Abstract
The transcription factor PU.1 is a master regulator of myeloid differentiation and function. On the other hand, only scarce information is available on PU.1-regulated genes involved in cell survival. We now identified the glycolytic enzyme hexokinase 3 (HK3), a gene with cytoprotective functions, as transcriptional target of PU.1. Interestingly, HK3 expression is highly associated with the myeloid lineage and was significantly decreased in acute myeloid leukemia patients compared with normal granulocytes. Moreover, HK3 expression was significantly lower in acute promyelocytic leukemia (APL) compared with non-APL patient samples. In line with the observations in primary APL patient samples, we observed significantly higher HK3 expression during neutrophil differentiation of APL cell lines. Moreover, knocking down PU.1 impaired HK3 induction during neutrophil differentiation. In vivo binding of PU.1 and PML-RARA to the HK3 promoter was found, and PML-RARA attenuated PU.1 activation of the HK3 promoter. Next, inhibiting HK3 in APL cell lines resulted in significantly reduced neutrophil differentiation and viability compared with control cells. Our findings strongly suggest that HK3 is: (1) directly activated by PU.1, (2) repressed by PML-RARA, and (3) functionally involved in neutrophil differentiation and cell viability of APL cells.
Introduction
Acute promyelocytic leukemia (APL or AML-M3) accounts for 5% to 8% of acute myeloid leukemia (AML) subtypes.1 APL is characterized by a balanced chromosomal translocation involving the promyelocytic leukemia (PML) gene on chromosome 15 and the retinoic acid receptorα (RARA) on chromosome 17 that results in expression of the oncogenic fusion gene PML-RARA.2 PML-RARA is a gain-of-function protein that promotes leukemic transformation by impairing the formation of functional PML nuclear bodies3 and repressing RARA target genes in a dominant-negative manner.4 This repression interferes with gene expression programs involved in differentiation, apoptosis, and self-renewal and leads to increased cell survival and inhibition of terminal differentiation, with an accumulation of promyelocytes. The block in differentiation that is observed in APL cells can be reverted with pharmacologic doses of ATRA by triggering PML-RARA degradation and thus restoring normal myeloid differentiation (reviewed by de Thé and Chen5).
The Ets-family member PU.1 is a master transcriptional regulator of myeloid differentiation,6,7 as evidenced in PU.1 null mice that die at birth because of the lack of functional myeloid cells.7 Inhibition of PU.1-induced myeloid differentiation may represent a critical step in APL leukemogenesis. On the one hand, PU.1 is inhibited at the transcriptional level by PML-RARA binding to its promoter region8; on the other hand, a recent study identified PML-RARA as a binding partner of PU.1,4 resulting in repression of PU.1-activated genes. A majority of PU.1 target genes are directly involved in myeloid differentiation and function, such as CD11b, CD45, the granulocyte/macrophage colony-stimulating factor receptor (GM-CSFR), myeloperoxidase, lysozyme, and neutrophil elastase (reviewed by Kastner and Chan9). Notably, PU.1 is not only involved in the regulation of myeloid genes but also of genes implicated in cell survival.10,11 Moreover, we showed that PU.1 directly binds to the DNA-binding and oligomerization domains of p53/p73 proteins, reducing their transcriptional activity and thus inhibiting the activation of genes important for cell cycle regulation and apoptosis.10 In this study, we aimed at identifying and characterizing novel PU.1 transcriptional targets involved in cellular viability. In our initial gene profiling of PU.1-null and restored 503 cells, we found that hexokinase 3 (HK3) is induced in PU.1-restored 503 cells. Hexokinases (HK; ATP:D-hexose 6-phosphotransferase) are enzymes that catalyze the first step of glycolysis, namely, the phosphorylation of glucose to glucose 6-phosphate (reviewed by Cardenas et al12). There are 4 hexokinase isozymes in mammals: HK1, HK2, HK3, and HK4. HK4, commonly known as glucokinase, is mainly expressed in the liver and pancreas where it plays a role as glucose sensor. HK1 to HK3 share extensive sequence homology but exhibit unique characteristics in terms of catalytic and regulatory properties, subcellular localization, and tissue distribution, suiting them for distinct physiologic roles (reviewed by Wilson13). HK3 is found at low levels in virtually all tissues with a moderate to high expression in lung, kidney, and liver.14,15 In particular, HK3 is the major hexokinase member present in granulocytes (70%-80% of total activity), with the remaining 20% to 30% of the activity provided by HK1.16
Here we describe that HK3 is significantly down-regulated in primary APL, and its expression is restored on ATRA therapy. We further identified HK3 as direct transcriptional target of PU.1 and PML-RARA. Moreover, we show that down-regulation of HK3 expression impairs neutrophil differentiation of APL cells and promotes cell death of APL cells treated with ATRA or anthracyclines.
Methods
Primary patient samples, cell lines, and culture conditions
Isolation of primary myeloid cells was done as described earlier.17 Protocols and the use of 67 human samples acquired in Bern were approved by the Cantonal Ethical Committee at the Inselspital. A cohort of 98 samples from patients with a diagnosis of primary AML were enrolled on HOVON/SAKK (Dutch-Belgian Hematology-Oncology/Swiss Group for Clinical Cancer Research Cooperative group) protocols -04, -04A, -29, and -42 (available at www.hovon.nl) between 1987 and 2006.18–21 All patients provided written informed consent in accordance with the Declaration of Helsinki. After informed consent was given, bone marrow aspirates or peripheral blood samples were taken at diagnosis. Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard) centrifugation and cryopreserved. The AML samples contained 80% to 100% blast cells after thawing, regardless of the blast counts at diagnosis. Mutational analyses were all performed as described previously.22 Our findings were validated in a second AML cohort of 181 patients from the Munich Leukemia Laboratory. Patient data from the different cohorts are summarized in supplemental Tables 1 and 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The human APL cell lines NB4, NB4-R2, and HT93, as well as the human non–small cell lung carcinoma cell line H1299, were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS (#S0615; Biochrom AG), 1% penicillin/streptomycin (P4333; Sigma-Aldrich). Cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C. The human embryonic kidney 293T cells were cultured in DMEM (Sigma-Aldrich) supplemented with 5% FBS, 1% penicillin/streptomycin, 1% HEPES (Sigma-Aldrich), and kept in a humidified atmosphere containing 7.5% CO2 at 37°C. For neutrophil differentiation experiments, cells were treated with 1μM all-trans retinoic acid (ATRA; Sigma-Aldrich) for 6 days. Successful granulocyte differentiation was evidenced by CD11b (#21279114; Immunotools) FACS analysis or by increased mRNA expression of CCAAT/enhancer binding protein ϵ (CEBPE). Cell viability upon ATRA, idarubicin (Sigma-Aldrich), or doxorubicin (Pfizer) treatment was measured using an Alamar Blue assay (Invitrogen). Caspase-3/caspase-7 activation was measured using Caspase-Glo 3/7 Assay according to the manufacturer's instructions (Promega AG).
