Abstract
Platelet-derived growth factor receptor-beta (PDGFRβ) is required for the development of mesenchymal cell types, and plays a diverse role in the function of fibroblasts in tissue homeostasis and regeneration. In this study, we characterized the expression of PDGFRβ in fibroblasts derived from human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), and showed that this expression is important for cellular functions such as migration, extracellular matrix production and assembly in 3D self-assembled tissues. To determine potential regulatory regions predictive of expression of PDGFRβ following differentiation from ESCs and iPSCs, we analyzed the DNA methylation status of a region of the PDGFRB promoter that contains multiple CpG sites, before and after differentiation. We demonstrated that this promoter region is extensively demethylated following differentiation, and represents a developmentally regulated, differentially methylated region linked to PDGFRβ expression. Understanding the epigenetic regulation of genes such as PDGFRB, and identifying sites of active DNA demethylation, is essential for future applications of iPSC-derived fibroblasts for regenerative medicine.
Key words: 3D tissues, DNA methylation, PDGFRβ, Fibroblasts, Human embryonic stem cells, Induced pluripotent stem cells
Introduction
Platelet-derived growth factor (PDGF) is a mitogen and chemoattractant that stimulates the recruitment of mesenchymal cells (Betsholtz, 2004; Heldin and Westermark, 1999), and facilitates the development and repair of several stromal tissue types (Andrae et al., 2008; Gao et al., 2005). Activation of the PDGF receptor plays a central role in modulating a broad range cellular functions, including extracellular matrix (ECM) production, angiogenesis, osteogenesis and mesengenesis in a variety of tissue environments (Ball et al., 2007; Trojanowska, 2008). Many of these functions are controlled by signaling pathways activated by the beta isoform of the PDGF receptor (PDGFRβ) (Abramsson et al., 2003; Gao et al., 2005), which is required for the normal embryonic development of mesenchymal cell types (Betsholtz, 2004). Beyond this role in mesenchymal lineage commitment, PDGFRβ-mediated signaling participates in the differentiation of cells to specific mesenchymal fates by directing fibroblasts to myofibroblasts that increase production of ECM proteins needed for repair (Betsholtz, 2004; Gao et al., 2005). Precise control of PDGFRβ expression and activity is required for proper remodeling during connective tissue repair because it is known that tissues lacking PDGFRβ show a deficiency in production of granulation tissue after wounding (Gao et al., 2005) and aberrant upregulation of PDGFRβ has been implicated in fibrotic diseases owing to over-production of ECM proteins (Trojanowska, 2008).
In light of these PDGFRβ-mediated cellular functions, elucidating how expression of this receptor is regulated during development of mesenchymal cell types is of potential therapeutic value in understanding the mechanisms involved in tissue regeneration (Majore et al., 2011). However, as a result of numerous shortcomings of cells currently used for tissue repair strategies, deriving mesenchymal cells that manifest PDGFRβ-related functions from novel sources, such as induced pluripotent stem cells (iPSCs) and human embryonic stem cells (ESCs), might provide a renewable source of therapeutic cells for such regeneration strategies (Barberi et al., 2005; Riazi et al., 2009). Recent findings have shown that fibroblasts differentiated from iPSCs restore their replicative potential (Suhr et al., 2009), improve their mitochondrial function (Suhr et al., 2010) and gain the ability to prevent ischemic injury (Lian et al., 2010). In addition, we recently found that ESC-derived fibroblasts exhibited a repair-promoting phenotype in 3D skin-equivalent tissues (Shamis et al., 2011). These findings suggest that fibroblasts derived from iPSCs or ESCs display phenotypic properties or repair potential that might exceed that of the cells from which they were initially reprogrammed. However, it is not known whether the regenerative properties of iPSC-derived fibroblasts can be modulated by PDGFRβ or what role regulation of PDGFRβ expression might play during differentiation of fibroblasts from iPSCs.
The establishment and maintenance of cellular identity following differentiation from pluripotent cell types is dependent on global epigenetic changes that ‘landscape the genome’ to streamline specific functional programs and maintain a specified cell-lineage fate (Khavari et al., 2010). Differentiation of ESCs and iPSCs towards stable mesenchymal lineages is dependent on changes in DNA methylation of CpG islands that result in characteristic methylation profile that can be used to distinguish specific cells from normal and diseased states (Fernandez et al., 2012). Changes in DNA methylation at specific regions of the genome, known as differentially methylated regions (DMRs), are required for cells to undergo differentiation, because in the absence of DNA methylation, pluripotent cell types will not differentiate and they retain their pluripotent status (Chen et al., 2003; Pawlak and Jaenisch, 2011). Recently published DNA methylation screens performed using iPSC-derived cells have revealed that specific DMRs within iPSCs might also be resistant to reprogramming (Lister et al., 2011), and these DMRs might lead to aberrant gene expression as cells are differentiated from them, which is linked to retention of an epigenetic memory of the somatic cell type from which they were reprogrammed (Ohi et al., 2011). However, less is understood about specific epigenetic changes during the process of differentiation of iPSCs to specific cell types, and the impact of specific changes in DNA methylation on the function of these differentiated cells is still poorly understood. Furthermore, it is unclear whether differences in DNA methylation of specific gene promoters between ESCs and iPSCs will result in variation in the function of cells differentiated from them. Therefore, analysis of DNA methylation at specific CpG sites within gene promoters of cells differentiated from iPSCs will be a vital step to better understand their phenotypic stability. Beyond this, comparing these DNA methylation signatures in iPSC- or ESC-derived cells with their adult counterparts will further establish their identity, and will elucidate the significance of these DNA methylation signatures in iPSC- or ESC-derived cells. Specifically, the identification of gene promoters involved in lineage specification and function of iPSC- and ESC-derived fibroblasts will allow discernment of the importance of epigenetic rearrangement and DNA methylation during differentiation from pluripotent stem cells.
We have studied changes in DNA methylation of the PDGFRB promoter upon differentiation from iPSCs or ESCs and concomitant changes in the function of the resultant fibroblasts derived from them. We have done this by performing detailed characterization of the methylation status of the PDGFRB promoter before and after differentiation from both ESCs and iPSCs, and by assaying PDGFRβ-mediated functions in the fibroblasts derived from them. We have previously shown that ESC- and iPSC-derived fibroblasts exhibit the function of stromal fibroblasts as demonstrated by their capacity to support the development of 3D skin equivalent tissues in vitro (Hewitt et al., 2009), and to enhance the repair of these tissues through their production of paracrine factors (Shamis et al., 2011). We now report that PDGFRβ expression is increased in ESC- and iPSC-derived fibroblasts, and knockdown of PDGFRβ impaired the ability of ESC- and iPSC-derived fibroblasts to assemble 3D stroma-like ECM, and limited their cellular migration in response to PDGF stimulation, which are both essential functions for tissue regeneration and maintenance. We also found that the PDGFRB gene, which is known to mediate pericyte and mesenchymal stem cell (MSC) function, is more than 95% demethylated at 12 CpG sites within its promoter. Thus, we have identified a novel developmentally controlled DMR where CpG sites upstream of the transcription start site (TSS) that is demethylated following differentiation. The presence of this DMR within the PDGFRB promoter might have predictive value in identifying cells that can undergo mesenchymal lineage commitment and provide mesenchymal cell functions upon their differentiation from ESC and iPSCs.
