Abstract
So far, genetic diversity among strains within Mycobacterium massiliense has rarely been studied. To investigate the genetic diversity among M. massiliense, we conducted phylogenetic analysis based on hsp65 (603-bp) and rpoB (711-bp) sequences from 65 M. massiliense Korean isolates. We found that hsp65 sequence analysis could clearly differentiate them into two distinct genotypes, Type I and Type II, which were isolated from 35 (53.8%) and 30 patients (46.2%), respectively. The rpoB sequence analysis revealed a total of four genotypes (R-I to R-IV) within M. massiliense strains, three of which (R-I, R-II and R-III) correlated with hsp65 Type I, and other (R-IV), which correlated with Type II. Interestingly, genotyping by the hsp65 method agreed well with colony morphology. Despite some exceptions, Type I and II correlated with smooth and rough colonies, respectively. Also, both types were completely different from one another in terms of MALDI-TOF mass spectrometry profiles of whole lipid. In addition, we developed PCR-restriction analysis (PRA) based on the Hinf I digestion of 644-bp hsp65 PCR amplicons, which enables the two genotypes within M. massiliense to be easily and reliably separated. In conclusion, two distinct hsp65 genotypes exist within M. massiliense strains, which differ from one another in terms of both morphology and lipid profile. Furthermore, our data indicates that Type II is a novel M. massiliense genotype being herein presented for the first time. The disparity in clinical traits between these two hsp65 genotypes needs to be exploited in the future study.
Introduction
Rapidly growing mycobacteria (RGM) are ubiquitous organisms increasingly emerging as important human pathogens. Recently, there have been more frequent reports of RGM infections in immunocompetent people as well as in people with predisposing factors or those who are immunosuppressed [1], [2]. In particular, among RGMs, Mycobacterium abscessus is commonly associated with wound infection and abscess formation and is the RGM that most frequently causes chronic lung disease. M. abscessus is also notable for its resistance to treatment and the poor clinical outcome of infection with the organism [2]. In South Korea, in contrast to countries such as the United States and Japan [3]–[5], infection of M. abscessus is the most prevalent RGM infection, and second only to the M. avium complex for nontuberculous mycobacterium (NTM) [6], [7].
Recent application of multilocus sequencing has broadened our knowledge about the diversity between the M. abscessus complex. Two new species of mycobacteria closely related to M. abscessus, M. massiliense and M. bolletii, have been described [8], [9]. A recent molecular epidemiologic study using 144 RGM isolates from Korean patients showed that two M. abscessus related species, M. abscessus and M. massiliense, were responsible for the most of the infections [M. abscessus (65/144 isolates, 51.2%) and M. massiliense (59/144 patients, 46.5%)]. However, M. bolletii has rarely been isolated from Korean patients (2/144 patients, 1.6%) [10]. It should be noted that the disparity between M. abscessus and M. massiliense infections in terms of clinical significance was reported by a recent paper using Korean patients infected with either M. abscesus or M. massiliense, thereby putting the stress on the separation between the two related species via molecular - based methods in the clinical aspects [11].
As alternatives to 16 S rRNA gene sequencing, several other gene targets have been effectively used for the NTM differentiation, demonstrating the limited value in the separation between some closely related NTM strains [12]–[16]. Among these gene targets, partial sequencing that targeted the rpoB or hsp65 has increasingly been used to differentiate between M. abscessus, M. massiliense, and M. bolletii [8], [10], [11], [17], [18].
Due to the lack of clinical or epidemiological information regarding M. massiliense, studies about genetic or phenotypic traits of M. massiliense have rarely, if ever, been introduced, as compared to M. abscessus. Thus, the present study aims to elucidate the genetic diversity between M. massiliense clinical isolates by two different chronometer molecules - hsp65 and rpoB - genes and to identify the relationships between the determined genotype and phenetic traits, particularly in terms of colony morphology.
Results
Identification by hsp65 Sequencing Analysis
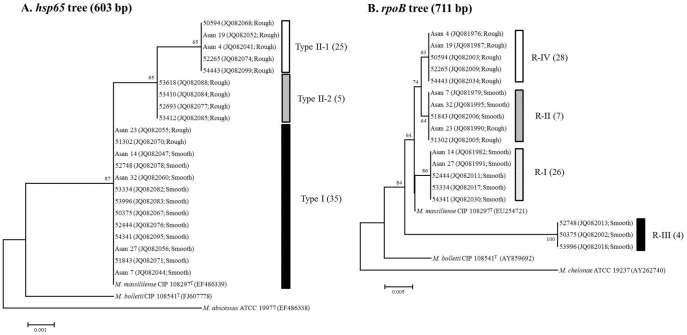
First, 109 M. abscessus complex strains from Asan medical center (AMC), which had been identified by rpoB PRA, were further analyzed using the hsp65 sequencing method, showing the prevalence of 44 M. abscessus (40.4%) and 65 M. massiliense strains (59.6%) (Table 1). However, no M. bolletii strains were found in our cohort (0%). Phylogenetic analysis based on the partial hsp65 sequence (603 bp) showed that there were two phylogenetic groups (Type I and II) within the 65 M. massiliense strains, composed of three sequevars (Figure 1 and 2A). Type I isolated from 35 patients (53.8%) included a single sequevar having the same 603-bp hsp65 sequences as M. massiliense CIP 108297T. But Type II, also isolated from 30 patients (46.2%), included two different sequevars, Type II-1 and Type II-2, which showed 2-bp (T444A and C714A) and 1-bp different sequences (T444A) from M. massiliense CIP 108297T, respectively (Table 2, Figure 1). Type I, Type II-1, and Type II-2 were isolated from 35 (53.8%), 25 (38.5%) and 5 patients (7.7%), respectively (Figure 2A).
Table 1. Separation of 109 M. abscessus related Korean strains used in this study into species or genotype level by sequence analysis based on the partial hsp65 gene sequence (603 bp).
| Species and group | No. (%) of strains |
| M. abscessus | 44 (40.4) |
| M. massiliense | 65 (59.6) |
| Type I | 35 (32.1) |
| Type II | 30 (27.5) |
| Total | 109 (100.0) |
Figure 1. Sequence polymorphisms between the three hsp65 sequevars of M. massiliense.
