010 Dec 1;70(23):9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):474–479. doi: 10.1073/pnas.0808930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001 Dec 15;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Meuwissen R, Linn SC, van der Valk M, Mooi WJ, Berns A. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene. 2001 Oct 4;20(45):6551–6558. doi: 10.1038/sj.onc.1204837. [DOI] [PubMed] [Google Scholar]
- 489.Kim WY, Perera S, Zhou B, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009 Aug;119(8):2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Jackson EL, Olive KP, Tuveson DA, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005 Nov 15;65(22):10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 491.Iwanaga K, Yang Y, Raso MG, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 2008 Feb 15;68(4):1119–1127. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Olive KP, Tuveson DA, Ruhe ZC, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004 Dec 17;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 493.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003 Sep;4(3):181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]