Results
PDGFRβ is expressed in ESC- and iPSC-derived fibroblasts
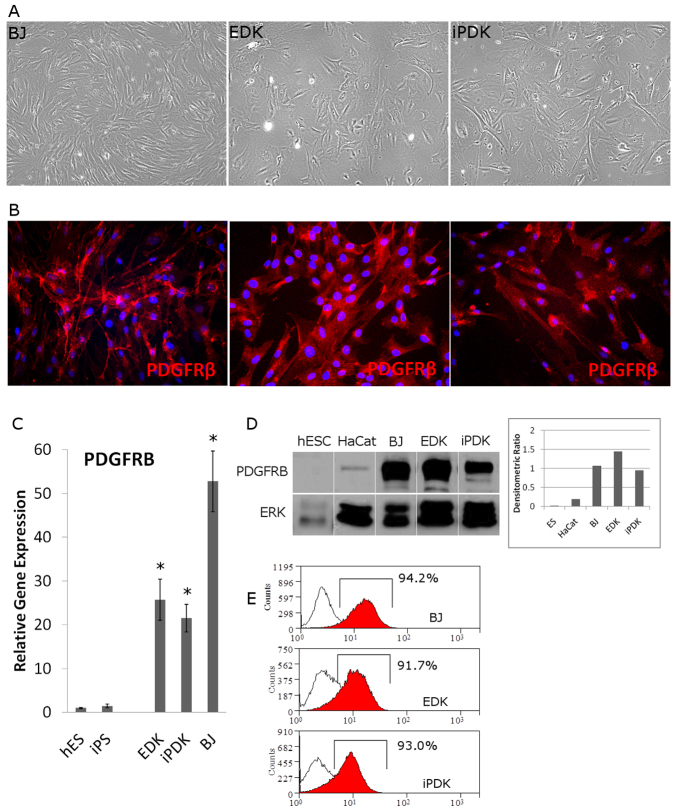
Several independent mesenchymal cell lines were derived from ESCs and iPSCs using a direct differentiation protocol. These ESC- and iPSC-derived cell lines demonstrated morphological features characteristic of stromal fibroblasts (Fig. 1A) and expressed the mesenchymal cell and pericyte marker PDGFRβ in a majority of ESC-derived (EDK) and iPSC-derived (iPDK) cells upon immunohistochemical staining (Fig. 1B). Foreskin-derived stromal fibroblasts (BJ), which were the parental, somatic cells from which iPSCs were initially reprogrammed, also expressed PDGFRβ in an intracellular staining pattern similar to EDK and iPDK cells (Fig. 1B). PDGFRβ expression in EDK and iPDK cells derived from three independent differentiation experiments was also analyzed by real-time RT-PCR and compared with that in ESC and iPSC cells before differentiation. All ESC- and iPSC-derived cell lines showed expression of PDGFRβ that was elevated by at least 20-fold when compared with ESCs and iPSCs, and expression was similar to that seen in BJ fibroblasts (Fig. 1C). In addition to gene expression changes, we also observed high levels of protein expression of PDGFRβ in ESC- and iPSC-derived cells that was undetectable in ESCs and had very low expression in unrelated HaCat epithelial cells (Fig. 1D). We next analyzed the surface protein expression by flow cytometry to determine the percentage of cells in which PDGFRβ was expressed following differentiation from ESCs and iPSCs. We found that at least 90% of EDK and iPDK cells expressed high levels of PDGFRβ that were similar to those seen in control BJ fibroblasts (Fig. 1E). These results indicated that expression of PDGFRβ was upregulated following the directed differentiation of fibroblasts from these pluripotent cells. The expression of PDGFRβ in ESC- and iPSC-derived cells supports previous findings establishing that PDGFRβ expression is a useful marker of mesenchymal fate in adult-derived MSCs and fibroblasts.
Fig. 1.
Expression of PDGFRβ in ESC- and iPSC-derived cells correlates with mesenchymal phenotype. Cells differentiated from ESCs (EDKs) and iPSCs (iPDKs) were morphologically similar to control fibroblasts (A) and expressed PDGFRβ as detected by immunofluorescence staining (B). (C) Analysis of mRNA levels of PDGFRB by real-time RT-PCR, showed that EDK, iPDK and BJ fibroblasts expressed significantly higher levels of PDGFRB compared with pluripotent cell types (ESCs and iPSCs). Error bars represent s.d. *P≤0.05 compared with ESCs. (D) Western blot analysis showed that PDGFRβ is similarly expressed in BJ, EDK and iPDK cells, is not expressed in ESCs and is expressed at very low levels in an unrelated epithelial cell line, HaCat. (E) These results were also confirmed by flow cytometry showing that greater than 90% of EDK and iPDK cells expressed detectable levels of PDGFRβ at the cell surface, similar to control fibroblasts (BJ).
PDGFRβ induces migration of ESC- and iPSC-derived fibroblasts
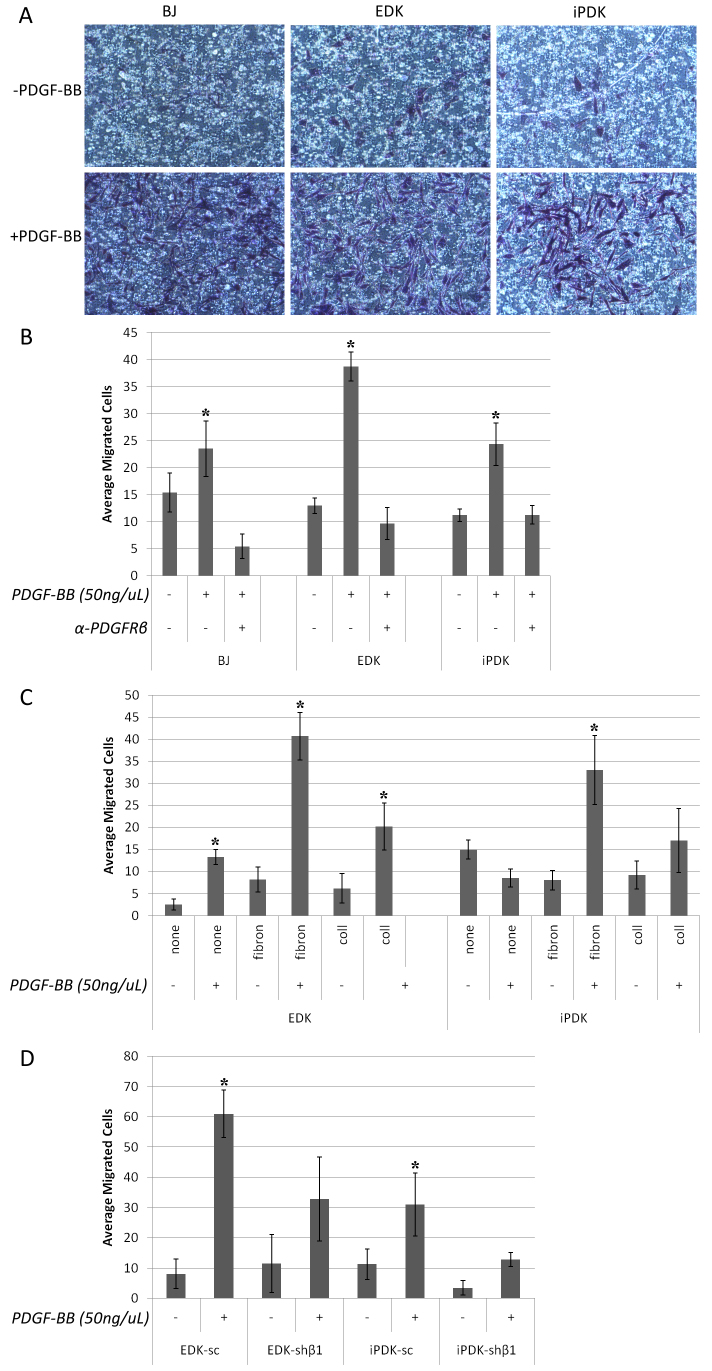
Because MSCs, pericytes and fibroblasts are known to respond to PDGFRβ by migrating towards a PDGF-BB gradient (Ball et al., 2007), we studied whether ESC- and iPSC-derived cells could also demonstrate cell migration in response to a PDGF-BB stimulus. We used a Boyden Chamber assay to compare the migration of ESC- and iPSC-derived cells to BJ fibroblasts in response to 50 ng/μl of PDGF-BB. We found that ESC- and iPSC-derived cells demonstrated enhanced migration across a fibronectin-coated membrane in response to PDGF-BB exposure (Fig. 2A). When we quantified the average number of cells that migrated across the membrane after 6 hours, we found that BJ, EDK and iPDK cells migrated at a similar rate in the absence of PDGF-BB stimulation (Fig. 2B). In response to PDGF-BB, we observed a threefold induction of migration of EDK cells, and a twofold induction in migration of iPDK cells (Fig. 2B). To establish that this migratory response was dependent on PDGFRβ, we pre-incubated these cells with a PDGFRβ blocking antibody and observed that the induction of migration to PDGF-BB was completely abrogated for all cell types (Fig. 2B). To determine whether the PDGF-BB-dependent migratory response of EDK and iPDK cells was specific for a fibronectin-coated surface, we compared the response to PDGF-BB on a fibronectin-coated membrane with that seen when cells were seeded on either uncoated membranes or membranes coated with type I collagen. We found that uncoated and collagen-coated membranes did not induce migration in response to a PDGF-BB stimulus for EDK, iPDK or BJ fibroblasts, indicating that the effect of PDGF-BB stimulation was dependent on the presence of fibronectin (Fig. 2C). These findings demonstrated that iPSC- and ESC-derived fibroblasts were similar to mature fibroblasts in their migratory response to PDGF-BB and that this migration was mediated through the PDGFRβ receptor in a substrate-specific manner.
Fig. 2.