Microarray analysis, TaqMan low density arrays and real-time quantitative RT-PCR
Chip microarray assays, analysis of the myeloid 503 cells, and the AML patient cohort from Munich were done as described.23 RNA extraction, RT-PCR, and low density array measurements as well as data analysis were done as described.26 TaqMan Gene Expression Assays for HMBS, ABL1, PU.1, and HK3 preloaded on low density arrays were Hs00609297_m1, Hs00245445_m1, Hs00231368_m1, and Hs00157923_m1 (Applied Biosystems), respectively. TaqMan gene expression assays for HK1, HK2, HK3, and CEBPE used in a 96-well format on the ABI PRISM 7700 Sequence Detection System were Hs00175976_m1, Hs00606086_m1, Hs01092839_m1, and Hs00357657_m1, respectively. Specific primers and probes for PU.1, HMBS as well as data analysis have been described.10 N-fold changes were calculated using the −ΔΔCt method of relative quantification. Data are mean plus or minus SD of at least triplicate experiments.
Generation of PU.1 and HK3 knockdown cell lines
pLKO1 lentiviral vectors expressing small hairpin (sh) RNAs targeting PU.1 (shPU.1_256: NM_003120.1-256s1c1/TRCN0000020536 and shPU.1_928: NM_003120.1-928s1c1/TRCN0000020538), HK3 (shHK3_560: NM_002115.1-560s1c1/TRCN0000037680, shHK3_1420: NM_002115.1-1420s1c1/ TRCN0000037679 and shHK3_1748 NM_002115.1-1748s1c1/ TRCN0000037683) or a nontargeting shRNA control (SHCOO2) were purchased from Sigma-Aldrich. All vectors contain a puromycin antibiotic resistance gene for selection of transduced mammalian cells. Lentivirus production and transduction were done as described.17
Immunoblotting
Cells were lysed in RIPA buffer (20 μL/5 × 106 cells) consisting of 50mM sodium phosphate, pH 7.0, 10mM Glc, 10mM thioglycerol, and 0.1% (volume/volume) Triton X-100 supplemented with Protease Inhibitor Cocktail (Roche). The lysates were incubated at 4°C in a shaker and then centrifuged at 13 000g for 10 minutes in a refrigerated microcentrifuge. Protein content of the supernatant was determined by a Bradford Assay (#500-0006; Bio-Rad) with BSA as a standard. A total of 80 μg of total protein was analyzed by electrophoresis on SDS–12% polyacrylamide gels. Primary antibodies used were anti-PU.1 (#2266; Cell Signaling), anti-GAPDH (MAB374; Millipore), and anti-K3 (#SAB1400124; Sigma- Aldrich). Secondary antibodies used were donkey anti–rabbit and sheep anti–mouse HRP-conjugated IgG (Amersham).
ChIP
Chromatin immunoprecipitation assay (ChIP) was performed using the ChIP-IT Express Chromatin Immunoprecipitation Kit (ChIP-IT Express, Active Motif, Rixensart, Belgium) according to the manufacturer's recommendation. Chromatin from 15 × 106 NB4 cells was fragmented to an average size of 500 bp using the provided enzymatic shearing cocktail. For immunoprecipitation, we added anti-PU.1, anti-PML, or anti-RARA (sc-352, sc-551, sc-5621X; Santa Cruz Biotechnology) antibodies. Immunoprecipitations with IgG (PP64B; Upstate) or an antiacetylhistone H3 antibody (Stratagene) were used as negative and positive control, respectively. Genomic regions containing putative PU.1 and PML-RARA binding sites were amplified by PCR using the following primers: A; forward 5′-TGAGCTCATTTTTGCCTGTGAAT-3′ and reverse 5′-ATGGTTTCAGCAGTCTTTCCAC-3′; B; forward 5′-GCCAGATAAAGAACTGAGGTCTGA-3′ and reverse 5′-TGGTCTGCTTATTGGTTCC-3′; C; forward 5′-CTGCCTGCTCCTTCCACAGCTG-3′ and reverse 5′-AGTGTCCACTTCCCCTGGTGTGA-3′. In addition, an unrelated sequence in the GAPDH gene was used as a negative control.25
Human HK3 promoter reporter assay
The HK3 promoter region was PCR amplified from genomic DNA of NB4 APL cells using the Pfu DNA polymerase (Promega) and cloned into the pGL4.10-basic luciferase vector (Promega) using the Cold Fusion kit (System Bioscience). For reporter assays, H1299 cells were transfected in triplicate using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were lysed 24 hours after transfection, and luciferase activity was measured using the Dual-Luciferase Reporter Plasmid System (Promega).
Statistical analysis and bioinformatics
Nonparametric Mann-Whitney U tests were applied to compare the difference between 2 groups using the program Prism Version 4 software (GraphPad). P values less than .05 were considered to be statistically significant. Promoter sequences were retrieved from online database (www.ncbi.nlm.nih.gov). Prediction of transcription factor binding sites was performed by MatInspector (www.genomatix.de).
Results
Repression of HK3 mRNA in primary AML patient samples
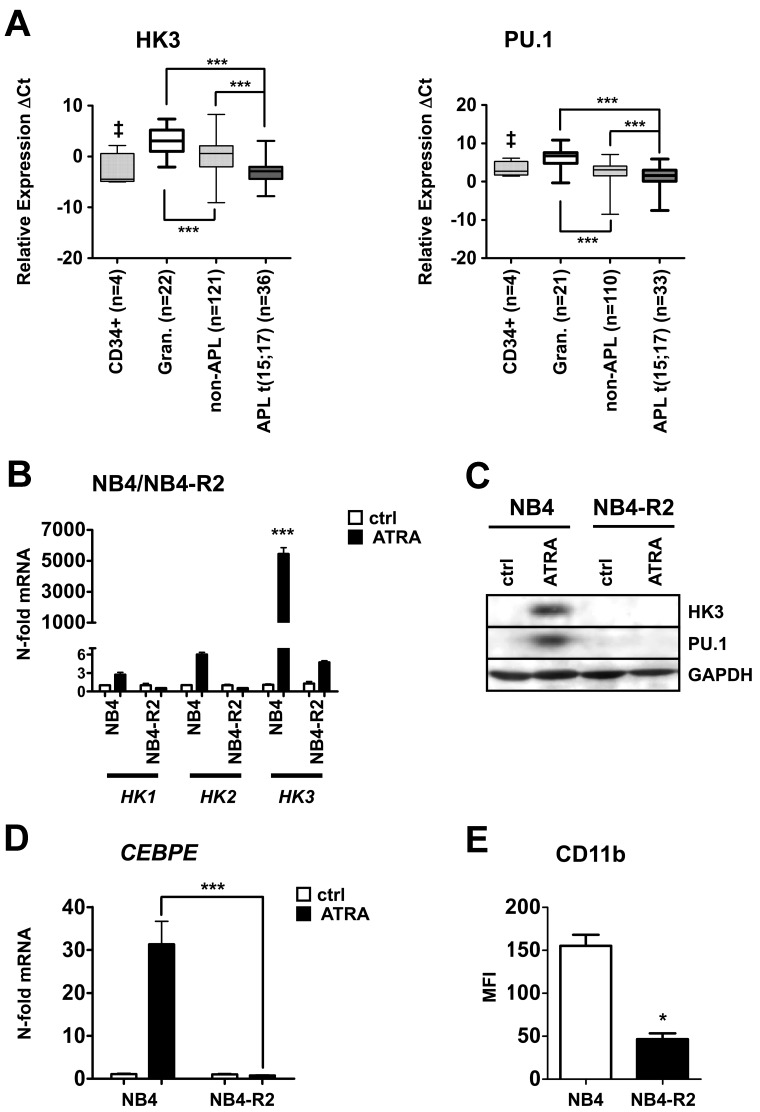
To identify novel PU.1-regulated genes, we assessed gene expression profiles of PU.1 restored versus 503 PU.1-null myeloid cells. Several known PU.1 target genes were up-regulated in the PU.1-rescued 503 cell line, thereby confirming successful restoration of PU.1.26,27 HK3 was among the most highly induced genes in PU.1-restored 503 cells. Interestingly, the hexokinase isozymes HK1 and HK2 did not show an induction on PU.1 restoration (Table 1). By bioinformatic analysis, we found that HK1 and HK2 are ubiquitously expressed in different cell types, whereas HK3 levels are particularly high in myeloid cells (supplemental Figure 1). We excluded HK4 from our analyses because its expression is limited to pancreatic and liver cells (reviewed by Postic et al28). We then focused on HK3 as a potential new PU.1-regulated gene. To investigate a possible role of HK3 in AML pathology, we first determined HK3 mRNA levels by quantitative RT-PCR in a total of 165 primary AML patient samples from the Bern and HOVON-SAKK studies, 4 CD34+ progenitor cells, and 22 granulocytes from healthy