Type I strains have the same sequence as the M. massiliense type strain; however, Type II-1 and Type II-2 strains differed from the M. massiliense type strain by 2-bp (T444A and C714A) and 1-bp (T444A), respectively. The nucleotide numbers correspond to those from the complete sequence of the heat shock protein 65 kDa (hsp65) gene of M. abscessuss ATCC 19977 (GenBank no. CU458896).
Figure 2. Phylogenetic trees based on the hsp65 gene (603 bp) and rpoB gene (711 bp) sequences.
Phylogenetic trees based on the (A) hsp65 gene (603 bp) and (B) rpoB gene (711 bp) sequences from M. massiliense clinical isolates, M. massiliense CIP 108297T, M. bolletii CIP 108541T, M. chelonae ATCC 19237, and M. abscessus ATCC 19977. These trees were constructed using the neighbor-joining method. The bootstrap values were calculated from 1,000 replications. Bootstrap values of <50% are not shown. The bars indicate numbers of substitutions per nucleotide position.
Table 2. The frequency of the two hsp65 genotypes (Type I and Type II) determined by hsp65 sequence analysis and Hinf I PRA methods, and the two colony morphotypes (rough and smooth) among 65 M. massiliense clinical strains.
| Hinf I PRA | Colony morphotype | ||||
| hsp65 genotypea | No. (%)b | 366, 278 | 280, 86, 278 | Rough | Smooth |
| Type I | 35 (30.1) | 35 (100.0) | 0 (0.0) | 12 (34.3) | 23 (65.7) |
| Type II | 30 (27.4) | 0 (0.0) | 30 (100.0) | 30 (100.0) | 0 (0.0) |
The hsp65 genotypes were determined by hsp65 sequence analysis (603-bp).
The percentage was calculated among M. massiliense strains.
rpoB Sequence Analysis of M. massiliense Strains
The partial rpoB sequences (711-bp) from the 65 M. massiliense strains were compared with each other to check the genetic heterogeneity between them. The rpoB based sequence analysis revealed the presence of four groups within the 65 M. massiliense strains, suggesting more genetic diversity within M. massiliense strains in the 711-bp rpoB sequence than in the 603-bp hsp65 sequence. All of the isolates showed different rpoB sequences than the M. massiliense CIP 108297T with sequence divergence ranging from 2-bp (R-I, R-II, and R-IV) to 14-bp (R-III). These groups were clearly separated by phylogenetic analysis based on rpoB gene sequences of the 65 M. massiliense strains (Figure 2B). Despite some minor exceptions, the genotype R-I, R-II, R-III and R-IV of four rpoB genotypes were related to the hsp65 Type I and Type II, respectively. The R-I were found most frequently in M. massiliense Type I (68.6%, 24/35 strains). The R-IV, which had an rpoB sequence that was 2-bp different rpoB sequence (T2760C and G2907A) from M. massiliense CIP 108297T were most frequently found in M. massiliense Type II (93.3%, 28/30 strains), but not in Type I (Table 3).
Table 3. The frequency of the four rpoB genotypes (R-I to R-IV) determined by rpoB sequence analysis (711-bp) among 65 M. massiliense clinical strains, polymorphisms of rpoB genotypes and relationships between rpoB and hsp65 genotypes.
| No. (%) | |||
| rpoB genotypea | hsp65 Type I | hsp65 Type II | P - value |
| R-I (T2484G and G2934A) | 24 (68.6) | 2 (6.7) | 0.010 |
| R-II (C2569T and T2851C) | 7 (20.0) | 0 (0.0) | 0.000 |
| R-III (C2475T, T2484C, T2835G, C2848G, A2849C, G2850C, C2853T, C2859T, A2861C, G2862A, G2868C, C2869A, A2870C, G2871C, G2874T, T2877G, C2880G, C2886T, C2988T, and C3022G) | 4 (11.4) | 0 (0.0) | 0.056 |
| R-IV (T2760C and G2907A) | 0 (0.0) | 28 (93.3) | 0.000 |
| Total | 35 (100.0) | 30 (100.0) | |
Polymorphisms which are different from the type strain of M. massiliense. The nucleotide numbers correspond to those from complete sequence of RNA polymerase beta subunit (rpoB) gene of M. abscessus ATCC 19977 (GenBank no. CU458896).
Relationships between hsp65 Genotypes and Colony Morphology
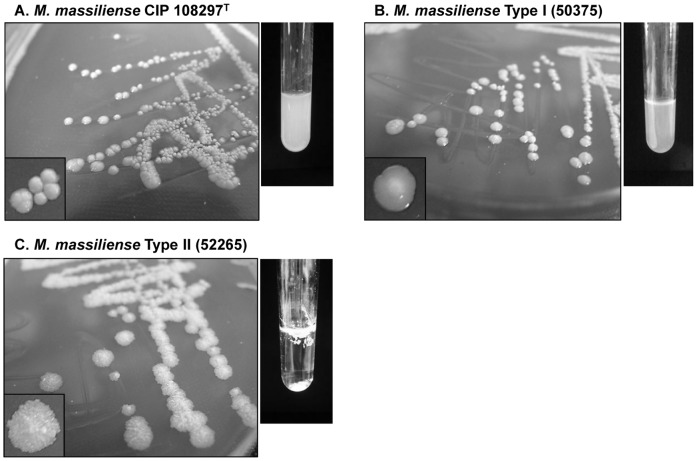
Sub-cultured colonies of the 65 M. massiliense strains identified by hsp65 sequence analysis and M. massiliense CIP 108297T were analyzed on 7H10 agar plates. Notably, substantial differences in colony morphology were found between two hsp65 genotypes, Type I and Type II (Figure 3). Interestingly, all the 30 of the isolates belonging to Type II (Type II-1 and Type II-2) showed rough colony without any exception. However, the Type I isolates showed both rough and smooth colony morphologies. In Type I, as shown in M. massiliense CIP 108297T, smooth morphology was more common than rough morphology [smooth vs. rough; 23/35 isolates (65.7%) vs. 12/35 isolates (34.3%)] (Table 2). In addition to the different colony morphologies in the agar plates, Type I of smooth morphotype and Type II of rough morphotype also differed in terms of the growth characteristics of 7H9 broth cultures. While Type I strain and M. massiliense CIP 108297T with smooth morphotype showed a dispersed growth pattern, the Type II isolate showed a typical aggregative pellicle growth that is confined to the surface of the medium, as shown in the M. tuberculosis clinical isolates (Figure 3). But, the Type I strain with rough morphotype was quite similar to Type II in terms of the growth pattern of the broth culture (data not shown).