Migration of ESC- and iPSC-derived fibroblasts towards PDGF stimulation is dependent on PDGFRβ activity and fibronectin. Cells were seeded on a polycarbonate membrane with 8 μM pores, and allowed to migrate for 6 hours towards either no stimulus or 50 ng/μl PDGF-BB in the lower chamber. (A) Migration of BJ, EDK and iPDK cells was determined by washing cells from the upper chamber, fixing cells in 4% paraformaldehyde, and staining cells that had migrated through the membrane with 0.1% Crystal Violet. (B) We quantified these results by counting and averaging migrated cells in three separate experiments and showed a significant increase in migration towards PDGF-BB, and this effect was abrogated with a PDGFRβ blocking antibody. (C) Fibronectin-coated membranes showed the highest level of migratory induction in response to PDGF-BB compared to uncoated or collagen-coated membranes. (D) EDK and iPDK cells with stable knockdown of PDGFRβ showed reduced migration compared to scrambled controls. Significant changes over control conditions were determined using a two-tailed t-test. *P≤0.05.
shRNA-mediated knockdown of PDGFRβ results in downregulation of PDGFRβ in EDK and iPDK cells and decreased cell migration
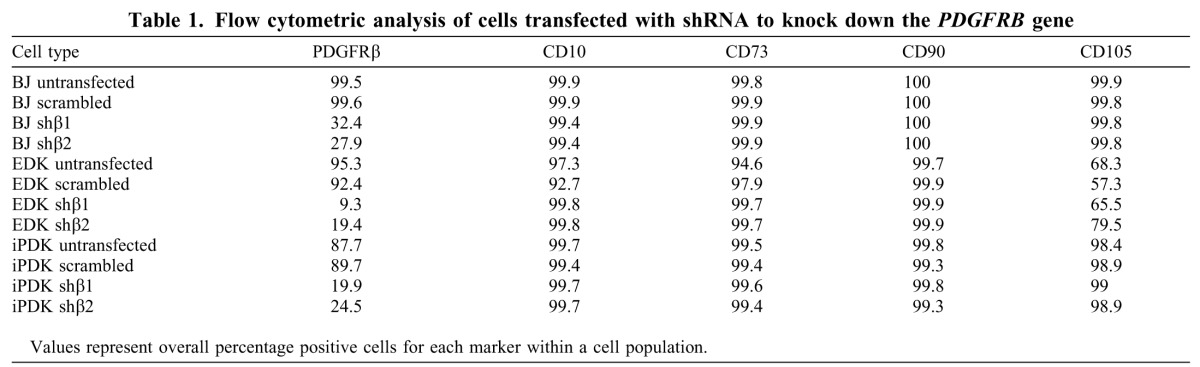
We next generated EDK and iPDK cells with decreased expression of PDGFRβ to confirm the specificity of expression of this receptor on their migratory properties. Parental BJ fibroblasts, EDK and iPDK fibroblast lines were infected with two distinct shRNA lentiviral constructs (shβ1 or shβ2) directed against the PDGFRB gene and the effect of this knockdown was compared with expression in cells infected with a non-specific, scrambled shRNA (scram). The extent of this knockdown in infected EDK and iPDK cell lines was determined by flow cytometric analysis, which demonstrated that levels of PDGFRβ on the cell surface using either shβ1 or shβ2 constructs were downregulated in at least 70% of EDK cells and 75% of iPDK cells (Table 1). Additionally, PDGFRβ knockdown did not significantly alter the mesenchymal identity of these cells, because we did not observe any changes in the CD profile typical of cells with a mesenchymal phenotype in knockdown cells (Table 1). To show that PDGFRβ knockdown could alter PDGFRβ-related function, we incorporated these cells into our Boyden Chamber migration assay. We found that PDGFRβ-knockdown cells showed a 50% decrease in their ability to migrate across a fibronectin-coated membrane towards a PDGF-BB stimulus in BJ, EDK and iPDK cells (Fig. 2D). These changes in migration were not related to changes in cell proliferation in knockdown cells because no significant differences were seen in BrdU incorporation when PDGFRβ-knockdown cells were compared with scram control cells grown on coverslips (supplementary material Fig. S1). These findings further demonstrate that PDGFRβ plays a role in mediating the migration of ESC- and iPSC-derived fibroblasts in a manner similar to that seen in parental, BJ fibroblasts.
Table 1.
Flow cytometric analysis of cells transfected with shRNA to knock down the PDGFRB gene
Knockdown of PDGFRB in ESC- and iPSC-derived cells reduces expression of genes involved in ECM protein production
In addition to cell migration towards a PDGF-BB stimulus, ECM production is an essential feature of stromal fibroblast function that is needed for tissue maintenance and repair. Our initial comparison revealed that both EDK and iPDK cell lines expressed high levels of genes involved in ECM production (Fig. 3A). Compared with BJ fibroblasts, EDK and iPDK cell lines expressed at least twice as much COL3A and fibronectin, whereas they expressed lower levels of tenascin C, and comparable levels of COL1A1 and COL1A2 (Fig. 3A). To determine whether PDGFRβ was important in the regulation of ECM production in EDK and iPDK cell lines, we analyzed ECM gene expression following shRNA-mediated knockdown of PDGFRβ with the shβ1 and shβ2 constructs described above. We confirmed by real-time RT-PCR that levels of PDGFRβ expression were downregulated at least fivefold in all cell types following their infection with each of the lentiviral shRNA constructs (shβ1 and shβ2) compared with knockdown with control scram shRNA (Fig. 3B). This degree of PDGFRβ knockdown resulted in significantly decreased expression of a broad spectrum of ECM proteins including type I collagen α1 chain (COL1A1), type I collagen α2 chain (COL1A2), type III collagen α1 chain (COL3A1), fibronectin (FN) and tenascin C (TENC) (Fig. 3B). Downregulation of type I collagen was most dramatic upon PDGFRβ knockdown as seen by the 50–60% decrease in expression in ESC-derived cells and 60–90% decrease in iPSC-derived cells. These findings demonstrated that PDGFRβ was important for the expression of a broad range of ECM proteins acquired upon the differentiation of iPDK and EDK fibroblasts from ESCs and iPSCs, and was similar to effects seen in control fibroblasts.
Fig. 3.
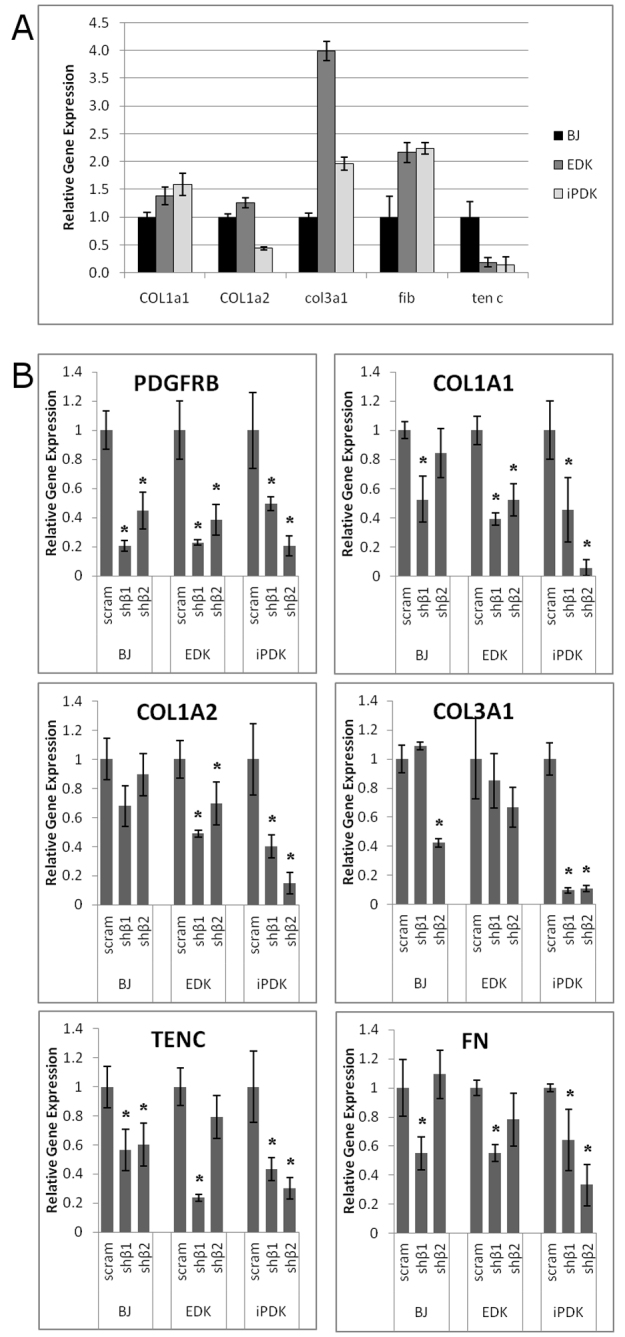
Knockdown of PDGFRβ results in decreased expression of matrix-related genes. EDK and iPDK cells were analyzed by real-time RT-PCR for RNA expression levels of genes involved in ECM production, and compared to control fibroblasts (BJ). (A) EDK and iPDK expressed similar levels of COL1A1, elevated levels of COL3A1 and FIB and reduced levels of TENC compared with BJ fibroblasts. (B) shRNA knockdown of PDGFRβ was confirmed in BJ, EDK and iPDK cells. Following shRNA knockdown, EDK and iPDK cells had reduced levels of ECM-related gene expression in a majority of knockdown cells using two different shRNA constructs. Error bars represent standard deviation from two independent experiments, and significance was determined compared with scrambled control-infected cells using a two-tailed t-test. *P≤0.05.
