Abstract
ADF/cofilin family proteins are essential regulators of actin cytoskeletal dynamics. Recent evidence also implicates cofilin in the regulation of mitochondrial function. Here, we identify new functional surfaces of cofilin that are linked with mitochondrial function and stress responses in the budding yeast Saccharomyces cerevisiae. Our data link surfaces of cofilin that are involved in separable activities of actin filament disassembly or stabilisation, to the regulation of mitochondrial morphology and the activation status of Ras, respectively. Importantly, charge alterations to conserved surfaces of cofilin that do not interfere with its actin regulatory activity lead to a dramatic increase in respiratory function that triggers a retrograde signal to upregulate a battery of ABC transporters and concurrent metabolic changes that support multi-drug resistance. We hypothesise that cofilin functions within a bio-sensing system that connects the cytoskeleton and mitochondrial function to environmental challenge.
Key words: ROS, Actin, Cofilin, Mitochondria, Multi-drug resistance, Yeast
Introduction
The actin cytoskeleton is a major cellular component that facilitates a plethora of essential functions. These include the generation and maintenance of cell morphology and polarity, endocytosis and vesicle trafficking, contractility and motility, and cell division (for a review, see Dominguez and Holmes, 2011). The dynamic status of actin is regulated by accessory proteins that facilitate rapid assembly and disassembly of filaments. Cofilin is a member of the ADF/cofilin family of small actin-binding proteins found in all eukaryotic cells which function to dynamise the actin cytoskeleton (Van Troys et al., 2008). Cofilin has the ability to destabilise and promote severing of actin filaments in a pH-dependent manner (Hawkins et al., 1993; Hayden et al., 1993) and promotes the recycling of actin monomers for further polymerisation events. Such severing activity can, depending on the local concentration of free G-actin, generate new free barbed ends, which might promote the polymerisation of new filaments (Maciver et al., 1998; Pope et al., 2000) or alternatively, increase the rate of depolymerisation (Hayden et al., 1993). The ability of cofilin to bind to actin can be negatively regulated by phosphorylation which can be further regulated by the control of kinase or phosphatase functions by upstream factors such Rho-GTPases (Van Troys et al., 2008) and cAMP–PKA signalling (Nadella et al., 2009). In this manner, the regulation of cofilin and downstream control of actin dynamics can be tethered to environmental sensing. Cofilin–actin interactions are also negatively regulated by interactions with phosphoinositides, the most significant of which is with phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2]. Recent evidence suggests that electrostatic interactions that rely on dispersed lysine residues on one side of the cofilin molecule mediate interactions with multiple PtdIns(4,5)P2 headgroups in a manner that can lead to a clustering effect (Zhao et al., 2010). Because PtdIns(4,5)P2 can be found within multiple compartments of the cell, these new data raise the distinct possibility that cofilin–phospholipid interactions could provide an important mechanism by which cytoskeletal dynamics and signalling properties might be compartmentalised.
The regulation of cofilin function is also recognised as a factor associated with a number of disease states. Cofilin has also been linked to the development of mypothies, for example, a single point mutation that destabilises CFL-2 has been linked to nemaline myopathy (Agrawal et al., 2007). Cofilin and actin aggregates termed ADF/cofilin rods (ACRs) are commonly observed within post mortem Alzheimer's disease (AD) brains (Minamide et al., 2000) and under conditions of stress. The formation of ACRs is generally reversible; however, irreversible damage to mitochondria can lead to their persistent formation (Minamide et al., 2000). Experimental data suggest the formation of ACRs within neurons under stress can protect against a reduction of mitochondrial membrane potential and ATP. However, although ACR formation might transiently protect neurons under conditions of stress, their persistence is postulated to contribute to the loss of cognitive function during early stages of neurodegeneration (Bernstein et al., 2006; Minamide et al., 2000).
Further links between cofilin and mitochondrial function have been revealed in recent studies that have focused upon mitochondria dependent apoptosis. Induction of apoptosis has been shown to lead to a translocation of cofilin to mitochondria (Chua et al., 2003). Recent data also suggest that the oxidation of cysteine residues upon oxidative stress imparted by Taurine chloramine leads to the dissociation of cofilin from actin and its translocation to mitochondria where it stimulates mitochondria-dependent, Bax- or Bak-independent apoptosis (Klamt et al., 2009). However, other authors report that cofilin might also function within Bax-dependent apoptotic mechanisms (Posadas et al., 2012).
The sequence and structural conservation that exists between cofilins allows us to use model systems, such as yeast, to discover new functions that might be applicable to higher organisms. Yeast posses a single and essential cofilin, encoded by the COF1 gene (Lappalainen and Drubin, 1997). We made use of a library of isogenic yeast strains that express mutant forms of cofilin as the sole source. Within these strains, clusters of charged residues on the surface of cofilin have been mutated to alanines (Lappalainen et al., 1997). We report that cofilin mutations that stabilise F-actin trigger the hyperactivation of Ras, loss of mitochondrial function and apoptosis in yeast. Our analysis also revealed that small changes to the surface charge of cofilin in regions that are not located at the actin or PtdIns(4,5)P2 binding faces result in pronounced changes in mitochondrial function. Finally, we identify that mutation of cofilin can lead to the Ras-independent elevation of respiratory function by a novel post-translational mechanism that triggers a retrograde signal to induce a PDR1-dependent multi-drug-resistance response. We hypothesise that cofilin functions as a biosensor, which can integrate cytoskeletal regulation with mitochondrial function and stress response.
