Abstract
Identifying new, effective biomarkers for diseases is proving to be a challenging problem. We have proposed that antibodies may offer a solution to this problem. The physical features and abundance of antibodies make them ideal biomarkers. Additionally, antibodies are often elicited early in the ontogeny of different chronic and infectious diseases. We previously reported that antibodies from patients with infectious disease and separately those with Alzheimer's disease display a characteristic and reproducible “immunosignature” on a microarray of 10,000 random sequence peptides. Here we investigate the physical and chemical parameters underlying how immunosignaturing works. We first show that a variety of monoclonal and polyclonal antibodies raised against different classes of antigens produce distinct profiles on this microarray and the relative affinities are determined. A proposal for how antibodies bind the random sequences is tested. Sera from vaccinated mice and people suffering from a fugal infection are individually assayed to determine the complexity of signals that can be distinguished. Based on these results, we propose that this simple, general and inexpensive system could be optimized to generate a new class of antibody biomarkers for a wide variety of diseases.
The effort to make medicine preventative should include the development of systems to detect disease before the appearance of major symptoms. The value of early detection is widely accepted and has been the spur to develop new biomarkers of disease that enable earlier diagnosis and treatment. Over 100,000 biomarkers have been reported in the literature to date 1 yet there are only 43 approved by the FDA 2 including 19 genomic markers 3. This low return on investment for biomarker discovery suggests that new approaches are needed. Here we characterize a method that has been proposed as an alternative strategy for biomarker discovery.
Discovery of biomarkers for early diagnosis of disease poses exceptional demands. For example, in the case of cancer, in order to detect the small number of cells in an early tumor one has to overcome the blood dilution problem. For example, if 106 initiating cancer cells release 1000 molecules each of a biomarker into five liters of blood at steady state, the concentration of this biomarker would only be 3 × 10−14 m. Clearly, it would be an advantage if the response to the biomarker could be amplified. Antibodies are ideal in this sense. An activated B cell produces 5000–20,000 antibodies per minute 4, 5 and the cell itself replicates every ∼70 h 6 with a lifespan of up to 4 ½ months 7, 8 leading to ∼1011 amplification of a specific signal in 1 week. Unpurified antibodies are stable in blood, unlike other biomarkers, opening up the possibility of testing historical samples 9.
There are three key issues relative to using antibodies as biomarkers of early disease. Do they respond to diseases other than infections? Do they respond early in the course of disease? Can these antibodies be identified with a simple and inexpensive detection system?
There are reports in diabetes 10, arthritis 11, and cancer 12 that the humoral response is activated specifically and early in these chronic diseases. A number of autoantibodies have been identified that appear months or years before the disease is first diagnosed 13–15. In the case of Type I diabetes, antibodies against GAD, IA2 and insulin are found in various combinations well before the onset of clinical disease 16. In patients with paraneoplastic syndrome, specific neurological symptoms appear years before a cancer is detected 17–19. The immune response to the nascent tumor reacts with neurons to elicit neurological symptoms 20 that correlate with future tumor appearance. These examples for cancer, diabetes and arthritis also address the second issue: is there an immune response among different individuals that appears early in patients with the same disease? The fact that the same autoantigens, or symptoms in the case of paraneoplastic syndrome, commonly occur indicates that antibodies might also be consistent across patients.
The third issue, and the one we address here, is how to detect the informative antibodies in an efficient and simple way. Most antibody biomarkers were the product of arduous research. Protein microarrays have facilitated this process 21 by immobilizing most of the proteins from a pathogen or human onto a glass slide, but these arrays are expensive, exclude non-transcribed antigens, and are pathogen or auto-antibody specific. The ProtoArrayTM v5 of Invitrogen currently has ∼9000 unique human proteins; these can detect autoantigens associated with a specific disease. However, only autoantigens can be discovered and the cost impedes epidemiology-sized studies. A more complicated approach has been to biochemically fractionated cellular proteins, spot and then react these fractions with patient sera 22. Although this method does use authentic material, it is limited by having no control over the relative amounts of proteins spotted, and it requires cells from the case subjects' own tissue.
Screening for antibody reactivity to random peptides has been generally successful when using phage or mRNA display of random 8–12 amino acid sequences. Pasqualini and Ruosiahti 23 panned a phage library against sera from cancer and healthy subjects to find phage that were preferentially bound by the cancer associated antibodies. This method is unbiased as to the nature of the antigen, and the antibody can eventually be captured (if arduously). Given that random sequence peptides can yield mimotopes of almost any type of antigen 24, any disease-associated antigen could theoretically be detected. However promising this method appeared, to date this approach has not produced disease biomarkers for a number of reasons. A serious limitation is that the recurrent panning of the phage is subject to many influences besides just binding of the antibody, it does not lend itself to large numbers of samples, and there is no simple way to measure intermediate or low binding.
In order to discover and display relevant antibodies contained in the 1010 antibody complexity in humans, we explored a technology (PCT/US2010/039269, “Compound Arrays for Sample Profiling”) for antibody biomarker discovery that combines simple, rapid and inexpensive assays from microarrays with the enhanced breadth of the ligand repertoire found in phage-based systems. We created microarrays with only 10,000 random sequence peptides but chose a relatively long length (17 amino acids + 3 residue linker) to allow each peptide to encompass much more complexity than typical epitope peptides. The random sequences allow an unbiased display of antibody binding; the length provides many possible epitope positions per peptide and (potentially) allows for some structural complexity. The array format allows the assay to be run without the biological complications of phage display and high-speed piezo printing onto commercially produced substrate allows thousands of microarrays to be consistently produced each month. We recently demonstrated the potential utility of these arrays by immunosignaturing vaccines, infections, and Alzheimer's disease 25,26. In order to utilize this technology as a clinical diagnostic, we must first characterize the physical and chemical properties of antibody binding to these peptide microarrays.
MATERIALS AND METHODS
Peptide Synthesis and Microarray Construction
The peptide microarray consists of 10,000 20-residue peptides of 17 random sequence amino acids, with a fixed C-terminal linker of Gly-Ser-Cys-COOH, synthesized by Alta Biosciences, Birmingham, UK. The synthesis scale was 2.5 μm (∼1 mg total at 75% purity) with 2% of the peptides tested at random by mass spectrometry. Dry peptide was brought up in 100% dimethyl formamide until dissolved, then diluted 1:1 with purified water pH 5.5 + 0.5 × phosphate-buffered saline (PBS) pH 7.2 to a final concentration of ∼1 mg/ml for printing. Gold Seal glass microscope slides were obtained from Fisher (Fair Lawn, NJ, cat# 3010) and treated with aminosilane, activated with sulfo-SMCC (Pierce Biotechnology, Rockford, IL) creating a maleimide-activated surface designed to react with the peptide's terminal cysteine. Spotting was initially done with an Arrayit Nanoprint 60 using 48 Telechem SMP2 style 946 titanium pins that deposit ∼500 pL of peptide per spot. The spotting environment is 25 °C, 55% humidity. Fluorescent fiducials are applied asymmetrically using Alexa-647 and Alexa-555-labeled bulk peptides. Slides are stored under argon at 4 °C until used. Currently arrays are piezo-printed at Applied Microarrays (Tempe, AZ). Quality control consists of imaging the arrays by laser scanner (Perkin-Elmer ProScanArray HT, Perkin Elmer, Wellesley, MA) at 647 nm to image the spot morphology. Print batches have <30% CV (coefficient of variance) average across all peptides. Data extraction uses GenePix Pro 6.0 (Molecular Devices Inc., Sunnyvale, CA), data analysis uses R and GeneSpring 7.2 (Agilent, Santa Clara, CA).