PDGFRβ knockdown decreases the production of type I collagen and fibronectin in a self-assembled ECM grown as a 3D stromal tissue
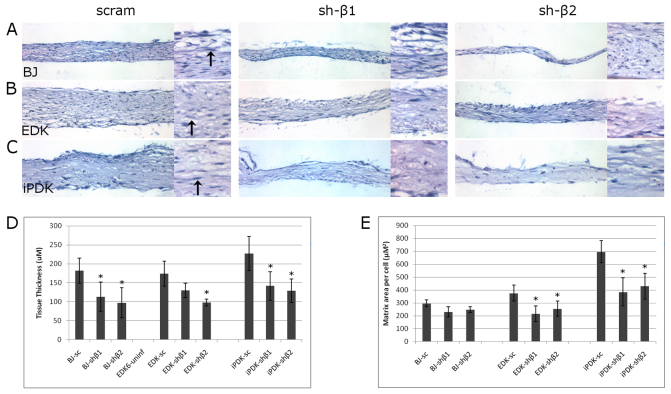
To assess the ability of ESC- and iPSC-derived cells to produce a self-assembled ECM in a 3D tissue microenvironment, and to determine whether the organization of this stroma-like tissue was dependent on PDGFRβ, we seeded EDK and BJ cells onto a PET membrane in the presence of ascorbic acid and cultured cells for 5 weeks. In this way, it was possible to assess the self-assembly of ECM produced by EDK, iPDK and BJ fibroblasts under more stringent experimental conditions than in 2D monolayer cultures alone. BJ, EDK and iPDK cell lines infected with either control shRNA or PDGFRβ-knockdown shRNA were examined for the degree to which they could organize well-structured, stromal architecture and for tissue thickness following hematoxylin and eosin (H&E) staining. Tissues self-assembled from EDK cells infected with scram, shβ1 or shβ2 developed a well-organized and compact, cellular tissue (Fig. 4B), and cells within these tissues demonstrated an elongated, fibroblast-like appearance (Fig. 4B, inset, arrows), similar to tissues assembled using iPDK cells (Fig. 4C). By contrast, tissues harboring BJ fibroblasts developed a more fibrillar connective tissue compared with those assembled by EDK cells (Fig. 4A). In addition, tissues generated with both PDGFRβ-knockdown vectors assembled stromal tissues with significantly reduced ECM production with both EDK and BJ cells, although the general organization of the matrix was comparable between scrambled and knockdown conditions for each cell type (Fig. 4A–C). To quantify these findings, we serially sectioned tissues from several independent experiments and measured tissue thickness, area and cellularity. Tissue thickness was significantly reduced in shβ1- or shβ2-infected cells by 50% in tissues harboring BJ fibroblasts, and by 40% in EDK and iPDK cell lines (Fig. 4D). When overall tissue production was quantified, by determining the average area of tissue deposited on a per cell basis, we observed a significant decrease in tissue production of 43% in shβ1- and 32% in shβ2-infected EDK cells, and 44% in shβ1-infected and 38% in shβ2-infected iPDK cells. Interestingly, we did not observe any change in tissue deposited on a per cell basis in control BJ fibroblasts, or in either shβ1- or shβ2-infected cells, although the tissues were significantly thinner (Fig. 4E). These differences in ECM deposition were not due to an increase in cell numbers as a result of proliferation, because there was no significant difference in proliferation between any of the three cell types in scram and PDGFRβ-knockdown tissues as determined by BrdU incorporation assays (supplementary material Fig. S1).
Fig. 4.
Knockdown of PDGFRβ reduces thickness of tissues generated in a 3D matrix-assembly assay. BJ, EDK and iPDK cells infected with either non-specific shRNA controls (scram) or shRNA directed against PDGFRβ (sh-β1 and sh-β2) were seeded on a 24-well Millicell (Millipore) hanging cell culture insert with 1.0 μM pore size at a density of 15,000 cells/insert. (A–C) Paraffin-embedded tissue sections of cells after 5 weeks in culture were analyzed by hematoxylin and eosin (H&E) staining to compare tissue thickness and architecture. At 20× magnification, we observed a reduction in the overall thickness of tissues infected with either sh-β1 or sh-β2. At 40× magnification (insets), overall tissue architecture remained unchanged with PDGFRβ knockdown. Arrows indicate characteristic elongated fibroblast morphology typical of healthy stromal tissue. (D,E) When we quantified the changes in matrix deposition between cell types with scram, sh-β1 or sh-β2 infections, we found a significant reduction in tissue thickness and overall tissue matrix deposited per cell in PDGFRβ-knockdown cells. Results represent mean ± s.d. of five sections per tissue, and four tissues per condition. Significant changes compared with scrambled control tissues were determine using two-tailed paired t-test. *P≤0.05.
In addition to overall matrix production, it is known that the organization and structure of stromal tissues is dependent on the levels of production of individual ECM proteins within that tissue. An important property of fibronectin during ECM development is its capacity to organize other ECM proteins such as type I collagen within developing stromal tissues (Sottile et al., 2007). To provide a more detailed assessment of the composition of self-assembled tissues with and without PDGFRβ knockdown, we analyzed protein levels of type I collagen and fibronectin by western blot analysis within self-assembled tissues following ECM self-assembly for 5 weeks. Protein lysates from these tissues revealed a distinct change in the composition of ECM proteins assembled by both BJ fibroblasts and EDK cell lines harboring PDGFRβ knockdown, as seen by the significantly decreased levels of Type I collagen and fibronectin in PDGFRβ-knockdown tissues (Fig. 5A). In addition, protein levels of fibronectin were 2- to 2.5-fold greater in tissues constructed with EDK cells than in tissues constructed with BJ fibroblasts (Fig. 5B). Taken together, these results indicate that PDGFRβ can regulate type I collagen and fibronectin and overall production of stroma-like tissues, further demonstrating that PDGFRβ expression plays an important role in the function of ESC- and iPSC-derived fibroblasts and is required for optimization of ECM production and organization.
Fig. 5.
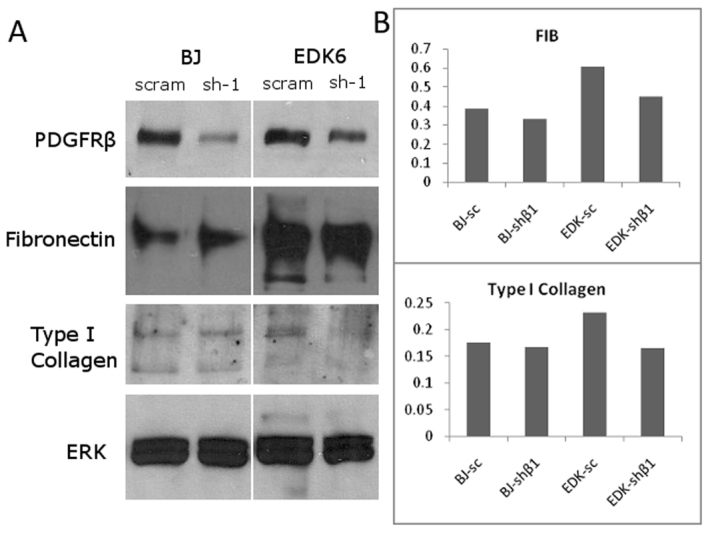
PDGFRβ knockdown reduces secretion of extracellular matrix genes in self-assembled tissues. Self-assembled tissues were homogenized by sonication and protein was extracted and run on a gradient 4–12% polyacrylamide gel. (A) Western blot analysis of tissue lysates after 5 weeks of growth confirmed the stability of the PDGFRβ knockdown in tissues, and also demonstrated reduction in the levels of production of fibronectin and type I collagen. (B) Densitometry analysis confirmed this decrease in production of fibronectin and type I collagen in EDK cells, and also showed that EDK cells with scrambled infections produced higher levels of both fibronectin and type I collagen compared with BJ scrambled controls.