donors (Figure 1A left panel). We detected HK3 expression in 157 of 165 AML patients and in all control cells. HK3 mRNA levels were significantly lower in non-APL patient samples and CD34+ progenitor cells compared with granulocytes from healthy donors. Importantly, HK3 expression was significantly lower in APL t(15;17) patients compared with non-APL and granulocyte samples (P < .0001). PU.1 mRNA expression, detectable in 143 of 165 AML patient samples, 21 of 22 granulocytes and 4 of 4 CD34+ progenitor cells, paralleled the expression pattern of HK3, with particularly low PU.1 levels in APL patients (Figure 1A right panel). To support our findings, we analyzed HK3 and PU.1 expression data from a microarray profiling experiment in the independent Munich validation cohort. Again, we found significantly lower expression of HK3 (P < .0001) and PU.1 (P < .05) in APL patients compared with non-APL patient samples (supplemental Figure 2A). HK3 expression in patients with different recurrent genetic abnormalities, such as inv(16) and t(8:21), with normal or complex karyotypes, were not significantly different from the other AML samples in this cohort. However, in a subpopulation of 67 patients from the Bern cohort that were molecularly characterized for CEBPA mutations, AML patients with mutated CEBPA displayed significantly lower HK3 mRNA expression compared with CEBPA wild-type AML patients (Mann-Whitney U test: P < .01; data not shown).
Table 1.
Induction of HK3, but not HK1 and HK2, mRNA levels in PU.1-restored murine 503 PU.1 knockout cells
| Probe set | Gene symbol | 503.PU.1/503 ctrl. |
|
|---|---|---|---|
| Experiment I | Experiment II | ||
| 1437974_a_at | HK1 | 0.43 | 0.39 |
| 1420901_a_at | HK1 | 0.24 | 0.22 |
| 1422612_at | HK2 | 0.27 | 0.23 |
| 1435490_at | HK3 | 55.56 | 58.20 |
| 1442798_x_at | HK3 | 151.83 | 161.95 |
Gene expression analysis of myeloid 503 PU.1-null and PU.1-restored 503 myeloid cells was done as described.26 Values are represented as fold regulation referred to 503 PU.1 knockout cells.
Figure 1.
Significantly reduced HK3 and PU.1 mRNA levels in primary AML patient samples and induction of HK3 expression during neutrophil differentiation of NB4 APL cells. (A) Primary AML blasts were isolated using a Ficoll gradient, total RNA was extracted, and HK3 (left panel) and PU.1 (right panel) mRNA levels were measured by real-time quantitative RT-PCR. Measured cycle threshold (Ct) values represent log2 expression levels. Values were normalized to the expression levels of the housekeeping genes HMBS and ABL1. ***P < .0001. ‡P < .05. (B) NB4 and NB4-R2 cells were differentiated with 1μM ATRA for 6 days. HK1, HK2, and HK3 mRNA were measured by real-time quantitative RT-PCR and given as n-fold changes compared with untreated SHCOO2 cells and normalized to the housekeeping gene HMBS. ***P < .0001. (C) PU.1 and HK3 protein expression was measured by Western blotting in NB4 and NB4-R2 treated as in panel B. GAPDH expression was used as loading control. (D) CEBPE mRNA expression was determined in NB4 cells treated as in panel B. ***P < .0001. (E) CD11b flow cytometric statistical analysis of NB4 or NB4-R2 cells treated as in panel B. *P < .01 (all P values Mann-Whitney U tests).
In line with our findings of low HK3 expression in APL patient samples, 7 APL patients undergoing ATRA therapy displayed markedly increased HK3 mRNA expression. In 2 APL patients under ATRA therapy, HK3 was induced 20- to 55-fold on day 5 and 7, respectively (supplemental Figure 2B). A similar induction of HK3 mRNA was seen in 5 APL patients after finishing ATRA therapy (mean 1.3 months; supplemental Figure 2C).
Taken together, we showed that HK3 mRNA levels were particularly low in APL patients, and we found evidence that HK3 expression can be restored in APL patients, who received ATRA therapy. Generally, low HK3 expression was paralleled by low PU.1 expression in AML patient samples.
Neutrophil differentiation of APL cells is associated with HK3 induction
Our findings of significantly decreased HK3 levels in APL patient samples prompted us to analyze HK3 expression during ATRA-induced neutrophil differentiation of APL cell lines. On neutrophil differentiation of NB4 APL cells, we found a 5400-fold HK3 mRNA induction compared with untreated cells (P < .0001; Figure 1B). Expression of the HK3 relatives, HK1 and HK2, during neutrophil differentiation of NB4 cells only increased 2.3- and 6.2-fold, respectively. To exclude that the induction of HK3 was the result of an unspecific stress response to ATRA treatment, we used ATRA-resistant NB4-R2 as a control. No significant induction of HK3, HK1, or HK2 mRNA was seen in these cells upon ATRA treatment. To confirm our results, we differentiated a second APL cell line, HT93, using ATRA. We found a 1000-fold induction of HK3 message during neutrophil differentiation, whereas HK1 and HK2 levels only showed minor regulation (supplemental Figure 3A). Next, we measured HK3 protein expression on ATRA treatment by Western blotting in NB4, NB4-R2, and HT93 cells. HK3 and PU.1 protein levels were clearly increased in NB4 and HT93 cells on neutrophil differentiation, whereas no change in HK3 or PU.1 protein expression was seen in NB4-R2 cells (Figure 1C; supplemental Figure 3B). Successful neutrophil differentiation of NB4 and HT93 APL cells was confirmed by a significant increase of CEBPE mRNA expression as well as CD11b surface expression (Figure 1D-E; supplemental Figure 3C-D). Our findings show that HK3, but not HK1 and HK2, expression levels are significantly increased during ATRA-induced neutrophil differentiation.