Binding Sample to Microarrays
Slides were blocked with 1 × PBS, 3% bovine serum albumin, 0.05% Tween 20, 0.014% β-mercaptohexanol for 1 h at 25 °C in a darkened humidified chamber, then sera or antibodies were diluted in 3% bovine serum albumin, 1 × PBS, 0.05% Tween 20 pH 7.2 to a 10 nm concentration for monoclonal antibodies or a 1:500 dilution for mouse and human sera, and allowed to bind for 1 h at 37 °C at 20 RPM rotation to the microarray surface. Later slides (Figs. 8 and 9) were processed using the Tecan HS4800 Pro Hybridization Station (Tecan AG, Männedorf, Switzerland) using custom programs that mirrored these manual steps. Slides were washed 3 × 5′ with 1 × tris-buffered saline, 0.05 Tween20 pH 7.2 followed by 3 washes with distilled water. The slides were dried by centrifugation and images were recorded using the Agilent ‘C’ Scanner at 100% laser power (SHG-YAG laser@532 nm or HeNe laser@633 nm), 70% PMT. For PepPerPrint microarrays (Fig. 5), incubations, scans and alignments were performed exactly as for the immunosignaturing microarrays with the exception that human serum was added at 1:50 concentration rather than 1:500, per manufacturer's recommendation.
Fig. 8.
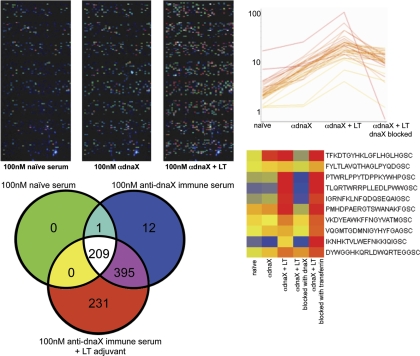
Immunological Testing of Random Peptide Binding. Nine BALB/c mice were genetically immunized with the coding region to dnaX, a DNA polymerase III subunit found in Chlamydophila abortus S26/329,30. 60 days following immunization immune sera was run on the immunosignature microarrays. Top left: images of microarrays as different immune serum is added. As the adjuvant and then the antigen are examined, the total measurable signal on the array increased even though the total amount of IgG remained measurably constant. The Venn diagram immediately below the array images indicates the overlap in the peptides that were 4 SD above background for each selection. As the immune response increased, the number of detectable peptides increased. Far right top: line graph shows those peptides that were significantly different between naïve and dnaX-immunized mice at p < 3.31 × 10−9. Recombinant dnaX protein produced in E. coli and an irrelevant human protein, Transferrin, were used to adsorb the immune sera from the dnaX + LT-vaccinated mice. Only the dnaX protein could adsorb the signal.
Fig. 9.
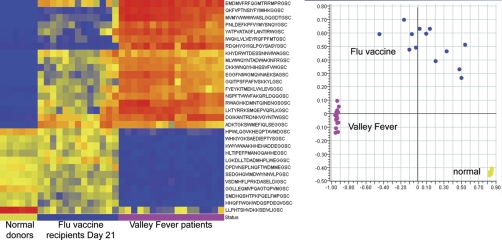
Infectious diseases tested on the immunosignature microarray. Left: Heatmap of 30 ANOVA-selected peptides (p < 1. 6 × 10−6) classify six healthy individuals, 13 day—21 flu vaccine recipients, and 17 Valley Fever patients with 0% misclassification rate using Linear Discriminate Analysis and leave-one-out cross validation. Right: Scatterplot of the first two principal components of the same 30 peptides shows the relative differences between disease states.
Fig. 5.
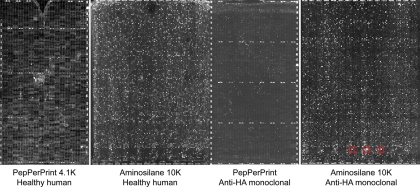
Comparison of PepPerPrint epitope microarray with immunosignaturing microarray. PepPerPrint GmbH (Heidelberg, Germany) manufactures epitope microarrays using in situ synthesis of peptides onto a proprietary surface. 4128 cysteine-free random-sequence 11mers + aspartic acid + PEG linker were assembled on the slide surface. Each slide is ringed by HA and FLAG control peptides. Above left: Image shows a portion of a PepPerPrint microarray on which was run a direct-labeled anti-HA monoclonal at 5 nm concentration plus 1:50 dilution of pooled healthy human serum, detected with a mouse anti-human secondary antibody. Middle-left: Image shows immunosignaturing microarray with 1:50 dilution of pooled healthy human serum. Healthy serum produced no discernible signal on the PepPerPrint microarrays, but numerous signals on the immunosignaturing microarray. Middle-right: PepPerPrint microarray on which was run only 5 nm anti-HA monoclonal. Far right: Immunosignaturing microarray on which was run 5 nm anti-HA monoclonal. Red boxes indicate HA epitope peptides.
Antibody Detection
Each antibody or IgG fraction was detected by biotinylated secondary antibody followed by streptavidin-conjugated Alexafluor 555 or 647 (see Table I for antibodies used). Secondary antibodies were incubated at a concentration of 5 nm, streptavidin at a concentration of 1 nm. Single-color experiments were performed exclusively, but dye choice depended on availability. Detection wavelength did not affect resolution, dynamic range, or reproducibility. Direct-labeled antibodies were created for the Fc competition experiment (Fig. 6) using Lightning-Link antibody labeling kit and Cy5 dye according to the manufacturer's protocol (Innovabioscience, Cambridge. UK).
Table I. List of antibodies used in Fig. 2, their epitope (if known), the isotype and source.
| Protein | Antibody | Epitope | Isotype | Company | Secondary |
|---|---|---|---|---|---|
| αTubulin | DM1A | AALEKD (387–392) | IgG1 kappa | Labvision | Invitrogen HL |
| p53 (Ab1) | PAb240 | RHSVV (212–217) | G1 | LabVision | Invitrogen HL |
| p53 (Ab8) | DO-7, BP53–12 | DLWKLL (21–26) | IgG2b, IgG2a | LabVision | Invitrogen HL |
| Interleukin2 | LNKB-2 | KPLEEVLNL (64–72) | IgG1 | Santa Cruz Bio | Invitrogen HL |
| MHC class I | MHC | 3D | IgG1 | MBL Int'l | Bethyl |
| H1N1coat protein | H1N1 1, 2 and 3 | Unknown | IgG1 | US Bio | Invitrogen HL |
| Transferrin | HTF-14 | N-term of transferrin | IgG1 | Abcam | Invitrogen HL |
| Transferrin | 11D3 | Unknown | IgG1 | Abcam | Invitrogen HL |
| Transferrin | 1C10 | Unknown | IgG1 | Abcam | Invitrogen HL |
| 2E4 | polyreactive | IgM | A. Notkins | Novus | |
| B78 | autoantibody | GAD65 protein | IgG1 | A. Notkins | Novus |
| B96 | autoantibody | GAD65 protein | IgG1 | A. Notkins | Novus |
| Herceptin | HER2-NEU | Unknown | IgG1 | Genentech | Novus |
| 8 pooled | 1C10, endorphin, IL2, TP, DM1A, p53AB1, p53Ab8, LNKB2 | ||||
Fig. 6.