Developmentally controlled DMR within the PDGFRB promoter is demethylated at 12 CpG sites during fibroblast differentiation from iPSCs and ESCs
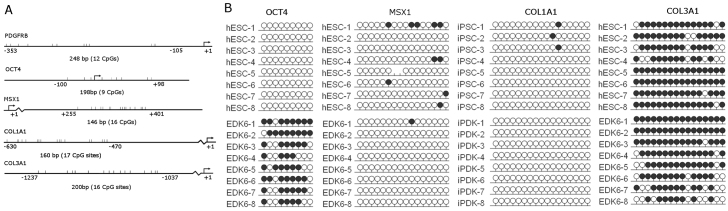
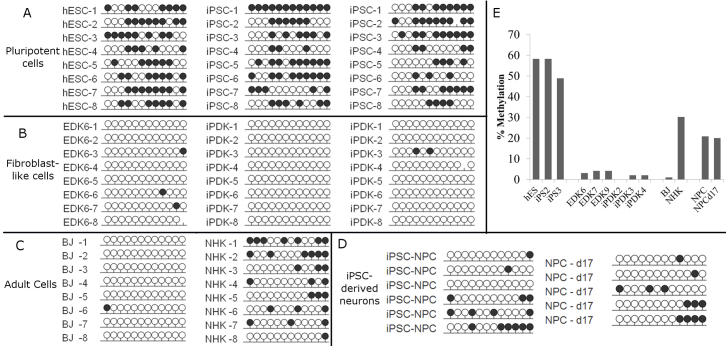
The functional importance of PDGFRβ in establishing the phenotypic properties of iPDK and EDK fibroblasts upon their differentiation from ESCs and iPSCs, directed us to study mechanisms that might regulate its expression upon differentiation. Because epigenetic regulatory elements are known to control differentiation and lineage commitment through the modulation of DNA methylation at CpG sites within the promoter, we performed a detailed characterization of the DNA methylation status of the PDGFRB promoter by bisulfite sequencing and compared iPSCs to iPDK, ESCs to EDK, and both iPDK and EDK cells to BJ fibroblasts. Because methylation patterns of this promoter have not been previously studied, PCR primers were designed against the upstream regulatory region of a specific region of the promoter, from position −353 to −105 relative to the TSS of the PDGFRB gene (Fig. 7A). Methylation patterns were determined for each cell population by analyzing the amplified PCR product of isolated DNA in eight bacterial clonal isolates, for one ESC line and each of two independently reprogrammed iPSC lines. We found that all of these pluripotent cell lines were methylated at multiple CpG sites within the PDGFRB promoter (Fig. 6A). By contrast, these CpG sites in the PDGFRB promoter in both iPDK and EDK cell lines were nearly entirely unmethylated in all eight clonal isolates studied from one EDK cell line and two iPDK cell lines, with most clones not demonstrating any methylation (Fig. 6B). Importantly, this pattern of demethylation was similar to that seen in BJ fibroblasts, yet differed greatly from that seen in primary keratinocytes (NHKs) (Fig. 6C). In addition, bisulfite sequencing of neural progenitor cells derived from iPSCs (iPSC-NPs), as well as differentiated neurons derived from these iPSC-NPs (supplementary material Fig. S2), revealed that 20% of CpG sites in the PDGFRB promoter were methylated (Fig. 6D). Thus, these neural progenitors demonstrated an intermediate level of methylation in this DMR that was greater than that seen in fibroblasts derived from iPSCs but less than methylation seen in the iPSCs. This suggests that the complete demethylation seen in the developmentally controlled DMR of the PDGFRB promoter was specific for cells with a fibroblast phenotype, because this did not occur during differentiation of other cell types from iPSCs. When the average overall methylation along 12 CpG sites in the promoter was quantified, it confirmed that although nearly 60% of CpG sites were methylated in pluripotent cells, on average fewer than 5% of CpG sites were methylated in fibroblast cell types (Fig. 6E). This pattern of demethylation seen in EDK, iPDK and BJ fibroblasts contrasted with the DNA methylation pattern within the PDGFRB promoter of NHK cells, where 30% of all CpG sites were methylated, and this population was 100% methylated at position 12 (Fig. 6C), indicating that the unmethylated state at this region of the PDGFRB promoter did not occur in unrelated cell types. These findings indicated that the specific patterns of DNA methylation and demethylation seen at the promoter region of PDGFRB represents a developmentally controlled DMR in cells with properties of stromal fibroblasts following their differentiation from ESCs and iPSCs. OCT4, which is known to be expressed specifically in ESCs and iPSCs, was methylated following differentiation, consistent with other reports following differentiation of ESCs and iPSCs (Fig. 7B). The nearly complete demethylation of the PDGFRB promoter following differentiation, establishes that the shift in the CpG methylation profile is a developmentally regulated epigenetic event that occurs during the differentiation of fibroblasts from pluripotent cells.
Fig. 7.
Promoter methylation profile of genes associated with mesenchymal lineage. (A) Bisulfite sequencing analysis of the OCT4, MSX1, COL1A1 and COL3A1 gene promoters were performed on ESC, iPSC, EDK and iPDK cells, and representative schematic images are shown. The position in the genome of the promoter sites analyzed relative to the TSS of each gene is shown, along with the number of CpG sites that were present in the amplified region. (B) OCT4 showed increased methylation upon differentiation, whereas MSX1 and COL1A1 remained unmethylated upon differentiation. COL3A1 was methylated in both ESC and EDK cells. Each dot represents one CpG site in the promoter region of PDGFRB and each row represents a clonal isolate of DNA fragments from indicated cell types. Methylated CpGs are indicated by a black circle, and unmethylated CpGs are indicated by a white circle.
Fig. 6.
DNA methylation profile of the PDGFRβ gene promoter in pluripotent and pluripotent-derived cells. Bisulfite sequencing analysis of the PDGFRβ gene promoter was performed on ESC, iPSC, EDK, iPDK, primary fibroblasts (BJ) and keratinocyte (NHK) cell types, as well as iPSC-derived neural progenitors (iPSC-NPCs) and neurons following 18 days of differentiation (NPC-d18). Each dot represents one CpG site in the promoter region of PDGFRβ and each row represents a clonal isolate of DNA fragments from indicated cell types. Methylated CpGs are indicated by a black circle, and unmethylated CpGs are indicated by a white circle. (A) PDGFRβ is methylated in pluripotent cell types h9-hES, iPS2 and iPS3. (B) PDGFRβ is unmethylated in hES-derived EDK, iPS2-derived iPDK and iPS3-derived iPDK. (C) In adult cell types, PDGFRB promoter was unmethylated in BJ fibroblasts, compared with primary keratinocytes (NHK), which are relatively highly methylated. (D) iPSC-derived neural progenitors (NPCs) and neurons following 18 days of differentiation (NPC d18), demonstrated an intermediate level of methylation at the PDGFRβ promoter. (E) Quantification of bisulfite sequencing results showed greater than 50% methylation in pluripotent cell types, and 0–5% methylation in pluripotent-derived EDK and iPDK cells, and adult fibroblasts, but not neurons derived from iPSCs.
To determine whether this pattern of developmentally regulated demethylation seen in these fibroblast lineages was specific for the PDGFRB promoter, we performed bisulfite sequencing to analyze DNA methylation patterns in the promoter regions of other genes linked to a mesenchymal cell phenotype following their differentiation from iPSCs and ESCs. Interestingly, DNA methylation patterns at multiple CpG sites in the promoter region of both the α1 chain of type I collagen (COL1A1) and the α1 chain of type III collagen (COL3A1) were unchanged following differentiation of iPSCs and ESCs to iPDK and EDK cells (Fig. 7B). Type I and type III collagen have previously been shown to be dramatically upregulated following differentiation of iPSCs to a fibroblast phenotype (Hewitt et al., 2011). These data indicate that this upregulation is probably not linked to the demethylation of their promoters. MSX1, a key regulator of limb formation and craniofacial morphogenesis during embryonic development, which is known to be expressed in adult mesenchymal cells, did not undergo demethylation and remained in a methylated state following differentiation (Fig. 7B). These results indicate that differentiation towards mesenchymal cell fate is linked to changes in DNA methylation of specific sites within the PDGFRB promoter that might be characteristic of its emerging fibroblastic phenotype, whereas other gene promoters associated with the mesenchymal phenotype remain unchanged following differentiation.