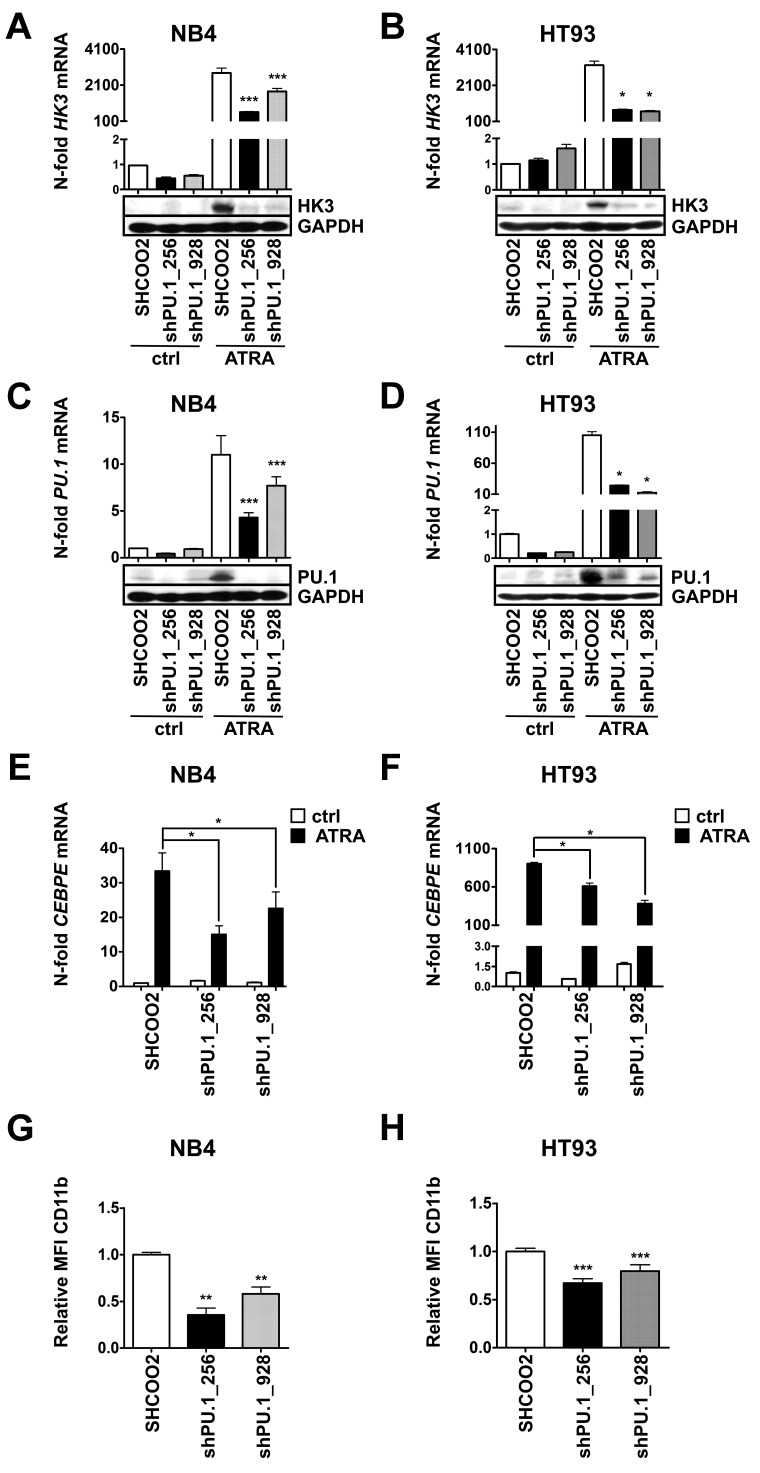
Down-regulation of PU.1 impairs ATRA-induced HK3 expression
Given our findings of HK3 induction on PU.1 restoration of PU.1-null myeloid cells, and the particularly low HK3 and PU.1 expression in APL cells, we aimed at knocking down PU.1 in APL cell lines to test for PU.1-dependent induction of HK3. To this end, we decreased PU.1 expression in NB4 and HT93 cells using lentiviral vectors expressing nontargeting shRNA (SHCOO2) or 2 different short hairpin shRNAs targeting PU.1. We observed significantly reduced HK3 mRNA (P < .001) and protein levels in both NB4 PU.1 knockdown cell lines during neutrophil differentiation compared with SHCOO2 control cells (Figure 2A; P < .0001). Similarly, knocking down PU.1 in HT93 APL cells resulted in significantly reduced HK3 mRNA and protein levels during ATRA treatment (Figure 2B). Both shRNA constructs significantly reduced PU.1 mRNA and protein expression in both NB4 and HT93 cell lines during neutrophil differentiation (Figure 2C-D). As previously reported, knocking down PU.1 reduced neutrophil differentiation of NB4 and HT93 cells, as seen by significantly reduced CEBPE mRNA and CD11b surface expression (Figure 2E-H). Overall, our results indicate that PU.1 is a crucial positive regulator of HK3 during neutrophil differentiation of APL cells.
Figure 2.
Transcriptional regulation of HK3 by PU.1 in NB4 and HT93 APL cell lines. NB4 and HT93 cells were stably transduced with nontargeting shRNA (SHC002) or shRNAs targeting PU.1 (shPU.1_256, shPU.1_928) and differentiated with 1μM ATRA for 6 days. HK3 and PU.1 mRNA expression as well as protein expression were measured in NB4 (A,C) or HT93 (B,D) cells by real-time quantitative RT-PCR and Western blotting. CEBPE mRNA expression was measured in NB4 (E) and HT93 (F) cells by real-time quantitative RT-PCR. CD11b flow cytometric analysis of NB4 (G) and HT93 (H) cells. ***P < .0001; **P < .01; *P < .05 (all P values Mann-Whitney U tests).
PU.1 and PML-RARA directly regulate the HK3 promoter
So far, we have shown that HK3 expression is down-regulated in APL patients compared with normal granulocytes, that ATRA therapy induces a massive up-regulation of HK3 gene expression in APL in vivo and in vitro, and that down-regulation of PU.1 reduces HK3 induction during ATRA-induced neutrophil differentiation of APL cells. These data suggest that HK3 might be directly regulated by PU.1 and PML-RARA.