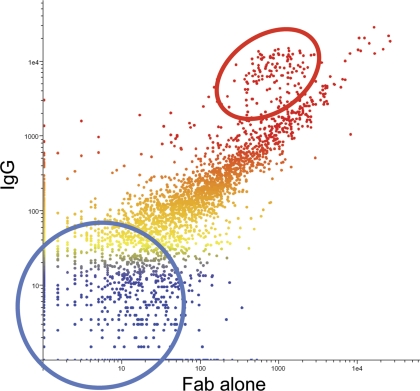
Fab fragment binds similarly to intact IgG. The Fab Fragment and the intact Ig of the same monoclonal (anti-HLA-G clone 87G) were used to probe the 10K immunosignature microarray. The signal intensity of the Fab is plotted on the x-axis against the signal of the intact Ig on the y-axis; peptides are colored by intensity with blue indicating low intensity, red indicating high intensity. The scatterplot indicates that most peptides exhibit similar binding to the monovalent and the bivalent forms of the antibody. The red circle highlights peptides with the highest pI, the blue circle contains peptides with the lowest pI values. The differences between the Ig and Fab appear to be driven by the charge of the peptide at pH 7.2.
Biochemistry
SPR methods of peptide/antibody affinity were conducted as in (27) with the following exceptions: rather than four different concentrations of peptide spotted onto the SPR chip, we used 1.0 μm only, and rather than using 1 μm of Gal80 protein we used 5 μm of the specific antibody tested. Calorimetry was conducted using an N-ITC III (Calorimetry Sciences Corporation, Linden, UT). Injections of 10 μl of 1 mg/ml peptide were added to 50 μm solution of antibody solution (20 mm PBS pH 6.8) in a total volume of 1.5 ml with stirring at 250 rpm.
Antibody Blocking with Recombinant Protein
Blocking for the dnaX experiment was done by pre-incubating anti-dnaX antisera with His-tagged recombinant dnaX protein for two hours at 25 °C. The immune complexes were removed from the solution by incubation of antibody-bound dnaX with nickel Sepharose (Amersham Biosciences, Piscataway, NJ) for two hours followed by centrifugation. The supernatant was quantitated for protein concentration and processed for binding to the microarrays. Negative control was anti-dnaX antisera incubated with an irrelevant His-tagged protein (a fusion of F1-V from Yersinia pestis).
Quality Control
Our microarray manufacturing process requires that three slides per 136-slide batch are quality checked using pooled human naïve serum. We examine batch-to-batch correlations across previously manufactured batches. Typical array-to-array correlations are >0.93 and batch-to-batch correlations >0.89 (see www.peptidearraycore.com).
Statistical Analysis
Statistical analysis of microarray data was performed using GeneSpring 7.3.1 and R by first importing image-processed data from GenePix Pro 6.0 as gpr text files. Preprocessing of raw data was done by first median normalizing values and then log10 transformation. Student's t test or 1-way ANOVA with 5% Family Wise Error Rate multiple testing correction were performed. The minimum detectable fold-change at 95%ile across three technical replicates averaged 1.3-fold 28.
Human Subjects
Human subjects were consented and deidentified according to IRB Protocol# 0905004024, Arizona State University. Blood samples of 5 ml were taken as noted in the manuscript.
Animal Care
Use of animals was approved by the Animal Care and Ethics Committee of Arizona State University (IACUC#10–1099R). All animals were anesthetized using isofluorane or a mixture of ketamine, xylazine and acepromazine. Animals were sacrificed with CO2.
Antibodies, Immunizations, Infections
For the dnaX vaccine (DNA polymerase III subunit gamma/tau [Chlamydophila abortus S26/3]; CAH63776), 9 CBA/J mice were immunized via genetic immunization29,30 with a plasmid construct encoding the open reading frame for dnaX with and without a genetic adjuvant of heat-labile enterotoxin (LTA/LTB). Immune serum was collected 60 days after a prime and two genetic boosts. For comparing naïve to PR8 infection in mice using the NSB slides, we followed the protocol in 25 exactly.
RESULTS
Our basic premise is that the antibody profile from an individual reflects their health status. If this profile can be displayed on a sufficiently complex array, the particular responses to chronic diseases will be apparent. We manufactured microarrays onto which 10,000 random-sequence 20-mer peptides were printed. Each peptide is (from NH3 to COOH termini) 17 residues of any amino acid except cysteine, followed by GSC as the linker. The GS amino acids offer rotational freedom and the C-terminal cysteine was used to attach the peptide to the surface through a maleimide linkage (Fig. 1, right) onto activated aminosilane slides, purchased from Schott, Inc. (Mainz, Germany). Because the peptides have no relationship to any natural sequence31, the same array can be used to profile any disease, any species without synthesizing a new library of peptides. The sample is diluted, applied to the array and allowed to bind. The array is washed and detected with a fluorescently labeled secondary antibody to the appropriate primary antibody isotype. The array is washed, dried, and scanned using a conventional microarray scanner. Fig. 1 left shows the image of a typical slide: on the left, a naïve individual and right, a day-21 post-seasonal flu vaccine recipient. The insets show peptides that bind differentially.
Fig. 1.
Image of the peptide arrays. The microarray is created in a 2-up format, with 10,000 peptides on top and bottom of each slide. In this false-color image, human naïve serum was applied to top the microarray (left), day 21 post-influenza vaccine serum was applied to the bottom (right). The yellow boxes in the small images indicate peptides that show differential binding. Spots are 120 μm in diameter with intensity values ranging from ∼100 to 65,000 relative fluorescence units. Correlation coefficients across technical replicates are typically 0.95 to 0.99. The attachment chemistry is shown on the right with an example peptide attached to the slide through the cysteine to a maleimide linker.
Monoclonal Antibody Profiles
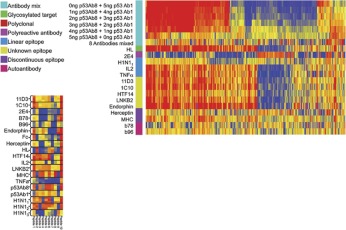
We first asked whether well-characterized antibodies produce discernible profiles on the array and whether those profiles were unique. It has been reported in the literature that monoclonal antibodies do bind to random peptide sequences31, 32, however we wished to systematically validate the underlying principals behind this observation. In Fig. 2 the relative binding of antibodies to a subset of the peptides on the array is portrayed in a heatmap, where blue is low binding and red represents high binding. 272 peptides were selected by ANOVA with a 5% Family Wise Error Rate at p < 1 × 10−12, representing peptides that were consistently different across the antibodies listed in the figure legend. Peptides and antibodies are arranged using hierarchical clustering with Euclidean distance as the measure of difference (GeneSpring 7.2.1, Agilent Technologies, Santa Clara, CA). A wide variety of commercial antibodies were used in this experiment including those to phosphorylated or glycosylated proteins, against both conformational and linear epitopes. The important interpretation of this heatmap is that monoclonal antibodies have reproducible, unique signatures of binding to random peptides and the eliciting antigen need not be linear or post-translationally unmodified. Linear Discriminate Analysis using these 272 peptides produced 0% cross-validation error suggesting that this method could be used to classify antibodies. This was an important finding and was critical in designing and understanding subsequent experiments. If this finding could be applied to complex mixtures of antibodies, perhaps one signature could be discernible from another when those two antibodies were physically mixed together. We diluted p53Ab1 into p53Ab8 (top of Fig. 2). The signatures shown here suggest that the p53Ab1 signature is discernible at a lower concentration than p53Ab8 but there are still a few peptides specific to p53Ab8 detectable at near equimolar concentrations. The lower heatmap in Fig. 2 shows the technical replicates of the 10 most differential peptides (x-axis) across these antibodies (y-axis), at p < 8.23 × 10−23. As few as 10 different peptides are able to produce a distinct and reproducible signature that distinguishes this group of antibodies from each other; reproducibility is extremely high.
Fig. 2.