Figure 3. Colony morphology and the growth patterns in 7H9 broth medium.
Colony morphology (left panel) and the growth patterns in 7H9 broth medium (right panel) of (A) M. massiliense CIP 108297T, (B) M. massiliense Type I strain, and (C) M. massiliense Type II strain.
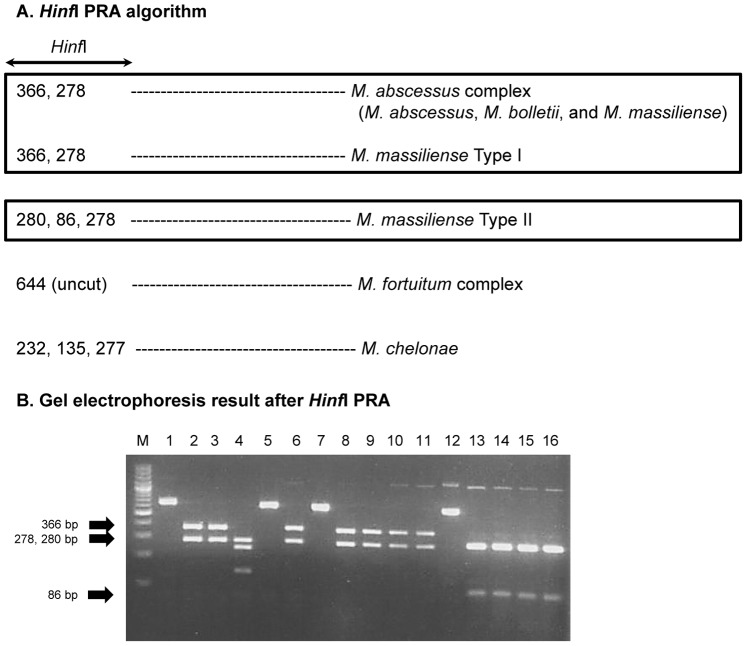
Differentiation between Two Genotypes of M. massiliense Strains by Hinf I PRA
To enable the simple separation of M. massiliense Type II strains from other related RGMs, we developed a novel Hinf I PRA algorithm. The results and the algorithm obtained by applying the Hinf I PRA method to the 65 M. massiliense-related strains and five reference strains are summarized in Table 2 and Figure 4A, respectively. As predicted, the M. massiliense Type II strains were clearly distinguished from other related RGMs, including M. massiliense Type I strains, producing distinct PRA patterns (280, 278, and 86-bp), although it was not possible to separate between the upper two bands, 280 and 278-bp (Figure 4B). When the results of both the Hinf I PRA and hsp65 sequencing methods were compared, the sensitivity and specificity of our Hinf I PRA method for separating between the two genotypes of M. massiliense were 100% and 100%, respectively (Table 2). The informations about rpoB and hsp65 genotypes, Hinf I PRA patterns and colony morphology of each isolate were shown in Table S1.
Figure 4. Identification of M. massiliense Type I and Type II strains by hsp65 PRA method.
(A) Hinf I PRA algorithm for differentiating of M. massiliense Type I and Type II strains. (B) Agarose gel electrophoresis after Hinf I PRA. Lanes: M, 100-bp ladder; 1, M. abscessus (uncut); 2, M. abscessus (Hinf I cut); 3, M. bolletii (Hinf I cut); 4, M. chelonae (Hinf I cut); 5, M. fortuitum (Hinf I cut); 6, M. massiliense (Hinf I cut); 7, 50375 (Type I, uncut); 8, 50375 (Type I, Hinf I cut); 9, 51302 (Type I, Hinf I cut); 10, 51843 (Type I, Hinf I cut); 11, 52352 (Type I, Hinf I cut); 12, 51048 (Type II, uncut); 13, 51048 (Type II, Hinf I cut); 14, 52008 (Type II, Hinf I cut); 15, 50594 (Type II, Hinf I cut); 16, 52012 (Type II, Hinf I cut).
Biochemical Tests and Drug Susceptibility Tests
The phenotypic characteristics of three reference strains, four M. massiliense Type I strains, and four M. massiliense Type II strains were analyzed and compared. With the exception of the colony morphology, no characteristic traits were not found to differentiate between Type I and Type II groups (Table S2). The results of our drug susceptibility test likewise showed no significant differences between Type I and Type II groups (Table S3). In addition, no point mutations were found at the adenine at position 2058 (A2058) or 2059 (A2059) in the peptidyltransferase region of the 23 S rRNA gene in any of the 65 M. massiliense strains (data not shown).