Discussion
In the present study, we have found that PDGFRβ is an important marker of mesenchymal lineage fate that is linked to the cellular function of fibroblasts following their differentiation from iPSCs and ESCs. We demonstrate that a region of CpG sites in the PDGFRB gene promoter underwent hypomethylation upon differentiation from these pluripotent cells as they acquired the properties of stromal fibroblasts. In doing so, we have identified a novel, differentiation-controlled, differentially methylated region (DMR) that is predictive of PDGFRβ expression during lineage commitment to fibroblasts. Beyond this, we have found that PDGFRβ is important for the function of iPSC- and ESC-derived fibroblasts, because shRNA-mediated knockdown of PDGFRβ expression decreased the capacity of these cells to produce ECM proteins and undergo cell migration. We also found an overlap in the methylation profiles in the region of the PDGFRB promoter studied between iPDK and EDK cell lines and foreskin fibroblasts, suggesting that this profile is a common feature of fibroblasts and that demethylation at these sites might be an important regulatory event in the specification and differentiation of fibroblasts during development.
The process of reprogramming fibroblasts to iPSCs is linked to changes in DNA methylation that are necessary to revert cells to an embryonic-like state (Maherali et al., 2007; Mikkelsen et al., 2008). Subsequent differentiation of specific cell types from iPSCs is dependent on epigenetic remodeling, including large changes in the DNA methylation, which restricts the expression of pluripotency genes and enables expression of tissue-specific genes needed to establish cellular fate and phenotype (Aranda et al., 2009; Gan et al., 2007). Furthermore, the process of DNA demethylation at certain promoters has been shown to be predictive in determining gene expression in a cell-type-specific manner (Wiench et al., 2011). By comparing the methylation profiles of CpG sites at specific gene promoters in cells differentiated from pluripotent stem cells to profiles seen in foreskin-derived fibroblasts, mechanisms involved in the establishment of lineage commitment and cell function of iPSC- and ESC-derived cell types can be determined (Aranda et al., 2009). Our results extend recent findings showing changes that occur in the methylome of pluripotent cells following differentiation (Hewitt et al., 2011; Laurent et al., 2010) by demonstrating that the changes in DNA methylation within a specific gene promoter that occurs upon their differentiation from pluripotent cells to fibroblast lineages are similar to methylation signatures seen in somatic fibroblasts.
Demethylation events similar to those we have characterized upon differentiation of iPSCs and ESCs to fibroblasts have also been found upon differentiation of adult progenitor cells to mesenchymal lineage fates. For example, gene promoters related to mesenchymal cell fate determination, such as LEP and PPARG2, demonstrate demethylation upon differentiation of mesenchymal cells but not other cell types or lineages (Noer et al., 2006). In addition, it has been shown that DNA demethylation in myoblasts can activate mesenchymal gene expression and induce trans-differentiation to osteogenic and adipogenic fates (Hupkes et al., 2011). Interestingly, it has recently been shown that differences exist in methylation at lineage-specific genes between adult MSCs and putative MSCs derived from ESCs that might account for differences in differentiation potential of MSC populations (Sørensen et al., 2010). In light of these findings, the CpG methylation status of lineage-specific promoters is likely to have consequences for mesenchymal cell function and fate upon differentiation from ESCs and iPSCs, thus highlighting the need for further characterization of the methylation profile of cell types derived from pluripotent stem cells that might be important in determining their lineage stability for future therapeutic applications. Interestingly, we did not observe any significant differences between ESC- and iPSC-derived cells in the CpG profile at the PDGFRB promoter. Whereas some reports indicate that iPSCs at early passage can retain epigenetic memory of their cell-of-origin, this is not seen in late-passage iPSCs, such as those used in these studies (Polo et al., 2010). Our findings on the epigenetic changes at the PDGFRB promoter during fibroblast differentiation demonstrate the potential use of DNA methylation as a marker of lineage commitment, although further work is necessary to show that specific DNA methylation events are crucial for cell lineage commitment during development.
The early events directing cell and tissue repair following injury are dependent on the spatially and temporally controlled production and deposition of ECM proteins by mesenchymal cells and through the recruitment of a variety of cell types through cell migration (Egging et al., 2006; Friedl and Bröcker, 2000). It is known that PDGFRβ plays a role during multiple stages of wound healing, because loss of PDGFRβ function impairs fibroblast migration (Gao et al., 2005) and stimulation of this receptor by recombinant PDGF-BB accelerates the healing of chronic wounds through deposition of ECM proteins (Pierce et al., 1992; Steed, 1995). Although it is known that PDGFRβ signaling activates a cascade of events that are crucial for cellular responses, such as ECM production and cell migration, inappropriate activation of this response has been linked to the pathogenesis of fibrotic conditions, including lung fibrosis and pulmonary arterial hypertension (Bonner, 2004; Trojanowska, 2008) that are, in part, due to transient collagen deposition linked to an acute fibrotic response (Yi et al., 1996). These aberrant responses signify the central role that PDGFRβ plays in maintaining the balance between ECM production and remodeling in the establishment and restoration of optimal tissue function. In this light, future therapeutic strategies using ESC- and iPSC-derived fibroblasts for repair and regeneration will require that regulation of PDGFRβ be better understood to reach optimal tissue outcomes.
Recently, the importance of PDGFRβ in MSC function has been recognized in many tissues, because it is known to be a marker of pericytes that are essential in directing these cells to blood vessels where they support neo-vascularization and vessel maturation (Veevers-Lowe et al., 2011). It has been proposed that pericytes are MSCs that migrate from the vessel wall following injury to become stromal constituents (da Silva Meirelles et al., 2008), where they provide paracrine support needed to direct tissue repair (Bernardo et al., 2009). Thus, in addition to the role PDGFRβ plays in ECM production and cell migration demonstrated in our study of ESC- and iPSC-derived fibroblasts, PDGFRβ plays a more ubiquitous role in repair by possibly directing the functional outcome of related cell types such as pericytes and MSCs. We suggest that DNA demethylation of the PDGFRB promoter correlates with PDGFRβ expression during differentiation, and might represent a marker of lineage specification towards a spectrum of mesenchymal cells with related functions. In this context, we also suggest that hypermethylation of the PDGFRB promoter in iPSCs following their reprogramming is important in establishing the pluripotent state, and demethylation is a requisite step for the establishment of lineage commitment.
The use of in vitro 3D tissues holds many advantages over 2D monolayer culture, because it provides a life-like microenvironment that is more highly predictive of cellular functions in tissues. It has previously been shown that self-assembled, ECM constructs harboring adult fibroblasts are composed primarily of type I collagen and fibronectin, and displayed features similar to human dermis (Pouyani et al., 2009). With this in mind, we have generated 3D, self-assembling stromal tissues as a way to assay ECM production and organization and provide a more physiologically relevant microenvironment to characterize the generation of stroma-like tissues from ESC- and iPSC-derived fibroblasts. Furthermore, using shRNA-knockdown strategies, we have demonstrated the importance of this system to assess functional tissue outcomes associated with PDGFRβ during tissue assembly. Beyond this, it appears that the proportions of fibronectin and type I collagen deposited by EDKs in these self-assembly assays are also regulated by PDGFRβ and might be important for tissue assembly and repair. One potential function linked to PDGFRβ suggested by these findings is that elevated levels of fibronectin produced by EDK fibroblasts might enhance interactions between fibronectin and type I collagen to generate more compact stromal tissues.
Understanding and directing the differentiation of mesenchymal cell types such as fibroblasts is of great interest in furthering therapeutic applications in regenerative medicine. First, pluripotent stem cells offer a plentiful source of cells, as a result of their unlimited replicative capacity, which can be reproducibly differentiated for cell-based therapies. Second, the ability to generate patient-specific cells upon differentiation of iPSCs to specified lineage fates might allow for improved, individualized cell-based therapies and development of human in vitro disease models (Bajada et al., 2008). Although it is known that adult-derived mesenchymal cells are capable of a wide range of physiologic functions that depend on their tissue type and context (English et al., 2010), these cells have limited proliferative potential. Understanding how to best harness their potential requires detailed characterization of key phenotypic modulators of cell behavior. Our findings extend previous observations that showed the importance of PDGFRβ on the function of MSCs and mature fibroblasts, by establishing that demethylation of the PDGFRB promoter is a common feature of ESC- and iPSC-derived fibroblasts. By studying additional changes in DNA methylation following differentiation, we hope to identify key modulators of events that can direct lineage fate and functional properties of ESC- and iPSC-derived cell types towards predictable outcomes. However, future work will require a more detailed understanding of methylation-mediated control of a broad spectrum of promoters important for fate and function determination.