Antibodies react with random sequence amino acids. Top: A heatmap represents relative binding of antibodies to 267 ANOVA-selected peptides at p < 1 × 10−12 (x-axis) relative to their associated antibody (y-axis). The colored boxes represent the class of epitope for the particular antibody (left). Blue and red in the heatmap indicate low and high binding respectively. The average of three technical replicates is shown per antibody, except IL2 which had 2. Data is median normalized per array and log10 transformed prior to plotting. Hierarchical clustering is used to group the peptides (x-axis), no grouping was done on the y-axis. Some proteins have more than one monoclonal antibody represented here (11D3, HTF14 and 1C10 are all against human Transferrin and p53Ab1 and p53Ab8 are both against human TP53). In the large heatmap we examined the outcome of mixing two different antibodies against human TP53: Ab1 and Ab8. The top row shows the p53Ab1 signature; the ratio between Ab1 and Ab8 is reversed until the sixth row which is only Ab8. Ab8 possesses a far less apparent signature than Ab1; there are but a few peptides that recognize Ab1 when any Ab8 is present (∼15 peptides to the far right). The next test was whether an eqimolar mixture of eight antibodies (IL2, LNKB2, 11D3, p53A1, H1N1, DM1A, TNFα, and 1C10) would yield a monotonic signature. The “8 Antibodies mixed ” row shows reduced signature complexity, but far from monotonic. The poly-reactive antibody 2E4 has low binding overall (40) but binds almost every peptide on the microarray at some level. The significance of these poly-specific antibodies is being investigated (31,41). Bottom: The small heatmap depicts the three technical replicates per antibody individually plotted using only the 10 most significant peptides at p < 8.23 × 10−23. This heatmap indicates the high reproducibility of the system and the small number of peptides needed to simultaneously discriminate 19 different antibodies with 0% misclassification. Antibodies used: 11D3, HTF14, and 1C10 are against human transferrin; IL2 and LNKB2 are against human Interleukin 2; p53Ab1 and p53Ab8 are against human TP53; b78 and b96 are monoclonal autoantibodies against GAD65; Herceptin is against human HER2/NEU; HL is against a glycosylated target in human cell line HL60; TNFα is against human TNF-alpha; MHC is against the native human MHC1 complex; H1N11, 2, 3 are polyclonals against mouse influenza strain PR8; Endorphin is against human endorphin; 2E4 is a poly-reactive antibody42; Fc is purified constant region from human IgG.
The signals from these microarrays span >3 logs of dynamic range; technical replicates had correlation coefficients from 0.92 to 0.99 with an average CV (coefficient of variation) of 14% and a minimum detectable fold-change of 1.3-fold/3 technical replicates at the 95th percentile. We believe the binding to these arrays is largely driven by the interaction of the variable region of the antibody and the peptides for two reasons. First, the binding pattern for each antibody was different, even those from the same species. Second, when a directly labeled monoclonal antibody (p53Ab1, IL2 and 11D3 were tested) was competed with 10-fold excess Fc protein, there was no effect on the immunosignature.
How do the Antibodies Bind the Array?
It may seem surprising that monoclonal antibodies would bind strongly to noncognate peptides. One would expect that random peptides would have low affinity to a monoclonal. We have measured the solution phase affinity of peptides to antibodies using SPR and calorimetry and generally find that the peptide-antibody affinities are in the range of 10–100 μM, in line with previous reports 24 and sharply contrasted with affinities of 1–10 fM for some natural antibody-antigen pairs. Thus, another affect must be responsible for these apparently strong interactions at the surface of the microarray.
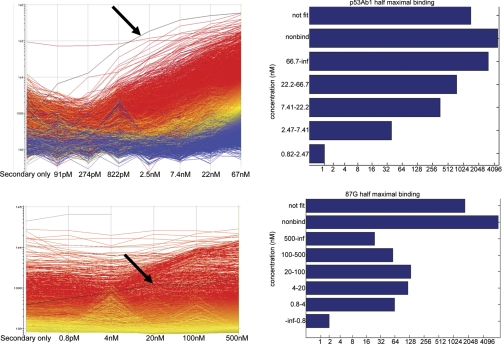
From Fig. 2 we determined that the p53Ab1 antibody appears to have high apparent affinity to many of the peptides on the array, even in the presence of a competing monoclonal. When the p53Ab1 was reacted with the array at various dilutions, one peptide in particular, ETRMIIKLAWETFVDHNGSC (arrow, Fig. 3, top) demonstrated half maximal binding of 6 nm, but it still produced signal detectable an order of magnitude above background at 91 pm. Most of the other peptides do not appear to be approaching saturation, so their apparent affinity is likely above 66 nm, but over 500 peptides had detectable binding at 822 pm. The bottom of Fig. 3 illustrates a similar experiment where we tested a mouse monoclonal anti-HLA-G (clone 87G). One peptide shows significant binding at 0.8 nm. As opposed to the p53 peptide, the HLA peptide had an estimated half maximal binding of 3.3 nm, suggesting that monoclonals may bind random sequence peptides with a dynamic range of several orders of magnitude, and the array allows detection of even relatively weak interactions. Although solution affinity of the random peptides for an antibody may be quite low, the apparent affinity for some peptides on the array is very high, presumably because of surface effects 33.
Fig. 3.
Dynamic Range of Antibody Binding. Top left: Serial dilution of the p53Ab1 monoclonal is shown on the x-axis, relative fluorescence on y-axis. Each line represents a single peptide colored by its signal at 67 nm with red indicating the highest signal, blue the lowest. One peptide (highlighted in black, ETRMIIKLAWETFVDHNGSC) is detected below 100 pm (estimated kDa). Arrays were log10 transformed. Top right: barchart shows the number of peptides that bind 2 stdev above background at each concentration. “Not fit ” contains peptides that could not be fit with an RSQ>0.8. Peptides that did not bind >2 SD above background are in the “not fit ” bin. Bottom left: Dilution series of mouse monoclonal anti-HLA-G (clone 87G). Unlike the p53Ab1, only a few peptides show significant binding above below 1 nm. An example of a peptide that shows significant binding at 0.8 nm is highlighted in black (SREDKDSNDQRKDEQDSGSC). This peptide has an estimated half maximal binding of 3.3 nm, suggesting strong apparent affinity. Bottom right: Histogram of half maximal binding concentration for all 10,000 peptides.
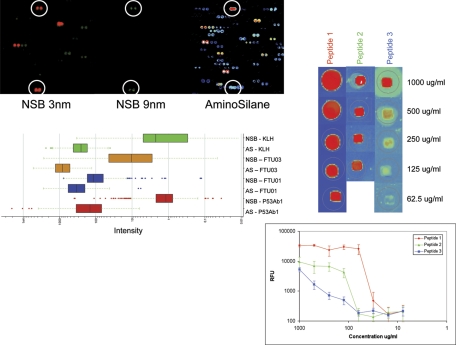
The most obvious surface effect that might contribute to the amplification of signal on the array would be the high local concentration of the peptides in each spot. We explored the effect of peptide concentration by spotting the peptides on a dendrimeric surface (NSB Postech, Seoul, Korea) where the reactive sites are spaced 9 nm apart (NSB27) or 3 nm apart (NSB9) 34. As seen in Fig. 4, the relative binding on the 3 nm surface is on average 30–1000-fold less than on our standard aminosilane surface whereas the 9 nm surface could not support a generally detectable signal. We calculate that in theory the peptides may be as close as ∼1 nm apart on the aminosilane-coated surface based on the density of binding sites of activated aminosilane-coated glass 35. We conclude that the peptide density in the spot is contributing to the high relative affinity. When we examined three naïve versus three PR8 influenza-infected C3H/HEJ mice at day 28, we were unable to statistically distinguish these diseases from each other implying that immunosignaturing needs the high signal strength obtained by close packed peptides in order to obtain sufficient discrimination between disease states. To test this further, we obtained microarray slides from PepPerPrint GmbH (Heidelberg, Germany) with 4128 random-sequence 11mer peptides printed in duplicate (8256 total spots/slide) plus HA and FLAG epitopes that ring the array. These microarrays have been thoroughly described in the literature36 and are extremely high-quality microarrays that have been optimized for epitope analysis. We tested their ability to distinguish human serum (Fig. 5, left) and a monoclonal antibody (right) and found that even at 10 × higher serum concentration than used on the immunosignature microarrays, there was little detectable signal and no way to distinguish disease status. The HA monoclonal bound the cognate epitope only with no detectable cross-reactivity to any other peptide. The spacing of the PEGMA/MMA polymer surface is estimated to be >9 nm (Volker Stadler, pers. comm.).