Comparison of HPLC and MALDI-TOF Mass Spectrometry Profiles between Two Genotypes of M. massiliense
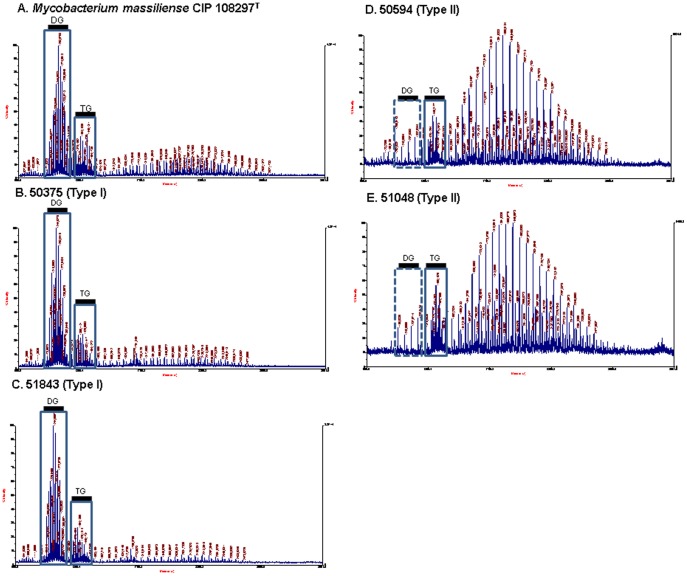
The profiles of HPLC and MALDI-TOF mass spectrometry of M. massiliense CIP 108297T, four M. massiliense Type I strains, and four M. massiliense Type II strains were analyzed and compared. Generally, the HPLC profiles of all the M. massiliense strains were virtually identical. However, both genotypes showed different profiles at the peak of about 1.91 retention time to be separated with each other. All four of the Type II strains showed a higher level of intensity at this peak than four Type I and M. massiliense CIP 108297T (Figure S1). The most substantial differences between the MALDI-TOF mass spectrometry profiles were found between the two genotypes. Usually, the MALDI-TOF mass spectrometry profiles have two distinct clusters of peaks ranging from m/z 1171 to m/z 1316 and from m/z 1360 to m/z 1464. The first cluster and the second cluster represent diglycosylated glycopeptidolipid (GPL) and triglycosylated GPL, respectively [19]. All four of the Type I strains and M. massiliense CIP 108297T showed the typical MALDI-TOF mass spectrometry profiles of two clusters, but all four of the Type II strains showed unusual profiles, with a significantly low intensity of the putative diglycosylated GPLs and diverse peaks of high intensity from m/z 1598 to m/z 2477, suggesting a disparity in the GPL nature between the two genotypes (Figure 5).
Figure 5. MALDI-TOF mass spectrometry analysis.
MALDI-TOF mass spectrometry analysis of extracted lipids from (A) M. massiliense CIP 108297T, (B) 50375 (Type I), (C) 51843 (Type I), (D) 50594 (Type II), and (E) 51048 (Type II). DG, diglycosylated GPLs; TG, triglycosylated GPLs.
Discussion
Following the recent taxonomic separation of three very closely related RGMs - M. abscessus, M. massiliense and M. bolletii - reports regarding human infections of M. massiliense have been increasing [20]–[23]. Thus, it has become more important to study the diversity between their interspecies or intraspecies [10], [24]. A recent report based on multilocus sequencing showed that M. massiliense is composed of strains with more diverse genetic heterogeneity than its closely related species, M. abscessus [24]. To investigate the intraspecies genetic diversity within M. massiliense, we studied 65 Korean strains by applying a gene-based sequencing approach consisting of two independent chronometers (hsp65 and rpoB gene).
Rather than 16 S rRNA gene-based analysis, which has been shown to have limited effectiveness in discriminating between mycobacterial strains, a 603-bp hsp65 sequencing analysis has been proven to be useful for identifying mycobacteria [25]–[27], particularly for the separating between the three related species of M. abscessus; M. abscessus, M. massiliense, and M. bolletii [11], [28]. Our hsp65-based sequence analysis showed that M. massiliense is more prevalent than M. abscessus (59.6% vs. 40.4%), supporting the previous report that, in South Korea, M. massiliense isolated with a relative higher frequency than in other areas [10]. So, this epidemiologic feature raised the possibility of the presence of a distinct M. massiliense strain in South Korea. Indeed, for the first time, we report the identification of a novel hsp65 genotype (Type II) that was found with a high frequency in our Korean M. massiliense isolates (46.2%, 30/65 patients). Since none of the hsp65 sequences in the NCBI databases perfectly match those of Type II, it seems that Type II may very well be a unique M. massiliense strain in Korea.
To investigate the phenotypic difference between two hsp65 genotypes, a number of phenetic traits were compared between both genotypes, including colony morphology. Although most traits could not provide any definitive criteria for differentiation, a strong correlation was discovered between the colony morphology of the two hsp65 genotypes. Specifically, while the majority of the hsp65 Type I genotype (65.7%, 23/35 strains) showed a smooth morphotype, all the 30 strains with hsp65 Type II showed a rough colony morphology. So, it is tempting to speculate that the rough morphotype of Type II, like M. tuberculosis, may be an innate trait that evolved from the smooth strain, rather than a trait acquired via induced mutations during the in vivo infection.
NTMs have long been recognized as having both rough and smooth colony phenotypes [29]. Several reports have found a correlation between colony morphology and virulence, with rough variants generally being more virulent than smooth variants [30], [31]. Particularly, in the M. abscessus strains, smooth morphotype has occasionally spontaneously reverted to rough morphotype after several passages on agar plates or via in vivo passage into mice due to the reduced expression of glycopeptidolipid (GPL) [30]. Although the exact mechanism still remains a mystery, the phenomena may be attributed to the loss of GPL resulting in excessive secretion of TNF-á from the macrophage [32]. Our MALDI-TOF MS analysis showed that the Type I strains with smooth morphotype and Type II strains had completely different lipid profiles. In particular, the peaks corresponding to putative diglycosylated GPL were reduced in the Type II strains as compared to Type I. Given that mycobacterial cell wall lipid is one of the most important factors determining virulence, leading to the modulation of the host immune response, it is likely that two hsp65 genotypes of M. massiliense would show different pathogenesis. However, the above hypothesis needs to be proven through detailed analysis of clinical data and in vivo and in vitro virulence studies in the future. In addition, we could not find any phenotypic or chemotaxonomic differences between the 11 strains showing hsp65 Type I genotype with rough phenotype and the 30 Type II strains. The question of whether the two genotypes with rough phenotype (Type I and II) acquire the rough phenotype in different ways should be addressed in a future study.