Materials and Methods
Cell culture
Human iPSCs and ESCs were maintained on a mouse embryonic fibroblast (MEF) feeder layer as described (Thomson et al., 1998). iPSCs were generated from human foreskin cells (BJ fibroblast cell line) (ATCC, Manassas, VA) using four (OSKM, iPS-1) or five (OSKMN, iPS-2) retroviral reprogramming vectors (Maherali et al., 2007). The H9 line of ESCs used in this study were purchased from the WiCell Institute (Madison, WI). Differentiation of cells was performed as described previously (Hewitt et al., 2009). Briefly, iPSCs (iPS-1, passage 12, iPS-2, passage 30) or ESCs were plated as aggregates onto fixed MEFs, and cultured in medium consisting of 3:1 DMEM:F12 (Invitrogen, Carlsbad, CA), 5% FCII (Hyclone, Logan, UT), 0.18 mM adenine, 0.5 μg/ml hydrocortisone, 10−10 M cholera toxin, 10 ng/ml EGF, 5 μg/ml insulin for 7 days and supplemented on days 4–7 with 0.5 nM BMP-4. Fibroblasts derived from ESCs (EDKs) or iPSCs (iPDKs) were then propagated for an additional 7 days on fixed MEFs, plastic and type I collagen (BD Biosciences, San Jose, CA). Fibroblast cell lines were maintained in DMEM medium containing 10% FBS (Hyclone, Logan, UT) and 0.18 mM adenine. All experiments were performed on these cells from passage 5–8.
Neural progenitors were derived from iPSC clone 8330-8 [derived from Coriell GM08330 fibroblasts and characterized as described (Sheridan et al., 2011)] expanded under feeder-free conditions to remove iMEFs by growth directly on Matrigel (BD Biosciences) in mTeSR1 culture medium (StemCell Technologies). Expandable neuronal progenitors were isolated by direct manual picking of neural rosette structures upon initiation of differentiation by overgrowth of the iPSC colonies. Isolated cells were expanded in neural expansion medium (70% DMEM (Invitrogen), 30% Ham's F-12 (Mediatech) supplemented with B-27 (Invitrogen), 20 ng/ml each EGF (Sigma) and bFGF (R&D Systems) on poly-ornithine (Sigma)- and laminin (Sigma)-coated culture plates. Terminal neural differentiation was achieved by plating expanded cells plated at a seeding density of 40,000 cells per cm2 on polyornithine-laminin plates as above in expansion medium lacking both EGF and bFGF, with medium replacement every 3–5 days for a total of 18 days. Cell samples were collected by scraping followed by pelleting and quick-freezing.
Real-time RT-PCR
Cells were lysed directly from a 10 cm plate, and RNA was isolated using an RNA purification kit (Qiagen, Valencia, CA). 0.5 μg RNA was used to generate cDNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. PCR reactions were done in triplicate with SYBRgreen Supermix and run on an iQ5 Real-Time PCR detection system (Bio-Rad, Hercules, CA). Error bars represent standard deviation of three biological replicates, and statistical significance was determined using a paired t-test with P≤0.05.
shRNA knockdown
Stable shRNA knockdowns of PDGFRβ in EDK, iPDK and BJ cells were generated by stable infection with lentivirus carrying predicted PDGFRβ-interacting sequences. Following infection, cells were selected by addition of 1 μg/ml of puromycin (Sigma, St Louis, MO) for 4 days. Viruses were obtained from MISSION shRNA Lentiviral Particles (Sigma). Out of five predicted sequences, two were selected (TRCN1999 and TRCN2000) based on degree of knockdown, verified by real-time PCR and flow cytometry.
Construction of self-assembled tissues
EDK, iPDK and BJ cells infected with lentivirus containing either non-specific control scrambled shRNA or shRNA directed against PDGFRβ (sh-β1 and sh-β2) were seeded onto 24-well polyethylene terephthalate (PET) inserts (Millipore, Billerica, MA) at a density of 16,000 cells per insert. Medium was supplemented with 10 μg/ml ascorbic acid, and cells were allowed to grow on the membrane for 5 weeks before analysis. Of six biological replicates, three were fixed in 4% paraformaldehyde, processed and paraffin-embedded for sectioning, whereas three were used for protein extraction and western blot analysis.
Western blotting
Self-assembled tissues were scraped off membranes, and solubilized in deoxycholate following five rounds of sonication to homogenize protein. Purified protein lysates were run on a 4–12% gradient polyacrylamide gel and transferred to a nitrocellulose membrane for staining and analysis. Primary antibodies used were against PDGFRβ (Abcam, Cambridge, MA), type I collagen (R&D Systems, Minneapolis, MN), Fibronectin (R&D Systems, Minneapolis, MN) and ERK (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunofluorescence
Cells were grown on coverslips until approximately 80% confluency, washed and fixed in 4% paraformaldehyde. Cells for analysis of proliferation using bromo-deoxyuridine (BrdU) labeling were pre-incubated with BrdU nucleotides for 6 hours before fixation. For staining, cells were permeabilized in 0.1% Triton X-100, blocked using 0.2% BSA, and incubated in primary antibodies against PDGFRβ (Abcam, Cambridge, MA) or BrdU (R&D Systems, Minneapolis, MN) for 1 hour. Secondary antibodies used were rabbit Alexa Fluor 594 and mouse Alexa Fluor 488. Coverslips were mounted using Vectashield DAPI-containing mounting medium (Vector Labs, Burlingame, CA) and visualized on a Nikon Eclipse 80i microscope.
Flow cytometry
EDK and iPDK cell lines were trypsinized, resuspended in 2% FBS in PBS, and stained with PE-conjugated antibodies against CD10, CD73, CD90, CD105, CD140b (PDGFRβ) and IgG1k (BD Pharmingen, San Jose, CA). Cells were incubated for 40 minutes at 4°C in the dark and washed with 2% FBS in PBS solution. All data were generated using a FACSCalibur and analyzed using Summit V4.3 software (Dako, Houston, TX). Analysis was performed on 200,000 cells per sample, and results are representative of two independent experiments.
Cell migration assay
Migration was assessed using a 96-well Transwell migration plate (Millipore, Bellerica, MA) with 8 μm pore size. Polycarbonate membranes were coated one day before seeding cells with fibronectin or type I collagen (BD Biosciences, Bedford, MA) and cells were prepared by serum starvation in serum-free medium overnight, trypsinized and incubated with or without an PDGFRβ neutralization antibody (AF385, R&D Systems) 1 hour before seeding. Recombinant PDGF-BB protein (BD Biosciences, Bedford, MA) in serum-free medium was added to the lower chamber at 50 ng/ml, and 1×104 cells per insert were added to the upper chamber. Cells were incubated at 37°C for 6 hours, washed, fixed with 2% paraformaldehyde, stained with 0.1% Crystal Violet for 10 minutes, and counted manually at 10× magnification. Data represent an average of three experiments and six technical replicates per experiment.
Bisulfite sequencing
DNA was purified and bisulfite treated using EZ DNA methylation (Zymo, Irvine, CA). Converted DNA was amplified by RT-PCR to include CpG islands within the promoter region of specified genes PDGFRB, OCT4, COL1A1, COL3A1 and MSX1. Amplification primers were designed to span regions of the promoter containing high CpG content using Methprimer (Bock et al., 2005) and methylation data is displayed to show methylated (black) or unmethylated (white) circles at each CpG site within the promoter.
Acknowledgements
We would like to thank Nimet Maherali and Konrad Hochedlinger for the BJ iPS cell line, Laurence Daheron for additional iPS cell lines, and Judith Edwards for help in preparation of this manuscript.