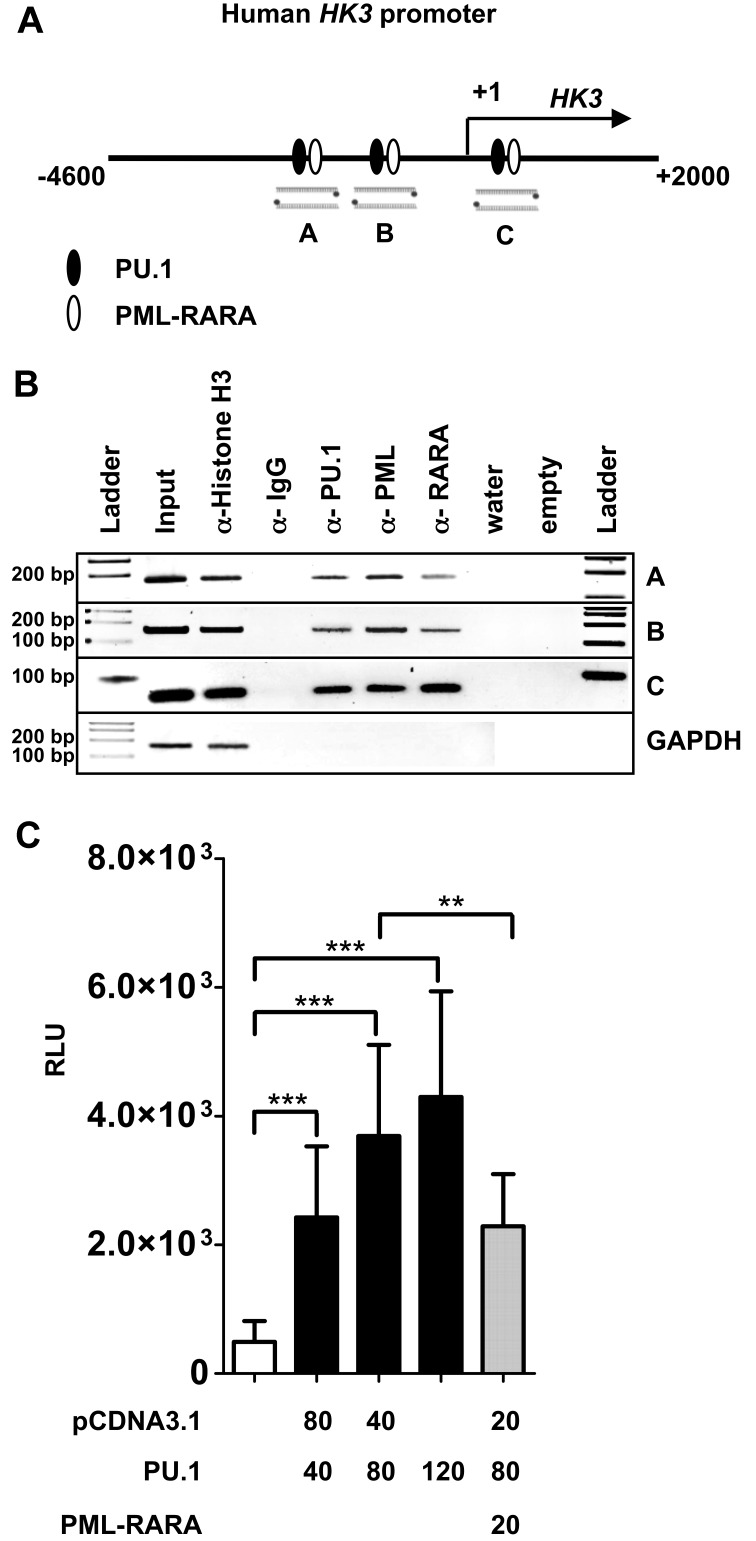
To further provide insights into HK3 transcriptional regulation, we analyzed 9.5 kb of the HK3 genomic region upstream of exon 1 (location 5q35.2, total range NC_000005.9-176′332′571…-176′323′000) for PU.1 and PML-RARA binding sites using MatInspector. We identified 3 putative PU.1 and PML-RARA binding sites in close proximity to each other in 3 different regions of the HK3 promoter (Figure 3A). To show direct binding of PU.1 and PML-RARA to these sites, we performed ChIP using anti-PU.1, anti-PML, and anti-RARA antibodies. PCR was done using primers encompassing the respective sites and primers amplifying an unrelated sequence in the GAPDH gene as negative control. As a positive and negative control for the immunoprecipitation, we used antihistone H3 and anti-IgG antibodies, respectively. Our data show in vivo binding of PU.1 and PML-RARA to each of these 3 sites as proven by ChIP experiments (Figure 3B).
Figure 3.
PU.1 and PML-RARA are direct regulators of the HK3 promoter. (A) Schematic representation of a 6.6-kb human HK3 promoter fragment. Lanes A, B, and C are 3 putative PU.1 binding sites (circles) found in the HK3 promoter as determined by MatInspector. (B) In vivo binding of PU.1 to the 3 binding sites was shown by ChIP in NB4 cells. ChIP was performed with antibodies against PU.1, PML, and RARA. Antibodies against acetyl-histone H3 and IgG served as positive and negative controls, respectively. As a negative control for the different pull-downs, absence of GAPDH amplification is shown. (C) HK3 promoter transactivation assays. H1299 cells were transiently transfected with 40 ng HK3 promoter reporter construct and PU.1 or PML-RARA expression vectors as indicated. Total amount of transfected DNA was adjusted with pcDNA3.1 empty vector as indicated. pRL-TK Renilla luciferase vector (40 ng) was cotransfected in each experiment as an internal control for transfection efficiency. After 24 hours, luciferase activity was measured. The promoter activity is shown as relative light units (RLU). Results are the mean ± SD of 5 independent experiments. ***P < .0001; **P < .001 (Mann-Whitney U tests).
We next investigated whether PU.1 activates the proximal human HK3 promoter upstream of the transcriptional start site including the PU.1 sites “A” and “B.” PU.1 coexpression with the HK3 promoter reporter resulted in a significant, dose-dependent activation of the HK3 promoter. Moreover, cotransfecting PML-RARA inhibited PU.1-mediated activation of the HK3 promoter (Figure 3C). Our data show direct regulation of the human HK3 promoter by PU.1 and PML-RARA.
Inhibition of HK3 impairs ATRA-induced neutrophil differentiation of APL cells
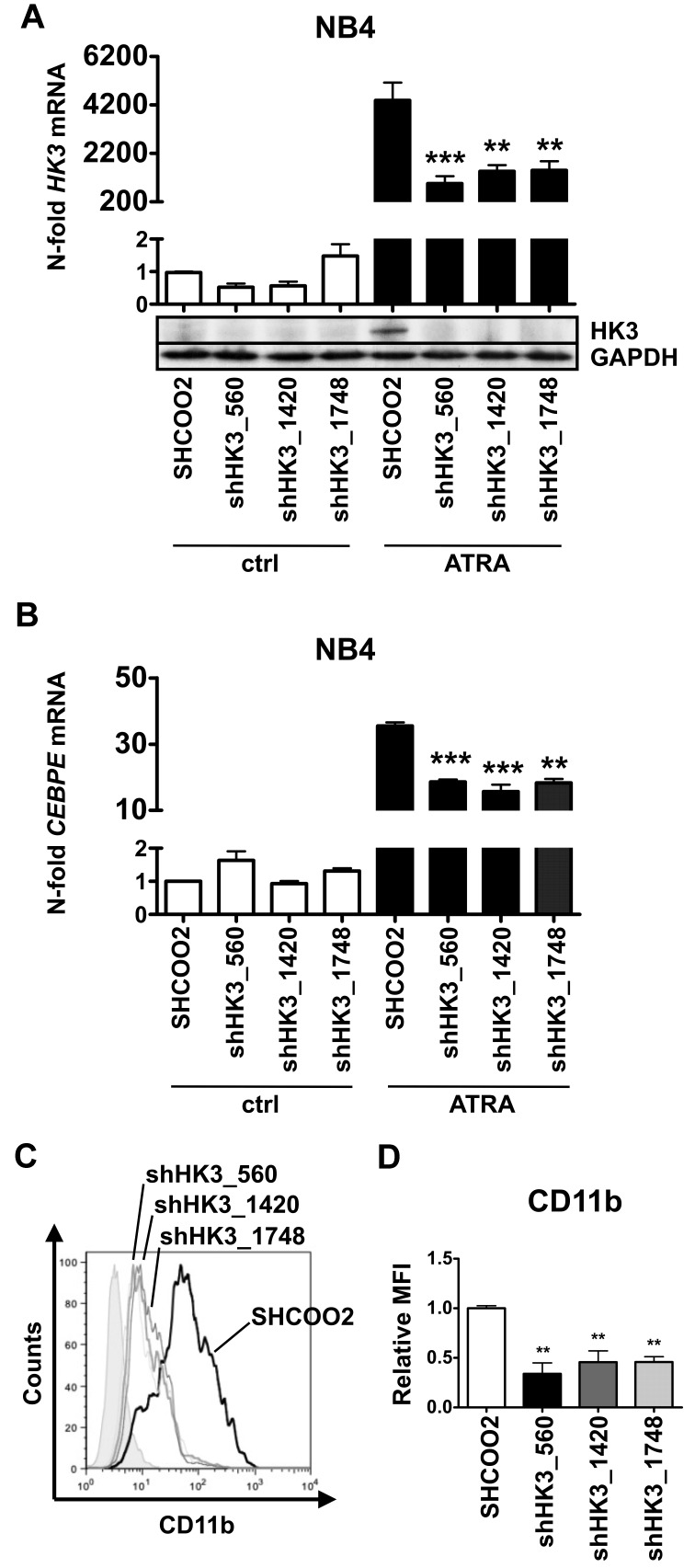
To test whether HK3 is functionally involved in ATRA-induced neutrophil differentiation of APL cells, we generated 3 independent NB4 HK3 knockdown cell lines using lentiviral vectors expressing shRNA targeting HK3. First, we determined HK3 knockdown efficiency by real-time quantitative RT-PCR and Western blotting. Each of the 3 NB4 shHK3 cell lines displayed more than 65% reduction in HK3 mRNA levels and clearly reduced protein expression compared with cells expressing a nontargeting shRNA on ATRA-induced neutrophil differentiation (Figure 4A). Most importantly, inhibiting HK3 in NB4 APL cells resulted in significantly diminished neutrophil differentiation compared with control cells as seen by a 50% reduction in CEBPE mRNA and CD11b surface expression (Figure 4B-D). We confirmed these results in HT93 APL cells (supplemental Figure 4A-B). Altogether, these experiments show that HK3-depleted NB4 APL cells are impaired in neutrophil differentiation.