A previous report showed that sequence analysis based on the 711-bp rpoB gene was better than hsp65 sequence based analysis in terms of discriminating between closely related mycobacterial strains, particularly, strains within M. abscessus or M. massiliense [12]. Our rpoB sequence analysis was also able to separate all 65 of the M. massiliense strains into four more diverse genotypes (R-I to R-IV) than hsp65 sequence analysis. However, while the previous report [10] had discordant results between hsp65 and rpoB-based sequence analysis in species determination in some M. massiliense strains, all 65 of the isolates identified as M. massiliense by the hsp65 method were also identified as M. massiliense by rpoB methods (Table 3, Figure 2B). Phylogenetic analysis based on the rpoB sequences showed that most hsp65 Type II strains also formed a monophyletic clade in a rpoB tree (Figure 2B), suggesting that hsp65 Type II may be composed of genetically identical strains. However, hsp65 Type I strains were divided into the three different clades in a rpoB tree, suggesting that hsp65 Type I may be composed of more diverse genetic groups. These results provide a likely explanation for the presence of phenotype divergence (the coexistence of both morphotypes) in type I over type II strains.
Although PRA is a previous century technique, replaced by sequencing analysis producing more accurate results, the PRA method has still been used for routine screening purposes in clinical settings for differentiating the mycobacterial strains, due to the its ease and rapidity [33]–[35]. So, we developed a novel PRA method using Hinf I enzyme targeting a 644-bp hsp65 genes for selective identification of M. massiliense Type II. A blind test for evaluating this PRA method proved that it can successfully identify all Type II strains from Type I, suggesting its feasibility for the diagnostic or epidemiologic purposes.
Our hsp65 and rpoB based methods failed to differentiate rough and smooth strains within hsp65 Type I. To overcome this limitation, methods with more discriminatory power such as variable-number tandem repeat (VNTR) [36]–[38] or whole genome sequencing [39]–[41] should be applied into hsp65 Type I strains for the future study.
In summary, through hsp65 sequencing analysis of 65 Korean M. massiliense strains, we found two hsp65 genotypes, Type I from 35 patients (53.8%) and Type II from 30 patients (46.2%), which were related to smooth and the rough colony phenotypes, respectively. Furthermore, the two hsp65 genotypes were also completely different in terms of their lipid profiles by MALDI-TOF MS. A total of four genotypes (R-I to R-IV) were also found by rpoB sequencing analysis, three of which (R-I, R-II, and R-III) were related to hsp65 Type I and the other (R-IV) which was related to hsp65 Type II. Our data indicates that the Type II hsp65 genotype, which also shows the R-IV rpoB genotype, is a novel M. massiliense group introduced for the first time in this study. In addition we developed a novel PRA method for selectively separating hsp65 Type II from other RGMs. Our data suggests that M. massiliense strains may be composed of genetically distinct diverse groups, of which pathogenic potentials need to be evaluated for future study.
Materials and Methods
Bacterial Strains
A total of 109 strains from sputa samples of different patients were provided by Tae Sun Shim, Asan Medical Center, South Korea were collected from January 2004 to June 2011. All the experiments were performed just on the extracted DNA the isolated strains, not directly sputa DNA. Furthermore, all the samples were collected in an anonymized manner, and any information about the personal details of the patients and any details about their clinical history were not supplied. In this case, the study could be under the waiver of informed consent. With the documentation for waiver of informed consent, this work was approved by the institutional review board of Seoul National University Hospital (C-1202-057-398) and Asan Medical Center (2012-0170). These samples were proved to be M. abscessus complex strains by rpoB PRA method [34]. For the further separation at the species level, these isolates were used for re-identification by direct sequencing protocol targeting the partial hsp65 gene [14]. The type strains of M. abscessus (ATCC 19977), M. massiliense (CIP 108297), and M. bolletii (CIP 108541) were used for comparison.
Colony Morphology and Broth Culture
To observe the colony morphology, the isolates cultivated on Ogawa media were sub-cultured on 7H10 agar medium supplemented by OADC at 37°C for 3 to 5 days [42]. To examine growth patterns on the broth culture, the clinical isolates and M. massiliense type strain were inoculated into 7H9 broth supplemented with ADC at approximately 1×104 CFU/ml and cultured at 37°C for 5 to 7 days [30], [42].
Biochemical Tests and Drug Susceptibility Tests
To determine their taxonomic relationships, M. massiliense CIP 108297T, four M. massiliense Type I strains (50375, 51843, 52352, and 52444), and four M. massililense Type II strains (50594, 51048, 52188, and 52265) were tested for biochemical and drug susceptibility profiles. These strains were cultured into Middlebrook 7H9 broth supplemented by ADC at 37°C. Colony morphology, pigment production in the dark condition, photo-induction, and the ability to grow at various temperatures (25, 37, and 45°C) were analyzed during six-week incubation on Middlebrook 7H10 agar plates. Acid-alcohol fastness was determined by Ziehl-Neelsen and auramine O staining. Other tests measured: niacin accumulation, nitrate reductase, arylsulfatase on days 3 and 14, heat-stable catalase (pH 7, 68°C), tellurite reductase, Tween 80 hydrolysis, urease and pyrazinamidase (PZA) [43]. Additional biochemical tests, such as to detect the activity of alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, and crystine arylamidase were conducted with the API ZYM kit (bioMerieux) as recommended by the manufacturer.
Inhibition tests including tolerance to thiophene-2-carboxylic acid hydrazide (TCH), p-nitrobenzoate (PNB), 5% sodium chloride, ethambutol (EMB) and picric acid were carried out, and the ability to grow on MacConkey agar without crystal violet was examined. Also, antimicrobial susceptibility was determined by the agar proportion method on 7H10 medium [43].
High-performance Liquid Chromatography (HPLC) Analysis
HPLC was used to analyze mycolic acids from M. massiliense CIP 108297T, the four aforementioned M. massiliense Type I strains, and the four aforementioned M. massililense Type II strains as previously described [44]. The Microbial Identification system (MIDI Inc.) and the HPLC mycobacterium library (available online at http://www.MycobacToscana.it) were used to identify mycolic acid patterns.
Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry Analysis
To analysis MALDI-TOF mass spectrometry, lipids were extracted with CHCl3/CH3OH (1∶1 v/v, adding 0.5 µl of 2,5-dihydroxybenzoic acid) from 30 ml 7H9 broth cultures of M. massiliense CIP 108297T, the four M. massiliense Type I strains, and the four M. massililense Type II strains. MALDI-TOF mass spectrometry was performed on the extracted samples with a Voyager DE-STR MALDI-TOF instrument (Perseptive Biosystems) equipped with a pulse nitrogen laser emitting at 337 nm as previously described [45].
DNA Extraction and PCR
Total DNAs were extracted from cultured colonies using the bead beater-phenol extraction method [14], and then used as templates for PCR. To study genetic variations of the M. massiliense related strains, two independent target genes - hsp65 and rpoB - were amplified. To amplify partial gene (644 bp), the hsp65 PCRs were applied to a total of 109 clinical isolates. A set of primers HspF3 (forward; 5′-ATC GCC AAG GAG ATC GAG CT-3′) and HspR4 (reverse; 5′-AAG GTG CCG CGG ATC TTG TT-3′) was used [14]. To amplify partial gene (1092 bp), the rpoB PCRs (with a slight modification of previous methods) [12], were applied to 65 clinical isolates, which had already been identified into M. massiliense strains by the hsp65 sequencing protocol. Primer sets of MycoF2 (forward; 5′-ATC GCC GAC GGT CCC TGC-3′) and MycoR2 (reverse; 5′-GAA CCG CTG GCC ACC GAA CT-3′) were used for PCR. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejeon, South Korea) containing one unit of Taq DNA polymerase, 250 µM of deoxynucleotide triphosphate, 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 1.5 mM MgCl2, and gel loading dye. The final volume was adjusted to 20 µl with distilled water, and the reaction mixture was then amplified as previously described [14], [46] using a model 9700 Thermocycler (Perkin-Elmer Cetus).
Hinf I PCR Restriction Fragment Length Polymorphism Analysis (PRA) Targeting 644 bp hsp65 gene
A PRA algorithm using the Hinf I enzyme to differentiate M. massiliense Type II from M. massiliense Type I, as well as other M. massiliense related species (M. abscessus, M. bolletii, M. chelonae, and M. fortuitum) was designed by using MapDraw (version 3.14; DNASTAR, Madison, Wis.). To verify the authenticity of the PRA algorithm for Type II separation, it was applied to six Type strains (M. abscessus ATCC 19977T, M. bolletii CIP 108541T, M. chelonae, M. fortuitum, M. massiliense CIP 108297T) and 65 M. massiliense clinical isolates. All of the samples were blind tested. Briefly, ten microliters of 644-bp hsp65 PCR products, 2 U of Hinf I restriction enzyme, and a restriction buffer were transferred into a microcentrifuge tube, and distilled water was added to a final volume of 20 µl. Digestion was performed for 2 h at 37°C in a water bath. After digestion, the mixtures were electrophoresed in 2% agarose gel with 100 bp ladder DNA marker.
Nucleotide Sequencing
PCR products were purified using Qiaex II gel extraction kits (Qiagen, Hilden, Germany) and then sequenced directly using forward and reverse primers with an Applied Biosystems automated sequencer (model 377) and BigDye Terminator cycle sequencing kits (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). Both strands were sequenced as a crosscheck.
Sequence Analysis
Determined partial rpoB (711-bp), and hsp65 (603-bp) sequences were aligned using the ClustalW algorithm in MEGA4 [47]. A phylogenetic tree based on the rpoB and hsp65 gene sequences was constructed by the neighbor-joining [48] and maximum-parsimony [49] methods within the MEGA 4 program [47]. The constructed neighbor-joining tree was evaluated by bootstrap value calculated from 1,000 replicates [13]. Separately, the 23 S rRNA gene sequences of 65 M. massiliense clinical isolates were analyzed to observe any point mutation at the adenine at position 2058 (A2058) or at A2059 in the peptidyltransferase region of the 23 S rRNA gene [50].
Nucleotide Sequence Accession Numbers
Determined rpoB and hsp65 sequences that were different from reference strains were deposited in GenBank under accession no. JQ081974 to JQ082103.
Statistical Analyses
Results were expressed as percentages. The differences between categorical variables were analyzed using the Chi-square test. For continuous variables the Student’s t-test was used when the data showed a normal distribution, or the Mann-Whitney U test was used when the data was not normally distributed. A p-value of <0.05 (two-tailed) was considered to be statistically significant.
Supporting Information
Mycolic acid profiles of M. massiliense strains. Comparison of mycolic acid profiles of (A) M. massiliense CIP 108297T, (B) 50375 (Type I), (C) 51843 (Type I), (D) 52444 (Type I), (E) 50594 (Type II), (F) 51048 (Type II), and (G) 52188 (Type II) obtained from HPLC analysis. The relative retention time is indicated for each peak. LMW, Low-molecular-weight standard; HMW, High-molecular-weight standard. The asterisks represent a unique peak in Type II HPLC profiles compared with M. massiliense CIP 108297T and Type I strains.
(TIF)
The rpoB and hsp65 genotypes, Hinf I PRA patterns and colony morphology of 65 M. massiliense clinical isolates.
(XLSX)
Details of cultural and biochemical characteristics. Cultural and biochemical characteristics of M. abscessus ATCC 19977T, M. bolletii CIP 108541T, M. massiliense CIP 108297T, Type I strains (50375, 51843, 52352, and 52444) and Type II strains (50594, 51048, 52188, and 52265). Details of biochemical and cultural results are shown in text. ++, good growth, +, positive/growth; −, negative/no growth; ±, variable. 1, M. abscessus ATCC 19977T; 2, M. bolletii CIP 108541T, 3, M. massiliense CIP 108297T; 4, 50375 (Type I); 5, 51843 (Type I); 6, 52352 (Type 1); 7, 52444 (Type I); 8, 50594 (Type II); 9, 51048 (Type II); 10, 52188 (Type II); 11, 52265 (Type II).