Fig. 4.
Peptide spacing impacts the binding of antibodies. Peptides were printed on dendrimer slides (NSBPostech, Seoul, Korea), which have either 3 nm (top left, NSB9 Amine Slide) or 9 nm (middle, NSB27 Amine Slide) spacing between peptide attachment points. The same position on standard aminosilane microarrays is shown on the far right. Colors reflect intensity where white and blue are high binding, green is mid-level binding and orange to red indicate low binding. The p53Ab1 antibody was allowed to bind to the same 10,000 peptides as on the standard array, but signals notably decreased by 30–1000-fold on the 3 nm spacing slide and were almost entirely absent from the 9-nm slide. Circles indicate the peptides where signal remained detectable across these three different slides. The barchart immediately below is the signal characteristics of those peptides from each of the slides that bound at >2 SD above background. AS = aminosilane, NSB = 3 nm spacing, no data met the cutoff criteria from the 9-nm NSB slides. P53Ab1 is a mouse monoclonal against human TP53, FTU01, and FTU03 are peptides from the 10 K array which were used to immunize BALB/c mice, and KLH represents mice immunized with only Keyhole Limpet Hemocyanin adjuvant. Right: three peptides from the 10,000 random peptides were selected and resynthesized to produce a small custom microarray, printed at dilutions from 1 mg/ml to 7.8 μg/ml. These small arrays were probed with sera from BALB/c mice immunized with that peptide. Image represents the detectable spots. Directly below this image is a log-log plot of the signals from three technical replicates. The signal drops proportionally with the dilution, but the rate of signal decrease is not constant across all peptides.
The high density of peptides could lead to high effective affinity by two nonexclusive mechanisms—cooperative binding or avidity. Cooperative binding could arise from two peptides binding one antibody through the interactions with each arm simultaneously. We tested whether bivalent binding was a significant binding mechanism by comparing the binding of an intact monoclonal to its Fab fragment. Overall binding was very similar between the Fab fragment of mouse monoclonal anti-HLA-G clone 87G and the intact IgG (Fig. 6). Based on this result we conclude that classic cooperativity plays a negligible role and avidity or antibody rebinding because of the high density of peptides may be sufficient to account for high relative local affinity. Our data suggests that this dominant effect is partly dependent on the exact peptide sequence and to a lesser degree on the peptide charge.
Distinguishing Signatures
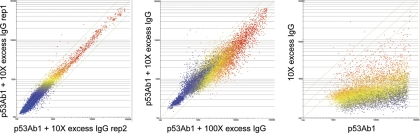
A fundamental question underlying this approach is how the mixture of antibodies in serum may interact, or actively compete, for binding to the random peptides on the array. The observation that each monoclonal antibody we tested binds many different random-sequence peptides implies by simple projection that a collection of antibodies would bind at a generally high level to most peptides on the array. This further suggests that it would be difficult to distinguish a signature of one antibody in a very large collection of different specificities, as in immune serum. If all antibodies recognized the sequence space represented by the random peptides on the array approximately equally, given 1010 specificities in the antibody repertoire, it would seem unlikely that specific antibodies would be recognized at all. An immunosignature of a disease or infection would only be evident if the antibodies produced in response to the disease/infection had higher affinity to the random peptides than the normal immunoglobulins in the sera of healthy people. To test this possibility, we diluted a high affinity commercial monoclonal antibody raised in mice against human TP53 (p53Ab1) into 10x and 100x excess immunolglobulin from healthy volunteers (Fig. 7). The left panel shows the baseline reproducibility (r = 0.97) of two technical replicates. The center panel shows that the p53Ab1 signature is apparent even when diluted by highly complex antibody mixtures, suggesting that antibodies in healthy serum are not directly competing for peptide binding sites of the p53 antibody at the relevant concentrations. The panel on the far right shows the contribution of IgG versus p53 antibody alone. When we mixed eight monoclonal antibodies together (Fig. 2) we saw a large number of high binding peptides; naïve human IgG seems to have reached a state where antibodies with strong affinity to specific random-sequence peptides are at a low concentration. This implies that high affinity antibodies, as would be produced against an infection or chronic disease, would stand out against the background binding of the bulk immunoglobulins in healthy people. This observation is important relative to the immunosignaturing concept and enables the analysis that follows.
Fig. 7.
Dilution experiment. Left panel shows the baseline reproducibility of the 10,000 peptides on the microarray when exposed to a direct-labeled mouse anti human p53Ab1 antibody + 10X excess naïve human IgG. Pearson's correlation coefficient is 0.97 across two technical replicates. The middle panel shows p53Ab1 + 100X excess IgG (x-axis) versus p53Ab1 + 10X excess IgG (y-axis), Pearson's correlation coefficient = 0.92. Far right panel shows p53Ab1 monoclonal (x-axis) versus 10X human IgG alone (y-axis), Pearson's correlation coefficient = 0.27.
Analysis of Immunosignatures in a Model System
In order to examine critical aspects of immunosignaturing, we employed a controlled mouse model. Five mice were bled before and after genetic immunization with a plasmid encoding a protein from Chlamydia abortus, dnaX. We had demonstrated earlier that this protein elicits a robust immune response administered as a gene vaccine 30. In addition, five other mice were immunized with the dnaX plasmid plus a plasmid encoding lethal toxin (LT), a powerful genetic adjuvant 37. The control mice were mock immunized with plasmid alone, not encoding an antigen. The dnaX-immunized mice on day 14 post immunization showed on average 210 peptides that had significantly (p < 3.31 × 10−9) more binding than control mouse serum. A representation of the differences in the arrays is presented in Fig. 8 (top left) and the Venn diagram (lower left). This difference was accentuated when the LT adjuvant was used. We note that total binding to the array increased upon immunization and even further with immunization with adjuvant, even though the total amount of immunoglobulin was held constant by measure of total IgG. The use of the adjuvant increased binding to peptides that were high binders from the dnaX vaccine alone, as well as a set of new peptides that met the significance cutoff over the controls. Our presumption was that most of this additional binding was driven by antibodies against dnaX, with some against LT alone. In order to test this, the serum from the dnaX immunized mice was adsorbed with beads bearing the dnaX protein and then applied to the array. The control was the same serum absorbed with an irrelevant protein (human Transferrin). As can be seen in Fig. 8 (top right), 35 of the 210 dnaX-specific peptides were reduced in intensity by the dnaX adsorption. This indicates that a specific immune signature induced by dnaX was actually to the dnaX antigen. The peptides that were bound by the dnaX serum but not reduced in intensity by the absorbed serum may have been against the LT adjuvant protein itself. Alternatively, the recombinant dnaX protein produced in E. coli might not contain the same epitopes as those made by the mouse cells in vivo. For example, antibodies elicited to post translational modifications in the mouse cell would not be present in the recombinant protein, thus it is possible that most if not all of the signature is to natural dnaX. This result indicates that a specific immune response can be discerned on the random array.