Figure 4.
Inhibition of HK3 by shRNA impairs neutrophil differentiation of APL cells. NB4 cells were stably transduced with nontargeting shRNA (SHC002) or shRNAs targeting HK3 (shHK3_560, shHK3_1420, shHK3_1748) and differentiated with 1μM ATRA for 6 days. HK3 (A) and CEBPE (B) mRNA levels were measured by real-time quantitative RT-PCR and are given as n-fold changes compared with untreated SHCOO2 cells and normalized to the housekeeping gene HMBS. HK3 protein expression was measured by Western blotting. GAPDH expression was used as loading control. (C-D) CD11b flow cytometric analysis of NB4 control (SHCOO2) and HK3 knockdown cells.***P < .0005; **P < .005
Down-regulation of HK3 expression by shRNA impairs APL cell viability
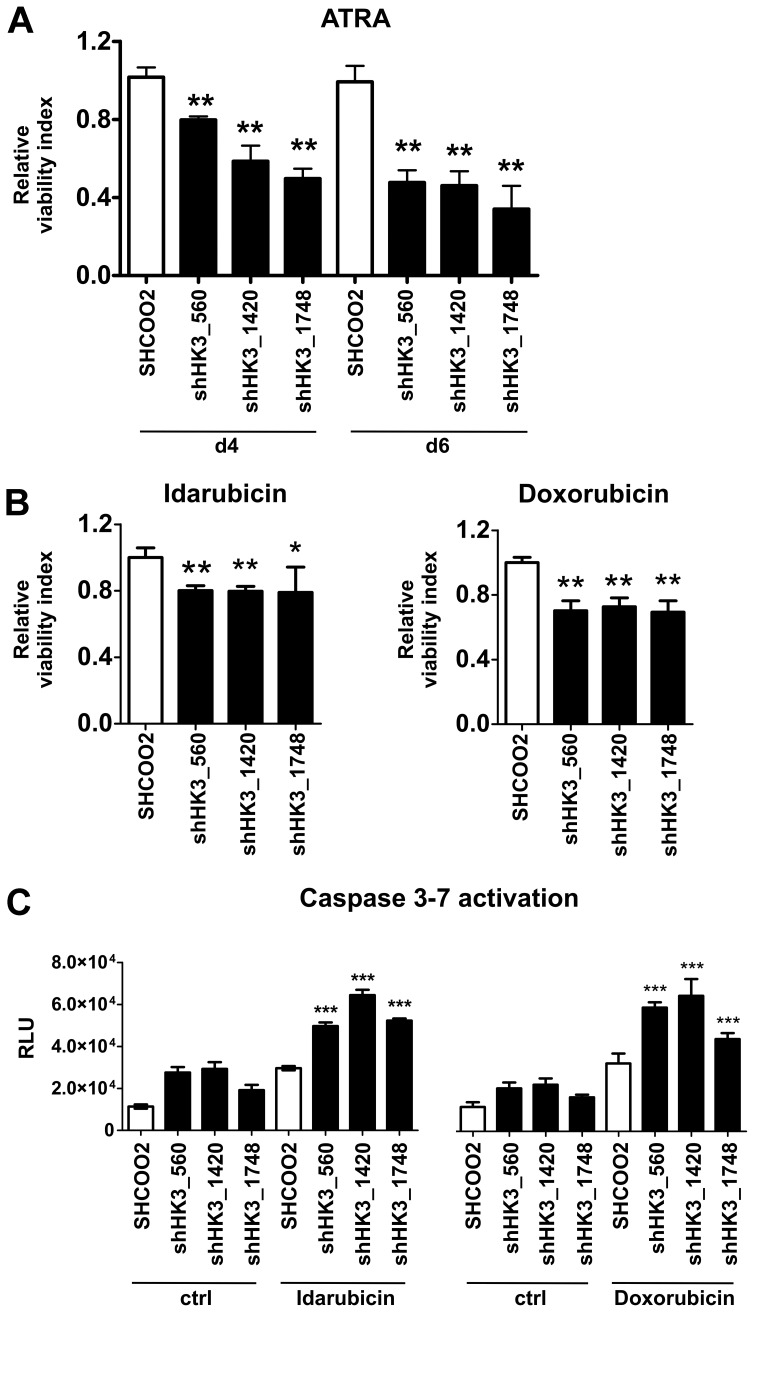
Given the role of HK1 and HK2 in cell survival,29 we then asked whether HK3 might affect APL cell viability during ATRA-induced neutrophil differentiation. Indeed, we found that cell viability of NB4 HK3 knockdown lines was 20% (shHK3_560) and 50% (shHK3_1420 and shHK3_1748) lower at day 4, and 50% (shHK3_560 and shHK3_1420) and 65% (shHK3_1748) lower at day 6 compared with SHCOO2 control cells (Figure 5A). Next, we asked whether HK3 also plays a role in resistance mechanisms to anthracyclins, cytostatic drugs routinely used in AML treatment. The most effective NB4 killing conditions were determined by conducting dose-response curves with idarubicin and doxorubicin. NB4 HK3 knockdown cells treated with idarubicin or doxorubicin displayed a 20% to 25% and 40% to 45% decrease in IC50 values compared with SHCOO2 cells, respectively (data not shown). We observed a significant 26% to 33% reduction of viable cells at 48 hours after idarubicin and at 72 hours after doxorubicin treatment in NB4 HK3 knockdown compared with SHCOO2 control cells (Figure 5B). Moreover, the reduction in cell survival of NB4 HK3 knockdown cells on anthracyclin treatment was associated with a significant activation of apoptosis as measured by the early activation of effector caspase-3 and caspase-7 (Figure 5C). These results indicate, for the first time, a role for HK3 in cellular protection of ATRA-differentiated APL cells, where HK3 is highly up-regulated. In addition, inhibiting HK3 in nondifferentiated APL cells increased sensitivity to anthracyclin treatment.