(DOCX)
Details of the antibiotic susceptibility profiles. Comparison of the antibiotic susceptibility test results among M. abscessus ATCC 19977T, M. bolletii CIP 108541T, M. massiliense CIP 108297T, Type I strains (50375, 51843, 52352, and 52444) and Type II strains (50594, 51048, 52188, and 52265). ‡ Ami, Amikacin; Cef, Cefoxitin; Cip, Ciprofloxacin; Cla, Clarithromycin; Dox, Doxycycline; Imi, Imipenem; Mox, Moxifloxacin; Rif, Rifampin; Sul, Sulfamethoxazole; Tob, Tobramycin; Emb, Ethambutol.
(DOCX)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grant A101205 from the Korean Healthcare Technology R&D project, Ministry for Health, Welfare & Family Affairs, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Han XY, De I, Jacobson KL. Rapidly growing mycobacteria: clinical and microbiologic studies of 115 cases. Am J Clin Pathol. 2007;128:612–621. doi: 10.1309/1KB2GKYT1BUEYLB5. [DOI] [PubMed] [Google Scholar]
- 2.Olivier KN, Weber DJ, Wallace RJ, Faiz AR, Lee JH, et al. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 3.CDC . Atlanta: Centers for Disease Control and Prevention; 1999. Nontuberculous mycobacteria reported to the Public Health Laboratory Information System by state public health laboratories in the United States, 1993–1996. pp. 1–51. [Google Scholar]
- 4.Petrini B. Non-tuberculous mycobacterial infections. Scand J Infect Dis. 2006;38:246–255. doi: 10.1080/00365540500444652. [DOI] [PubMed] [Google Scholar]
- 5.Pulcini C, Vandenbussche E, Podglajen I, Sougakoff W, Truffot-Pernot C, et al. Hip prosthesis infection due to Mycobacterium wolinskyi. J Clin Microbiol. 2006;44:3463–3464. doi: 10.1128/JCM.02685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006;129:341–348. doi: 10.1378/chest.129.2.341. [DOI] [PubMed] [Google Scholar]
- 7.Choi G-E, Jo Y, Shin SJ. Current Understanding of Mycobacterium abscessus Infection. JOURNAL OF BACTERIOLOGY AND VIROLOGY. 2012;42:17–28. [Google Scholar]
- 8.Adekambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006;56:133–143. doi: 10.1099/ijs.0.63969-0. [DOI] [PubMed] [Google Scholar]
- 9.Adekambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, et al. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol. 2004;42:5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HY, Kook Y, Yun YJ, Park CG, Lee NY, et al. Proportions of Mycobacterium massiliense and Mycobacterium bolletii strains among Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J Clin Microbiol. 2008;46:3384–3390. doi: 10.1128/JCM.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011;183:405–410. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 12.Adekambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003;41:5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Kim SH, Shim TS, Kim MN, Bai GH, et al. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int J Syst Evol Micr. 2005;55:1649–1656. doi: 10.1099/ijs.0.63553-0. [DOI] [PubMed] [Google Scholar]
- 15.Soini H, Viljanen MK. Diversity of the 32-kilodalton protein gene may form a basis for species determination of potentially pathogenic mycobacterial species. J Clin Microbiol. 1997;35:769–773. doi: 10.1128/jcm.35.3.769-773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takewaki S, Okuzumi K, Manabe I, Tanimura M, Miyamura K, et al. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol. 1994;44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 17.Kim HY, Yun YJ, Park CG, Lee DH, Cho YK, et al. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J Clin Microbiol. 2007;45:3127–3130. doi: 10.1128/JCM.00608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh WJ, Kwon OJ, Lee NY, Kook YH, Lee HK, et al. First case of disseminated Mycobacterium bolletii infection in a young adult patient. J Clin Microbiol. 2009;47:3362–3366. doi: 10.1128/JCM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripoll F, Deshayes C, Pasek S, Laval F, Beretti JL, et al. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics. 2007;8:114. doi: 10.1186/1471-2164-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmon KE, Pounder JI, Greene JN, Walsh F, Anderson CM, et al. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J Clin Microbiol. 2007;45:1978–1980. doi: 10.1128/JCM.00563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho AY, Kim YS, Kook YH, Kim SO, Back SJ, et al. Identification of Cutaneous Mycobacterium massiliense Infections Associated with Repeated Surgical Procedures. Ann Dermatol. 2010;22:114–118. doi: 10.5021/ad.2010.22.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakanaga K, Hoshino Y, Era Y, Matsumoto K, Kanazawa Y, et al. Multiple cases of cutaneous Mycobacterium massiliense infection in a “hot spa” in Japan. J Clin Microbiol. 2011;49:613–617. doi: 10.1128/JCM.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tortoli E, Gabini R, Galanti I, Mariottini A. Lethal Mycobacterium massiliense sepsis, Italy. Emerg Infect Dis. 2008;14:984–985. doi: 10.3201/eid1406.080194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, et al. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol. 2009;47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim BJ, Jeong J, Lee SH, Kim SR, Yu HK, et al. Mycobacterium koreense sp. nov., a novel slowly growing nonchromogenic species closely related to Mycobacterium triviale. Int J Syst Evol Microbiol. 2011. [DOI] [PubMed]