Immunosignatures in Human Serum
An inbred mouse may have a much simpler repertoire of antibodies than a human. It is possible that this immune complexity in humans would hide the signature of a health-affecting event. To test this possibility we compared the immunosignatures of people with confirmed Valley Fever (elicited by Coccidiodes immitis) to the immunosignatures of uninfected control individuals. As seen in Fig. 9, individuals with Valley Fever have peptides (p < 1.6 × 10−6) that are significantly more or less reactive against serum IgG in uninfected controls (“normal donors”) or persons who received a seasonal flu vaccine (“day 21 flu vaccine”). These data indicate that despite the complexity of the immunoglobulins in humans, it is possible to detect a specific immune response to a health disturbance.
Distinguishing a Simulated Multiple Infection
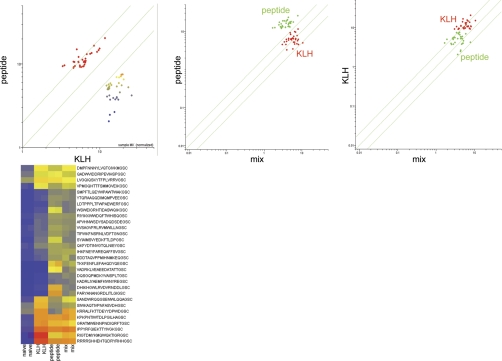
Lastly, we tested the concept raised in the monoclonal experiments, but with a more complex polyclonal response to a vaccine. Could we distinguish two different disease immunosignatures from the same physical sample? This is an important practical consideration since people may have several simultaneous conditions, such as two infections or chronic or autoimmune disease at the same time. Fig. 10 demonstrates the ability to distinguish a mixture of two complex “disease states,” simulated here by two different but separate vaccinations in BALB/c mice, and then a physical pooling of equal volumes from each cohort. A double vaccination was not done due to complex interplay within the host, and the desire to rigorously test only the sensitivity parameters of the microarray without imposing additional variances. Either KLH or a random-sequence peptide (PARYANANGRDLITLGIGSC) were used to vaccinate two different groups of three mice each. The 6-week immune serum for each was incubated on the 10K microarray, and an additional array was tested with a 50:50 mix. The scatterplots in Fig. 10 show 30 peptides from KLH (p < 1.04 × 10−8 versus naïve serum) and 30 peptides from PARY-immunized peptide (p < 8.68 × 10−11 versus naïve). Each scatterplot represents the average of the three mice, one microarray per mouse. The heatmap on the bottom is a visualization of the trend using only the top 30 of the 60 total peptides that by ANOVA discriminate the disease classes (p < 5.24 × 10−18).
Fig. 10.
Immune sera mixing experiment. Different immune sera were mixed together, simulating conditions where patients have simultaneous infections. Top scatterplots: 3 BALB/c mice were each immunized with either KLH (Keyhole Limpet Hemocyanin) or the peptide PARYANANGRDLITLGIGSC. Six-week post immune serum is used. Peptides that discriminated either KLH-immunized serum (30 peptides at p < 1.04 × 10−8 versus naïve serum) or PARYANANGRDLITLGIGSC-immunized serum (30 peptides at p < 8.68 × 10−11) are shown. Far left: The y-axis shows the PARY-immunized sera, the x-axis shows the KLH-immunized sera, peptides are colored by intensity. The two sera were physically mixed and compared with the average for each serum sample sequentially. Center: The PARY peptide immune sera (y-axis, green) was compared with the mixed sera (x-axis). KLH-immunized mice (red) are still distinguishable from PARY-immunized mice. Far right: the KLH-immunized immune sera (y-axis, green) was compared with the mixed sera (x-axis). The PARY-immunized mice (green) are still distinguishable from the KLH-immunized mice. The heatmap on the bottom represents a more conventional visualization of the trend: these 30 peptides (p < 5.24 × 10−18) were used to plot the values from the two naïve, two KLH, two PARY, and two mixed sera microarrays. The arrays can still distinguish the diseases as distinct using hierarchical clustering. Linear Discriminate Analysis with leave one out cross validation yields 0% misclassification.
DISCUSSION
We have examined several basic aspects of the immunosignaturing concept using an array of 10,000 relatively long peptides of random sequence. We first showed that all types of monoclonal antibodies tested produced a distinct pattern of binding to these random peptides. This effect has a number of clinical implications, but rather than base a diagnostic on a phenomenon, we investigated the mechanism of binding, providing evidence that the antibody signal we observe is enhanced due to the high peptide density. A notable finding if this technology were to be used as a diagnostic was that the signature of a high affinity antibody was unchanged in the presence of excess immunoglobulin from healthy people. This implies that it may be possible to discern newly developing, high affinity immune responses - responses that evolve presymptomatically in many cases. To test this we compared the serum of mice immunized with a gene vaccine for dnaX to controls and found that there was a clear immunization signal. This general effect has been demonstrated by Merbl et al. 38 in mice with implanted cancer cells, but not on the scale we have demonstrated; their effect was observed but not characterized. A portion of the dnaX immunization signature we identified was due to antibodies against the dnaX protein, a critical consideration when considering possible sources of noise. We demonstrated that signatures could be detected in human sera, showing that people with Valley Fever infections have immunosignatures distinct from non-infected individuals. Finally, we demonstrated that two different disease signatures could theoretically be distinguished in the same person.
It has previously been demonstrated that antibodies bind to peptides of random amino acid sequence on arrays 31, 32, 38. In general, the feature complexity of arrays reported to date has been less than half the complexity of the peptide microarray used in these studies, in many cases far less. The 17aa variable region of 10,000 peptides could encode all possible 3 mers and 4 mers and 20% of all 5-mers if designed with non-overlapping sequences. We are unable to ascertain whether significant secondary or tertiary structures would exist in these peptides, but in silico prediction suggest that the majority of these peptides are not folded or are only transiently non-linear. Even more importantly, these peptides have no significant regions of similarity to any peptides in on-line sequence databases. A number of 5-mer motifs from our peptides appear in Swissprot and NCBI, but most antibodies bind regions of 6–11aa. It would be improbable that epitope-like recognition would be detectable. It is unsurprising then, that each antibody would have only weak affinity to a particular peptide, as we have found. Yet, we demonstrated that most of the 18 monoclonal antibody tested bind hundreds of peptides on the array with a dynamic range near 3 logs with high reproducibility. When an actual monoclonal epitope is printed on the array, the cognate antibody binds that peptide extremely well (Fig. 5) but it does not bind that peptide alone. Despite the weak solution-phase affinity of antibodies to random sequence peptides, the random peptides on our microarray bind in unique and reproducible patterns to most any immunoglobulin molecule.
A number of different classes of monoclonals and affinity purified polyclonal antibodies were tested on the peptide microarray. This included antibodies raised to sugars, proteins modified with phosphates, conformational epitopes and haptens. All tested antibodies produced a distinct and reproducible pattern of binding. Polyreactive antibodies were unusual in that they tended to bind thousands of peptides at a moderate level, while most other monoclonals bound less than 200 different peptides but with relatively high intensity. We investigated whether protein microarrays would work the same way; we tested 11 different monoclonals on a commercial human protein microarray (Protoarray® from Invitrogen, Carlsbad, CA) and in each case the cognate protein or a near-identical family member had the strongest signal (27). The peptide microarray described here produces unique and discernible signals for antibodies to every antigen tested so far.