Footnotes
Funding
This work was support by the National Institute of Craniofacial and Dental Research [grant number #DE017413-01A1 to J.A.G.] and the National Institute of Mental Health [grant number R33MH087896 to S.D.S. and S.J.H.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.099192/-/DC1
References
- Abramsson A., Lindblom P., Betsholtz C. (2003). Endothelial and none-ndothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J. Clin. Invest. 112, 1142-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J., Gallini R., Betsholtz C. (2008). Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda P., Agirre X., Ballestar E., Andreu E. J., Román-Gómez J., Prieto I., Martín-Subero J. I., Cigudosa J. C., Siebert R., Esteller M., et al. (2009). Epigenetic signatures associated with different levels of differentiation potential in human stem cells. PLoS ONE 4, e7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajada S., Mazakova I., Richardson J. B., Ashammakhi N. (2008). Updates on stem cells and their applications in regenerative medicine. J. Tissue Eng. Regen. Med. 2, 169-183 [DOI] [PubMed] [Google Scholar]
- Ball S. G., Shuttleworth C. A., Kielty C. M. (2007). Mesenchymal stem cells and neovascularization: role of platelet-derived growth factor receptors. J. Cell. Mol. Med. 11, 1012-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T., Willis L. M., Socci N. D., Studer L. (2005). Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2, e161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo M. E., Locatelli F., Fibbe W. E. (2009). Mesenchymal stromal cells. Ann. N. Y. Acad. Sci. 1176, 101-117 [DOI] [PubMed] [Google Scholar]
- Betsholtz C. (2004). Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev. 15, 215-228 [DOI] [PubMed] [Google Scholar]
- Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. (2005). BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21, 4067-4068 [DOI] [PubMed] [Google Scholar]
- Bonner J. C. (2004). Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev. 15, 255-273 [DOI] [PubMed] [Google Scholar]
- Chen T., Ueda Y., Dodge J. E., Wang Z., Li E. (2003). Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23, 5594-5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Meirelles L., Caplan A. I., Nardi N. B. (2008). In search of the in vivo identity of mesenchymal stem cells. Stem Cells 26, 2287-2299 [DOI] [PubMed] [Google Scholar]
- Egging D. F., van Vlijmen I., Starcher B., Gijsen Y., Zweers M. C., Blankevoort L., Bristow J., Schalkwijk J. (2006). Dermal connective tissue development in mice: an essential role for tenascin-X. Cell Tissue Res. 323, 465-474 [DOI] [PubMed] [Google Scholar]
- English K., French A., Wood K. J. (2010). Mesenchymal stromal cells: facilitators of successful transplantation? Cell Stem Cell 7, 431-442 [DOI] [PubMed] [Google Scholar]
- Fernandez A. F., Assenov Y., Martin-Subero J. I., Balint B., Siebert R., Taniguchi H., Yamamoto H., Hidalgo M., Tan A. C., Galm O., et al. (2012). A DNA methylation fingerprint of 1,628 human samples. Genome Res. 22, 407-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Bröcker E.-B. (2000). The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 57, 41-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q., Yoshida T., McDonald O. G., Owens G. K. (2007). Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells 25, 2-9 [DOI] [PubMed] [Google Scholar]
- Gao Z., Sasaoka T., Fujimori T., Oya T., Ishii Y., Sabit H., Kawaguchi M., Kurotaki Y., Naito M., Wada T., et al. (2005). Deletion of the PDGFR-beta gene affects key fibroblast functions important for wound healing. J. Biol. Chem. 280, 9375-9389 [DOI] [PubMed] [Google Scholar]
- Heldin C. H., Westermark B. (1999). Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283-1316 [DOI] [PubMed] [Google Scholar]
- Hewitt K. J., Shamis Y., Carlson M. W., Aberdam E., Aberdam D., Garlick J. A. (2009). Three-dimensional epithelial tissues generated from human embryonic stem cells. Tissue Eng. Part A 15, 3417-3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt K. J., Shamis Y., Hayman R. B., Margvelashvili M., Dong S., Carlson M. W., Garlick J. A. (2011). Epigenetic and phenotypic profile of fibroblasts derived from induced pluripotent stem cells. PLoS ONE 6, e17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupkes M., van Someren E. P., Middelkamp S. H., Piek E., van Zoelen E. J., Dechering K. J. (2011). DNA methylation restricts spontaneous multi-lineage differentiation of mesenchymal progenitor cells, but is stable during growth factor-induced terminal differentiation. Biochim. Biophys. Acta 1813, 839-849 [DOI] [PubMed] [Google Scholar]
- Khavari D. A., Sen G. L., Rinn J. L. (2010). DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9, 3880-3883 [DOI] [PubMed] [Google Scholar]
- Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C. T., Low H. M., Kin Sung K. W., Rigoutsos I., Loring J., et al. (2010). Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q., Zhang Y., Zhang J., Zhang H. K., Wu X., Zhang Y., Lam F. F., Kang S., Xia J. C., Lai W. H., et al. (2010). Functional mesenchymal stem cells derived from human induced pluripotent stem cells attenuate limb ischemia in mice. Circulation 121, 1113-1123 [DOI] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Kida Y. S., Hawkins R. D., Nery J. R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55-70 [DOI] [PubMed] [Google Scholar]
- Majore I., Moretti P., Stahl F., Hass R., Kasper C. (2011). Growth and differentiation properties of mesenchymal stromal cell populations derived from whole human umbilical cord. Stem Cell Rev. 7, 17-31 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noer A., Sørensen A. L., Boquest A. C., Collas P. (2006). Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol. Biol. Cell 17, 3543-3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., Guo T., Qi Z., Downey S. L., Manos P. D., Rossi D. J., et al. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M., Jaenisch R. (2011). De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev. 25, 1035-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce G. F., Tarpley J. E., Yanagihara D., Mustoe T. A., Fox G. M., Thomason A. (1992). Platelet-derived growth factor (BB homodimer), transforming growth factor-beta 1, and basic fibroblast growth factor in dermal wound healing. Neovessel and matrix formation and cessation of repair. Am. J. Pathol. 140, 1375-1388 [PMC free article] [PubMed] [Google Scholar]
- Polo J. M., Liu S., Figueroa M. E., Kulalert W., Eminli S., Tan K. Y., Apostolou E., Stadtfeld M., Li Y., Shioda T., et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyani T., Ronfard V., Scott P. G., Dodd C. M., Ahmed A., Gallo R. L., Parenteau N. L. (2009). De novo synthesis of human dermis in vitro in the absence of a three-dimensional scaffold. In Vitro Cell. Dev. Biol. Anim. 45, 430-441 [DOI] [PubMed] [Google Scholar]
- Riazi A. M., Kwon S. Y., Stanford W. L. (2009). Stem cell sources for regenerative medicine. Methods Mol. Biol. 482, 55-90 [DOI] [PubMed] [Google Scholar]
- Shamis Y., Hewitt K. J., Carlson M. W., Margvelashvilli M., Dong S., Kuo C. K., Daheron L., Egles C., Garlick J. A. (2011). Fibroblasts derived from human embryonic stem cells direct development and repair of 3D human skin equivalents. Stem Cell Res. Ther. 2, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S. D., Theriault K. M., Reis S. A., Zhou F., Madison J. M., Daheron L., Loring J. F., Haggarty S. J. (2011). Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE 6, e26203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen A. L., Timoskainen S., West F. D., Vekterud K., Boquest A. C., Ahrlund-Richter L., Stice S. L., Collas P. (2010). Lineage-specific promoter DNA methylation patterns segregate adult progenitor cell types. Stem Cells Dev. 19, 1257-1266 [DOI] [PubMed] [Google Scholar]
- Sottile J., Shi F., Rublyevska I., Chiang H. Y., Lust J., Chandler J. (2007). Fibronectin-dependent collagen I deposition modulates the cell response to fibronectin. Am. J. Physiol. Cell Physiol. 293, C1934-C1946 [DOI] [PubMed] [Google Scholar]
- Steed D. L. (1995). Clinical evaluation of recombinant human platelet-derived growth factor for the treatment of lower extremity diabetic ulcers. J. Vasc. Surg. 21, 71-78 discussion 79-81 [DOI] [PubMed] [Google Scholar]
- Suhr S. T., Chang E. A., Rodriguez R. M., Wang K., Ross P. J., Beyhan Z., Murthy S., Cibelli J. B. (2009). Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE 4, e8124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr S. T., Chang E. A., Tjong J., Alcasid N., Perkins G. A., Goissis M. D., Ellisman M. H., Perez G. I., Cibelli J. B. (2010). Mitochondrial rejuvenation after induced pluripotency. PLoS ONE 5, e14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145-1147 [DOI] [PubMed] [Google Scholar]
- Trojanowska M. (2008). Role of PDGF in fibrotic diseases and systemic sclerosis. Rheumatology (Oxford) 47 Suppl 5, v2-v4 [DOI] [PubMed] [Google Scholar]
- Veevers-Lowe J., Ball S. G., Shuttleworth A., Kielty C. M. (2011). Mesenchymal stem cell migration is regulated by fibronectin through α5β1-integrin-mediated activation of PDGFR-β and potentiation of growth factor signals. J. Cell Sci. 124, 1288-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiench M., John S., Baek S., Johnson T. A., Sung M. H., Escobar T., Simmons C. A., Pearce K. H., Biddie S. C., Sabo P. J., et al. (2011). DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 30, 3028-3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi E. S., Lee H., Yin S., Piguet P., Sarosi I., Kaufmann S., Tarpley J., Wang N. S., Ulich T. R. (1996). Platelet-derived growth factor causes pulmonary cell proliferation and collagen deposition in vivo. Am. J. Pathol. 149, 539-548 [PMC free article] [PubMed] [Google Scholar]