- 26.Lee H, Lee SA, Lee IK, Yu HK, Park YG, et al. Mycobacterium paraterrae sp. nov. recovered from a clinical specimen: novel chromogenic slow growing mycobacteria related to Mycobacterium terrae complex. Microbiol Immunol. 2010;54:46–53. doi: 10.1111/j.1348-0421.2009.00184.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee HK, Lee SA, Lee IK, Yu HK, Park YG, et al. Mycobacterium paraseoulense sp. nov., a slowly growing, scotochromogenic species related genetically to Mycobacterium seoulense. Int J Syst Evol Microbiol. 2010;60:439–443. doi: 10.1099/ijs.0.012054-0. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, Lee H, Lee MK, Lee SA, Shim TS, et al. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J Clin Microbiol. 2010;48:3073–3080. doi: 10.1128/JCM.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fregnan GB, Smith DW. Journal of Bacteriology 83: 819–&; 1962. Description of Various Colony Forms of Mycobacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard ST, Rhoades E, Recht J, Pang X, Alsup A, et al. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology. 2006;152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 31.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile JF, et al. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun. 2007;75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhoades ER, Archambault AS, Greendyke R, Hsu FF, Streeter C, et al. Mycobacterium abscessus Glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-alpha by preventing interaction with TLR2. J Immunol. 2009;183:1997–2007. doi: 10.4049/jimmunol.0802181. [DOI] [PubMed] [Google Scholar]
- 33.Devallois A, Goh KS, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Park HJ, Cho SN, Bai GH, Kim SJ. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000;38:2966–2971. doi: 10.1128/jcm.38.8.2966-2971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Kim SH, Shim TS, Kim MN, Bai GH, et al. PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp Heat Shock Protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J Microbiol Methods. 2005;62:199–209. doi: 10.1016/j.mimet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Blauwendraat C, Dixon GL, Hartley JC, Foweraker J, Harris KA. Eur J Clin Microbiol Infect Dis; 2012. The use of a two-gene sequencing approach to accurately distinguish between the species within the Mycobacterium abscessus complex and Mycobacterium chelonae. [DOI] [PubMed] [Google Scholar]
- 37.Choi GE, Chang CL, Whang J, Kim HJ, Kwon OJ, et al. Efficient differentiation of Mycobacterium abscessus complex isolates to the species level by a novel PCR-based variable-number tandem-repeat assay. J Clin Microbiol. 2011;49:1107–1109. doi: 10.1128/JCM.02318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris KA, Kenna DT, Blauwendraat C, Hartley JC, Turton JF, et al. Molecular Fingerprinting of Mycobacterium abscessus Strains in a Cohort of Pediatric Cystic Fibrosis Patients. J Clin Microbiol. 2012;50:1758–1761. doi: 10.1128/JCM.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim BJ, Choi BS, Lim JS, Choi IY, Lee JH, et al. Complete Genome Sequence of Mycobacterium intracellulare Clinical Strain MOTT-02. J Bacteriol. 2012;194:2771. doi: 10.1128/JB.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim BJ, Choi BS, Lim JS, Choi IY, Lee JH, et al. Complete Genome Sequence of Mycobacterium intracellulare Strain ATCC 13950T. J Bacteriol. 2012;194:2750. doi: 10.1128/JB.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim B-J, Choi B-S, Lim J-S, Choi I-Y, Kook Y-H, et al. Journal of bacteriology; 2012. Complete genome sequence of Mycobacterium intracelllulare clinical strain MOTT-64, belonging to the INT1 genotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, Ryoo S. Exploitation of Culture Medium for Mycobacterium tuberculosis. JOURNAL OF BACTERIOLOGY AND VIROLOGY. 2011;41:237–244. [Google Scholar]
- 43.Kent PT, Kubica GP. Atlanta: Centers for Disease Control and Prevention; 1985. Public health mycobacteriology: a guide for the level III laboratory. [Google Scholar]
- 44.Butler WR, Thibert L, Kilburn JO. Identification of Mycobacterium-Avium Complex Strains and Some Similar Species by High-Performance Liquid-Chromatography. J Clin Microbiol. 1992;30:2698–2704. doi: 10.1128/jcm.30.10.2698-2704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez E, Constant P, Lemassu A, Laval F, Daffe M, et al. Characterization of three glycosyltransferases involved in the biosynthesis of the phenolic glycolipid antigens from the Mycobacterium tuberculosis complex. J Biol Chem. 2004;279:42574–42583. doi: 10.1074/jbc.M406246200. [DOI] [PubMed] [Google Scholar]
- 46.Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 49.Fitch WM. Toward Defining Course of Evolution - Minimum Change for a Specific Tree Topology. Syst Zool. 1971;20:406. [Google Scholar]
- 50.Meier A, Kirschner P, Springer B, Steingrube VA, Brown BA, et al. Identification of mutations in 23 S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mycolic acid profiles of M. massiliense strains. Comparison of mycolic acid profiles of (A) M. massiliense CIP 108297T, (B) 50375 (Type I), (C) 51843 (Type I), (D) 52444 (Type I), (E) 50594 (Type II), (F) 51048 (Type II), and (G) 52188 (Type II) obtained from HPLC analysis. The relative retention time is indicated for each peak. LMW, Low-molecular-weight standard; HMW, High-molecular-weight standard. The asterisks represent a unique peak in Type II HPLC profiles compared with M. massiliense CIP 108297T and Type I strains.
(TIF)
The rpoB and hsp65 genotypes, Hinf I PRA patterns and colony morphology of 65 M. massiliense clinical isolates.
(XLSX)
Details of cultural and biochemical characteristics. Cultural and biochemical characteristics of M. abscessus ATCC 19977T, M. bolletii CIP 108541T, M. massiliense CIP 108297T, Type I strains (50375, 51843, 52352, and 52444) and Type II strains (50594, 51048, 52188, and 52265). Details of biochemical and cultural results are shown in text. ++, good growth, +, positive/growth; −, negative/no growth; ±, variable. 1, M. abscessus ATCC 19977T; 2, M. bolletii CIP 108541T, 3, M. massiliense CIP 108297T; 4, 50375 (Type I); 5, 51843 (Type I); 6, 52352 (Type 1); 7, 52444 (Type I); 8, 50594 (Type II); 9, 51048 (Type II); 10, 52188 (Type II); 11, 52265 (Type II).
(DOCX)
Details of the antibiotic susceptibility profiles. Comparison of the antibiotic susceptibility test results among M. abscessus ATCC 19977T, M. bolletii CIP 108541T, M. massiliense CIP 108297T, Type I strains (50375, 51843, 52352, and 52444) and Type II strains (50594, 51048, 52188, and 52265). ‡ Ami, Amikacin; Cef, Cefoxitin; Cip, Ciprofloxacin; Cla, Clarithromycin; Dox, Doxycycline; Imi, Imipenem; Mox, Moxifloxacin; Rif, Rifampin; Sul, Sulfamethoxazole; Tob, Tobramycin; Emb, Ethambutol.
(DOCX)