It has been reported that aligning random sequence peptides bound by a monoclonal antibody against the protein immunogen could deduce the epitope that elicited the antibody 39. We have also found that this is possible, in some circumstances, even with only 10,000 random sequence peptides. By aligning the peptides bound from the dnaX serum using CLUSTAL, it was possible to map discontinuous (3–4 residues) portions of the immunogenic peptides onto the dnaX protein. This method works best when a fairly small protein is used as the immunizing antigen, or if the search space of possible target proteins can be restricted by size, species, or protein family. A monoclonal antibody may bind mimotopes on the array as well as or better than its cognate sequence (Fig. 3 in 31), which complicates this process. This article shows that, even with substantial noncognate targets, exact epitopes can occasionally be found. However, the absence of direct information about the antigen that raised the immune response is a limitation of the immunosignaturing approach. Although arduous, it is feasible to use the high-binding peptides from the microarray to affinity purify antibodies from patient serum which can then be used to isolate the antigen from tissue lysate. We accomplished this using a simple murine influenza virus model (25). We also demonstrated that the dnaX protein adsorbed the antibodies that made up the dnaX immunosignature (Fig. 8). This same approach could be used to test candidate antigens for any immunosignature. Additional information is contained in isotypes. We have found that certain peptides bind to immunoglobulin isotypes specifically. Typically we use pan-isotype secondary antibodies, but we also examined peptides that bound serum from day 21 immunized dnaX mice and found that the IgG1 and IgG2a ratios differed across a fixed range. The IgG1/IgG2a ratio can be a marker for Th1 and Th2 lymphocytes, and other isotypes can be tested simultaneously (25) with multiple fluorophores. IgM, IgA or IgE have the capacity to be more discerning than IgG in some disease states.
Relative to using these arrays for assaying human serum, we present a preliminary example, but we have found that the approach works well for many types of diseases. We show that people infected with Coccidiodes immitis (Valley Fever) have signatures distinct from healthy controls and from people that received the seasonal flu vaccine (Fig. 9). A biomarker study typically requires thousands of cases and controls for validation, but these results suggest that immunosignatures of infected persons are highly consistent even across varied genetic backgrounds and HLA types, reducing the required cohort considerably. This technology may lend itself to very large scale studies since the arrays are relatively inexpensive to print and can be processed in standard automated systems at ∼$1.50 per sample for reagents.
A second favorable feature is that the technology is amenable to utilizing archived samples because of the stability of antibodies and the small amount (∼1 ul) of serum required. The human samples used in this study were all from frozen samples. We tested fresh serum, fresh plasma, frozen serum and frozen plasma from the same volunteer with almost no discernible differences (correlation coefficient >0.96, Chase et al., in revision). The most high-throughput application might be screening a population with a known disease, selecting the most discriminating peptides, and printing those in a 24-up format (http://arrayit.com/Products/Microarray_Tools/Multi-Well_Microarrays/multi-well_microarrays.html) enabling extremely low per-assay cost.
We have noted an unexpected aspect of the immunosignatures: sera from infected individuals demonstrate generally higher reactivity for some peptides, but some peptides indicate less reactivity relative to normal controls (Fig. 8). Such a difference would not be detectable in standard ELISA assays, but may be illustrating an immunologically relevant effect. In mouse models of infection this phenomenon is due to decreased humoral reactivity over time. In the human samples where only one time point is available we cannot readily determine if the lack of some portion of total humoral immune reactivity is a product of the disease or a precondition for the disease. Either case is testable, and would be of considerable interest to disease specialists.
A primary focus of this manuscript is to describe why immunosignaturing works the way it does. We examined slides which allowed spacing peptides at a fixed distance from one another on a microarray surface (POSTech, Seoul, Korea). We found that increasing the peptide distance caused a pronounced fall-off in detectable signal. At the widest spacing of 9 nm, we completely lost the ability to distinguish flu-infected and naïve mice, implying that a certain percentage of peptides need to respond above background levels. When we tested a commercial microarray known for high-quality epitope peptides (PepPerPrint, Heidelberg, Germany), we found that the random-sequence peptides constructed by PepPerPrint had virtually no cross-reactivity, even to the same monoclonal that bound hundreds of random sequence peptides on the immunosignature microarray. We surmise that the density of peptide on the surface of the microarray creates a combination of local avidity, rebinding and trapping that enhances the “immunosignature effect.” Peptide arrays which are optimized for minimal cross-reactivity may be unsuitable for immunosignaturing, but are best for epitope mapping.
We envision several potential applications of these arrays. First, identification of peptides that detect disease-specific biomarker antibodies for clinical applications: the diagnostic peptides for a given disease could be used in a printed array format, on SPR surfaces, or for ELISA. Alternatively, disease-specific peptides could be used to purify the antibodies that bind to them. Those antibodies could then pull down the original antigen from a tissue lysate. That antibody could then be the biomarker. It may even be possible to use the microarray described here to continuously monitor healthy individuals for a change in health status in an unbiased manner. We have shown that a person's healthy signature, while often quite different from other healthy signatures, is remarkably self-consistent over time until that person becomes ill or receives a vaccine (data not shown, publication in preparation). Once a person is immunized, or becomes ill, the immunosignatures become quite homogenous, reflecting that canonical disease signature.
In summary, we present fundamental aspects of immunosignaturing: a simple, inexpensive technology for profiling antibody complexity in blood. We anticipate that this format will have broad applicability in research and diagnostics.
Acknowledgments
Serum samples were obtained as a generous gift from the University of Arizona Valley Fever Center for Excellence. We thank Muskan Kukreja for the mixing experiments and PepPerPrint (www.pepperprint.com) and Volker Stadler for the in situ synthesized microarrays. The immunosignature microarrays are available for purchase from www.peptidearraycore, and have NCBI accession number GPL14921.
Footnotes
* This work was supported by TRIF funds from the state of Arizona.