Figure 5.
Inhibition of HK3 decreases cell viability of ATRA-differentiated APL cells and renders APL cells more sensitive to anthracyclin therapy. (A) NB4 control (SHCOO2) or HK3 knockdown (shHK3_560, shHK3_1420, shHK3_1748) APL cells were grown in the presence or absence of 1μM ATRA for 4 or 6 days. Cell viability was measured by an Alamar Blue assay in 3 independent experiments. Cell viability is shown as percent change compared with untreated SHCOO2 control cells. **P < .005, shHK3 vs SHC002 control (Mann-Whitney U test). (B) NB4 control (SHC002) or HK3 knockdown (shHK3_560, shHK3_1420, shHK3_1748) APL cells were incubated with 0.01μM idarubicin for 48 hours (left panel) or 0.03μM doxorubicin for 72 hours (right panel), and cell viability was measured as in panel A. **P < .01; *P < .05 (Mann-Whitney U tests). (C) Caspase activation of NB4 cells treated with 0.1μM idarubicin for 24 hours (left panel) or 0.1μM doxorubicin for 48 hours (right panel) was measured by a luminescence assay. ***P < .0001 (Mann-Whitney U test).
Discussion
We previously reported a role for PU.1 in myeloid cell survival by directly activating the antiapoptotic BCL2 family member BCL2A1 or by inhibiting the transcriptional activity of the p53 tumor suppressor. In the latter case, PU.1 expression prevented the activation of antiproliferative p53 targets, such as p21CIP1 or Bax. In an attempt to characterize additional PU.1 targets involved in cell survival of myeloid cells, we investigated the regulation and function of HK3, a gene we found strongly up-regulated in PU.1-restored 503 PU.1 knockout myeloid cells. We found that HK3 levels are significantly repressed in primary AML blast cells that are blocked in their differentiation. Interestingly, the lowest HK3 expression levels were found in APL patients. In support of our findings, Payton et al described that low HK3 expression was clearly associated with a gene expression signature that is specific to APL.30 Moreover, low HK3 expression was paralleled by low PU.1 expression, and induction of HK3 during neutrophil differentiation of APL cell lines was clearly PU.1-dependent as HK3 activation was significantly dampened in PU.1 knockdown APL cells during neutrophil differentiation. We found direct activation of the HK3 promoter by PU.1 as evidenced by ChIP and promoter reporter assays. This observation links PU.1, for the first time, to HK3 regulation and glycolysis. Generally, the transcriptional regulation of HK3 is not well characterized. It was found that its regulation by the Oct-1 transcription factor is necessary for basal Hk3 expression in rat neuronal cells.31 The induction of OCT-1 on neutrophil differentiation of APL cell lines and its role as a direct transcriptional activator of PU.132 suggests that HK3 expression is controlled by OCT-1 either directly or indirectly via PU.1 during neutrophil differentiation. Furthermore, our findings of particular low HK3 levels in APL cells indicate that HK3 transcription is further repressed by PML-RARA. Indeed, we found that PML-RARA binds to the HK3 promoter and represses PU.1-mediated HK3 transcription. Alternatively, low HK3 levels in APL cells might be the result of direct PML-RARA–mediated repression of its positive regulator PU.1. On the other hand, a common feature in APL is that PML-RARA specifically targets PU.1-controlled genes via binding to the PU.1 protein, which itself binds to the DNA of its target genes.4 Because we found that PU.1 and PML-RARA bind to the same HK3 promoter regions using ChIP experiments, we cannot exclude that PML-RARA indirectly binds to the HK3 promoter and inhibits its activity via PU.1 binding. Lastly, our data showing significantly lower HK3 expression in AML patient samples with inactivating CEBPA mutations point to a possible regulation of HK3 by a second myeloid transcription factor. In summary, the regulation of HK3 controlled by transcription factors primarily involved in myeloid differentiation would explain the myeloid associated expression of HK3.
Apart from their role in the first step of glycolysis, HK1 and HK2 protect cells from cell death. Binding of HK1 and HK2 to mitochondria, for example, results in decreased cell death, and overexpression of these enzymes leads to cellular protection (reviewed by Mathupala et al29). Because HK3 lacks the hydrophobic N-terminal domain that is critical for binding to mitochondria but still exhibits prosurvival functions,33 different mechanisms leading to increased cell viability may be operative. In line with this second function of hexokinases, we observed significantly increased cell death of APL HK3 knockdown cells during myeloid differentiation or on treatment with anthracyclins. The majority of reports dealing with a role for hexokinases in cell survival analyzed HK1 or HK2. For example, dissociation of HK2 from the mitochondria results in the release of apoptosis-inducing factor ultimately leading to cell death in HL60 AML cells.34 Accordingly, ectopic expression of HK2 caused increased cell viability and resistance to the cytostatic drug cisplatinum in hepatocellular carcinoma cell lines, whereas knocking down HK2 in colon cancer cells increased sensitivity to 5-fluorouracil.35,36 Similarly, HK1 attenuates the caspase-driven cell death in a range of human cell lines, including hematopoietic cells.37 In line with our findings, it was shown that expression of HK3 protects HEK293 cells from oxidant-induced cell death.38
Granulocytes primarily depend on anaerobic glycolysis involving hexokinases to supply the necessary energy for locomotion and chemotaxis in low oxygen conditions.39 Thus, PU.1-dependent induction of HK3 in maturing neutrophils may be needed for these adaptive responses. In summary, we propose a new mechanism whereby PU.1 directly activates HK3 transcription during neutrophil differentiation, supporting short-term cell survival of mature neutrophils. This transcriptional activation of HK3 is primarily impaired in APL, in vivo and in vitro, because of direct or indirect PML-RARA repression. Our data link, for the first time, PU.1 to the HK3 survival pathway during neutrophil differentiation of APL cells.
Supplementary Material
Acknowledgments
The authors thank Deborah Shan (Experimental Oncology/Hematology, Department of Clinical Research, University of Bern, Bern, Switzerland) for excellent technical support.
This work was supported by the Swiss National Science Foundation (grant 3100A0-118276, M.P.T.); the Werner and Hedy Berger-Janser Foundation of Cancer Research (M.F.F. and M.P.T.); the Bernese Foundation of Cancer Research, the Marlies-Schwegler Foundation, the Ursula-Hecht-Foundation for Leukemia Research (M.F.F.); and the National Institutes of Health (R01HL091219, B.E.T.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.A.F. performed the experimental research, analyzed the data, and drafted the article; P.J.M.V., T.H., and B.L. provided patient samples, analyzed patient data, and revised the article; B.E.T. provided array data and PU.1 reagents and revised the article; M.F.F. instigated the experimental design and revised the drafted article; and M.P.T. designed the project, prepared lentiviral vectors, wrote the article, and gave final approval of the submitted manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario P. Tschan, Department of Clinical Research, Experimental Oncology and Hematology (MEM E829), University of Bern, Murtenstrasse 35, CH-3010 Bern, Switzerland; e-mail: mtschan@dkf.unibe.ch.