REFERENCES
- 1. Kurian S. M., Heilman R., Mondala T. S., Nakorchevsky A., Hewel J. A., Campbell D., Robison E. H., Wang L., Lin W., Gaber L., Solez K., Shidban H., Mendez R., Schaffer R. L., Fisher J. S., Flechner S. M., Head S. R., Horvath S., Yates J. R., Marsh C. L., Salomon D. R. (2009) Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS ONE 4, e6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amur S., Frueh F. W., Lesko L. J., Huang S. M. (2008) Integration and use of biomarkers in drug development, regulation and clinical practice: a US regulatory perspective. Biomarkers Med. 2, 305–311 [DOI] [PubMed] [Google Scholar]
- 3. Food and Drug Administration (2010) Table of valid genomic biomarkers in the context of approved drug labels. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm
- 4. Sulzer B., van Hemmen J. L., Neumann A.U., Behn U. (1993) Memory in idiotypic networks due to competition between proliferation and differentiation. Bull. Math. Biol. 55, 1133–1182 [DOI] [PubMed] [Google Scholar]
- 5. Cenci S., Sitia R. (2007) Managing and exploiting stress in the antibody factory. FEBS Lett. 581, 3652–3657 [DOI] [PubMed] [Google Scholar]
- 6. Cooperman J., Neely R., Teachey D. T., Grupp S., Choi J. K. (2004) Cell division rates of primary human precursor B cells in culture reflect in vivo rates. Stem Cells 22, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 7. Förster I., Rajewsky K. (1990) The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc. Natl. Acad. Sci. U. S. A. 87, 4781–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao Z., Rajewsky K. (2001) Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194, 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geijersstam V, Kibur M., Wang Z., Koskela P., Schiller J., Lehtinen M., Dillner J. (1998) Stability over time of serum antibody levels to human papillomavirus Type 16. J. Inf. Dis. 177, 1710–1714 [DOI] [PubMed] [Google Scholar]
- 10. Bonifacio E., Lampasona V., Bernasconi L., Ziegler A. G. (2000) Maturation of the humoral autoimmune response to epitopes of GAD in preclinical childhood Type 1 diabetes. Diabetes 49, 202–208 [DOI] [PubMed] [Google Scholar]
- 11. Thurlings R., Vos K., Gerlag D., Tak P. (2006) The humoral response in rheumatoid arthritis and the effect of B-cell depleting therapy. Nederland Tijdschr Geneeskd 150, 1657–1661 [PubMed] [Google Scholar]
- 12. Stockert E., Jäger E., Chen Y. T., Scanlan M. J., Gout I., Karbach J., Arand M., Knuth A., Old L. J. (1998) A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J. Exp. Med. 187, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scofield R. H. (2004) Autoantibodies as predictors of disease. Lancet 363, 1544–1546 [DOI] [PubMed] [Google Scholar]
- 14. Hampton T. (2003) Autoantibodies predict Lupus. J. Am. Med. Assoc. 290, 3186 [Google Scholar]
- 15. Arbuckle M., McClain M., Rubertone M., Scofield R. H., Dennis G., James J., Harley J. (2003) Development of autoantibodies before the clinical onset of systemic Lupus Erythematosus. N. Engl. J. Med. 349, 1526–1533 [DOI] [PubMed] [Google Scholar]
- 16. Leslie R., Atkinson M., Notkins A. L. (1999) Autoantigens IA-2 and GAD in Type I (insulin-dependent) Diabetes. Diabetologia 42, 3–14 [DOI] [PubMed] [Google Scholar]
- 17. Darnell R. B., Posner J. B. (2003) Paraneoplastic syndromes involving the nervous system. N. Engl. J. Med. 349, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 18. Gultekin S., Rosenfeld M., Voltz R., Eichen J., Posner J., Dalmau J. (2000) Paraneoplastic limbic encephalitis: neurological symptoms, immunological findings and tumour association in 50 patients. Brain 123, 1481–1494 [DOI] [PubMed] [Google Scholar]
- 19. Voltz R. (2002) Paraneoplastic neurological syndromes: an update on diagnosis, pathogenesis, and therapy. Lancet Neurol. 1, 294–305 [DOI] [PubMed] [Google Scholar]
- 20. Elrington G., Murray N. M., Spiro S. G., Newson-Davis J. (1991) Neurological paraneoplastic syndromes in patients with small cell lung cancer: A prospective survey of 150 patients. J. Neurosurg. Psychiatry 54, 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roche S., Dauvilliers Y., Tiers L., Couderc C., Piva M., Provansal M., Gabelle A., Lehmann S. (2008) Autoantibody profiling on high-density protein microarrays for biomarker discovery in the cerebrospinal fluid. J. Immunol. Methods 338, 75–78 [DOI] [PubMed] [Google Scholar]
- 22. Hanash S. (2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 23. Pasqualini R., Ruosiahti E. (1996) Organ targeting in vivo using phage display peptide libraries. Nature 380, 364–366 [DOI] [PubMed] [Google Scholar]
- 24. Adda C. G., Anders R. F., Tilley L., Foley M. (2002) Random sequence libraries displayed on phage: identification of biologically important molecules. Combinatorial Chem. High Throughput Screening 5, 1–14 [DOI] [PubMed] [Google Scholar]
- 25. Legutki J. B., Magee D. M., Stafford P., Johnston S. A. (2010) A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine 28, 4529–4537 [DOI] [PubMed] [Google Scholar]
- 26. Restrepo L., Stafford P., Magee D. M., Johnston S. A. (2011) Application of immunosignatures to the assessment of Alzheimer's disease. Ann. Neurol. doi: 10.1002/ana.22405 [DOI] [PubMed] [Google Scholar]
- 27. Diehnelt C. W., Shah M., Gupta N., Belcher P. E., Greving M. P., Stafford P., Johnston S. A. (2010) Discovery of high-affinity protein binding ligands backwards. PLoS ONE 5, e10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stafford P., Brun M. (2007) Three methods for optimization of cross-laboratory and cross-platform microarray expression data. Nucleic Acids Res. 35, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang D. C., DeVit M., Johnston S. A. (1992) Genetic immunization is a simple method for eliciting an immune response. Nature 356, 152–154 [DOI] [PubMed] [Google Scholar]
- 30. Stemke-Hale K., Kaltenboeck B., DeGraves F. J., Sykes K. F., Huang J., Bu C. H., Johnston S. A. (2005) Screening the whole genome of a pathogen in vivo for individual protective antigens. Vaccine 23, 3016–3025 [DOI] [PubMed] [Google Scholar]
- 31. Halperin R. F., Stafford P., Johnston S. A. (2011) Exploring antibody recognition of sequence space through random-sequence peptide microarrays. Mol. Cell. Proteomics 10, M110.000786 doi:10.1074/mcp.M110.000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reineke U., Ivascu C., Schlief M., Landgraf C., Gericke S., Zahn G., Herzel H., Volkmer-Engert R., Schneider-Mergener J. (2002) Identification of distinct antibody epitopes and mimotopes from a peptide array of 5520 randomly generated sequences. J. Immunol. Methods 267, 37–51 [DOI] [PubMed] [Google Scholar]
- 33. Giraudi G., Rosso I., Baggiani C., Giovannoli C. (1999) Affinity between immobilised monoclonal and polyclonal antibodies and steroid-enzyme tracers increases sharply at high surface density. Analyt. Chim. Acta 381, 133–146 [Google Scholar]
- 34. Park J. W. (2007) NSB Amine Slides Characterization. http://www.nsbpostech.com/2007/order/order3_detail.html?g_no=34
- 35. Kurth D. G., Bein T. (1993) Surface reactions on thin layers of silane coupling agents. Langmuir 9, 2965–2973 [Google Scholar]
- 36. Breitling F., Nesterov A., Stadler V., Felgenhauer T., Bischoff F. R. (2009) High-density peptide arrays. Mol. BioSyst. 5, 224–234 [DOI] [PubMed] [Google Scholar]
- 37. Bowman C. C., Clements J. D. (2001) Differential biological and adjuvant activities of cholera toxin and Escherichia coli heat-labile enterotoxin hybrids. Infection Immunity 69, 1528–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merbl Y., Itzchak R., Vider-Shalit T., Louzoun Y., Quintana F. J, Vadai E., Eisenbach L., Cohen I. R. (2009) A systems immunology approach to the host-tumor interaction: large-scale patterns of natural autoantibodies distinguish healthy and tumor-bearing mice. PLoS ONE 4, e6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burritt J. B., DeLeo F. R., McDonald C. L., Prigge J. R., Dinauer M. C., Nakamura M., Nauseef W. M., Jesaitis A. J. (2001) Phage display epitope mapping of human neutrophil flavocytochromeb 558. J. Biol. Chem. 276, 2053–2061 [DOI] [PubMed] [Google Scholar]
- 40. Chase B., Johnston S. A., Legutki J. B. (2012) Evaluation of biological sample preparation for immunosignature based diagnostics. Clin. Vaccine Immunol. doi:10.1128/CVI.05667–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Notkins A. L. (2004) Polyreactivity of antibody molecules. Trends Immunol. 25, 174–179 [DOI] [PubMed] [Google Scholar]
- 42. Dimitrov J. D., Planchais C., Kang J., Pashov A., Vassilev T. L., Kaveri S. V., Lacroix-Desmazes S. (2010) Heterogeneous antigen recognition behavior of induced polyspecific antibodies. Biochem. Biophys. Res. Commun. 398, 266–271 [DOI] [PubMed] [Google Scholar]
- 43. Zhou Z. H., Zhang Y., Hu Y. F., Wahl L. M., Cisar J. O., Notkins A. L. (2007) The broad antibacterial activity of the natural antibody repertoire is due to polyreactive antibodies. Cell Host Microbe 1, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]