References
- 1.Matutes E. Acute myeloid leukaemia with recurrent genetic abnormalities. 2001. [Accessed July 13, 2011]. http://publications.icr.ac.uk/103.
- 2.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature. 1990;347(6293):558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 3.Dyck JA, Maul GG, Miller WH, et al. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76(2):333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Wang P, Shi J, et al. PML/RARalpha targets promoter regions containing PU.1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer Cell. 2010;17(2):186–197. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 5.de Thé H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10(11):775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- 6.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 7.McKercher SR, Torbett BE, Anderson KL, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 8.Martens JHA, Brinkman AB, Simmer F, et al. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell. 2010;17(2):173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int J Biochem Cell Biol. 2008;40(1):22–27. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Tschan MP, Reddy VA, Ress A, et al. PU.1 binding to the p53 family of tumor suppressors impairs their transcriptional activity. Oncogene. 2008;27(24):3489–3493. doi: 10.1038/sj.onc.1211004. [DOI] [PubMed] [Google Scholar]
- 11.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331(6153):277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 12.Cárdenas ML, Cornish-Bowden A, Ureta T. Evolution and regulatory role of the hexokinases. Biochim Biophys Acta. 1998;1401(3):242–264. doi: 10.1016/s0167-4889(97)00150-x. [DOI] [PubMed] [Google Scholar]
- 13.Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206(12):2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 14.Katzen HM. The multiple forms of mammalian hexokinase and their significance to the action of insulin. Adv Enzyme Regul. 1967;5:335–356. doi: 10.1016/0065-2571(67)90025-8. [DOI] [PubMed] [Google Scholar]
- 15.Wilson JE. Hexokinases. Rev Physiol Biochem Pharmacol. 1995;126:65–198. doi: 10.1007/BFb0049776. [DOI] [PubMed] [Google Scholar]
- 16.Rijksen GSG, Rijksen G, Staal GE, et al. Compartmentation of hexokinase in human blood cells: characterization of soluble and particulate enzymes. Biochim Biophys Acta. 1982;719(3):431–437. doi: 10.1016/0304-4165(82)90230-6. [DOI] [PubMed] [Google Scholar]
- 17.Tschan MP, Fischer KM, Fung VS, et al. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J Biol Chem. 2003;278(44):42750–42760. doi: 10.1074/jbc.M307067200. [DOI] [PubMed] [Google Scholar]
- 18.Breems DA, Boogaerts MA, Dekker AW, et al. Autologous bone marrow transplantation as consolidation therapy in the treatment of adult patients under 60 years with acute myeloid leukaemia in first complete remission: a prospective randomized Dutch-Belgian Haemato-Oncology Co-operative Group (HOVON) and Swiss Group for Clinical Cancer Research (SAKK) trial. Br J Haematol. 2005;128(1):59–65. doi: 10.1111/j.1365-2141.2004.05282.x. [DOI] [PubMed] [Google Scholar]
- 19.Löwenberg B, Boogaerts MA, Daenen SM, et al. Value of different modalities of granulocyte-macrophage colony-stimulating factor applied during or after induction therapy of acute myeloid leukemia. J Clin Oncol. 1997;15(12):3496–3506. doi: 10.1200/JCO.1997.15.12.3496. [DOI] [PubMed] [Google Scholar]
- 20.Löwenberg B, van Putten W, Theobald M, et al. Effect of priming with granulocyte colony-stimulating factor on the outcome of chemotherapy for acute myeloid leukemia. N Engl J Med. 2003;349(8):743–752. doi: 10.1056/NEJMoa025406. [DOI] [PubMed] [Google Scholar]
- 21.Ossenkoppele GJ, Graveland WJ, Sonneveld P, et al. The value of fludarabine in addition to ARA-C and G-CSF in the treatment of patients with high-risk myelodysplastic syndromes and AML in elderly patients. Blood. 2004;103(8):2908–2913. doi: 10.1182/blood-2003-07-2195. [DOI] [PubMed] [Google Scholar]
- 22.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118(4):1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 23.Haferlach T, Kohlmann A, Wieczorek L, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28(15):2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschan MP, Shan D, Laedrach J, et al. NDRG1/2 expression is inhibited in primary acute myeloid leukemia. Leuk Res. 2010;34(3):393–398. doi: 10.1016/j.leukres.2009.08.037. [DOI] [PubMed] [Google Scholar]
- 25.Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci U S A. 2002;99(1):95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenal M, Batliner J, Reddy VA, et al. The anti–apoptotic gene BCL2A1 is a novel transcriptional target of PU.1. Leukemia. 2010;24(5):1073–1076. doi: 10.1038/leu.2010.26. [DOI] [PubMed] [Google Scholar]
- 27.Batliner J, Mancarelli MM, Jenal M, et al. CLEC5A (MDL-1) is a novel PU.1 transcriptional target during myeloid differentiation. Mol Immunol. 2011;48(4):714–719. doi: 10.1016/j.molimm.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postic C, Shiota M, Magnuson MA. Cell-specific roles of glucokinase in glucose homeostasis. Recent Prog Horm Res. 2001;56:195–217. doi: 10.1210/rp.56.1.195. [DOI] [PubMed] [Google Scholar]
- 29.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: Cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2000;25(34):4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payton JE, Grieselhuber NR, Chang L-W, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. 2009;119(6):1714–1726. doi: 10.1172/JCI38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastian S, White JA, Wilson JE. Characterization of the rat type III hexokinase gene promoter: a functional octamer 1 motif is critical for basal promoter activity. J Biol Chem. 1999;274(44):31700–31706. doi: 10.1074/jbc.274.44.31700. [DOI] [PubMed] [Google Scholar]
- 32.Mueller BU, Pabst T, Fos J, et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107(8):3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preller A, Wilson JE. Localization of the type III isozyme of hexokinase at the nuclear periphery. Arch Biochem Biophys. 1992;294(2):482–492. doi: 10.1016/0003-9861(92)90715-9. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Zhang H, Lu W, Huang P. Role of mitochondria-associated hexokinase II in cancer cell death induced by 3-bromopyruvate. Biochim Biophys Acta. 2009;1787(5):553–560. doi: 10.1016/j.bbabio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn KJ, Hwang HS, Park JH, et al. Evaluation of the role of hexokinase type II in cellular proliferation and apoptosis using human hepatocellular carcinoma cell lines. J Nucl Med. 2009;50(9):1525–1532. doi: 10.2967/jnumed.108.060780. [DOI] [PubMed] [Google Scholar]
- 36.Peng Q-P, Zhou J-M, Zhou Q, et al. Downregulation of the hexokinase II gene sensitizes human colon cancer cells to 5-fluorouracil. Chemotherapy. 2008;54(5):357–363. doi: 10.1159/000153655. [DOI] [PubMed] [Google Scholar]
- 37.Schindler A, Foley E. A functional RNAi screen identifies hexokinase 1 as a modifier of type II apoptosis. Cell Signal. 2010;22(9):1330–1340. doi: 10.1016/j.cellsig.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt E, Wu R, Rabeh W, et al. Regulation and cytoprotective role of hexokinase III. PLoS One. 2010;5(11):e13823. doi: 10.1371/journal.pone.0013823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisdorf D, Craddock P, Jacob H. Glycogenolysis versus glucose transport in human granulocytes: differential activation in phagocytosis and chemotaxis. Blood. 1982;60(4):888–893. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.