Results
Cofilin surface charge alterations affect mitochondrial function
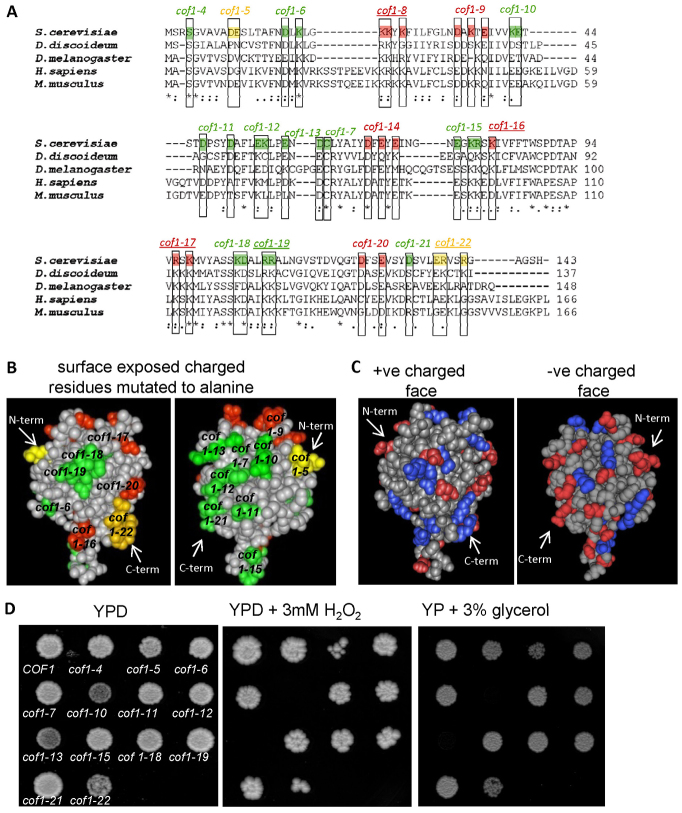
A previous study generated an isogenic series of yeast strains expressing cofilin alleles in which surface charged residues were replaced with alanine (Lappalainen et al., 1997). The actin regulatory role of cofilin is essential, and charged residues that are considered crucial for actin regulation, and thus lethal when mutated to alanine, are highlighted in red in Fig. 1A,B. The loss of charge at further residues, namely D10A, E11A in the cof1-5 allele and E134A,R135A,R138A in the cof1-22 allele (labelled in yellow in Fig. 1A,B) also lead to effects upon the actin cytoskeleton, but these can be tolerated in vivo (Lappalainen et al., 1997). Significant conservation exists with regard to both primary amino acid sequence and structural folding amongst cofilin proteins (Fig. 1A). Residues that have been identified as important for PtdIns(4,5)P2 binding in yeast (Ojala et al., 2001) are highlighted by underlining the allele name above highlighted residues in Fig. 1A. They are located at the positively charged face of the cofilin molecule (Fig. 1B,C). No functions have yet been assigned to many conserved charged residues that are located outside the known actin and PtdIns(4,5)P2 binding interfaces of cofilin (highlighted in green in the left panel of Fig. 1B and in blue and red in the left panel of Fig. 1C). The purpose of this study was primarily to search for new roles for cofilin that could be assigned to these previously unmapped surfaces.
Fig. 1.
Analysis of recognised actin and non-actin regulatory surfaces of cofilin family proteins. Alignment of yeast cofilin (Saccharomyces cerevisiae NCBI accession; AAA13256.1), slime mould cofilin (Dictyostelium discoideum; NCBI accession BAA07198.1), twinstar from fruit fly (Drosophila melanogaster; NCBI accession NP_477034.1), human cofilin (Homo sapiens; NCBI accession CAA64685.1) and mouse cofilin (Mus musculus; Accession: AAA37433.1). Residues essential for actin binding and for viability in yeast are coloured red. Those that affect actin dynamics but which are not essential in yeast are coloured in yellow and those that do not affect yeast growth are labelled in green. The names of cofilin mutant alleles that impair normal PtdIns(4,5)P2 binding abilities of cofilin are underlined. (A) Space-filling models of the 3.00 Å yeast cofilin crystal structure (Protein databank reference 1QPV) with residues involved in actin binding (red and yellow) and not involved in actin regulation (green) highlighted. (B) Surface charged residues are depicted in red (−ve) and blue (+ve) on the yeast cofilin space-filling model. (C) Wild-type (COF1) and cofilin mutant strains were grown as colonies on agar plates containing YPD, YPD + 3 mM H2O2 or YP + 3% glycerol for 3 days (D).
Cofilin might play a role in the regulation of mitochondria (Bernstein et al., 2006). We therefore investigated whether the alteration of cofilin surface charge led to phenotypes indicative of altered mitochondrial function. Initially, mutant strains were spotted onto solid medium containing the oxidant H2O2 (Fig. 1D). Although all strains grew on rich YP medium containing 2% glucose, mutants expressing the cof1-5 and cof1-22 alleles exhibited sensitivity to H2O2, whereas the cof1-10 and cof1-13 strains failed to grow (Fig. 1D). Interestingly, the pattern of growth on glycerol-containing plates, which requires functional mitochondria, was identical to that seen under conditions of oxidative stress (Fig. 1D).
Cofilin plays a role in the regulation of respiration and ROS production
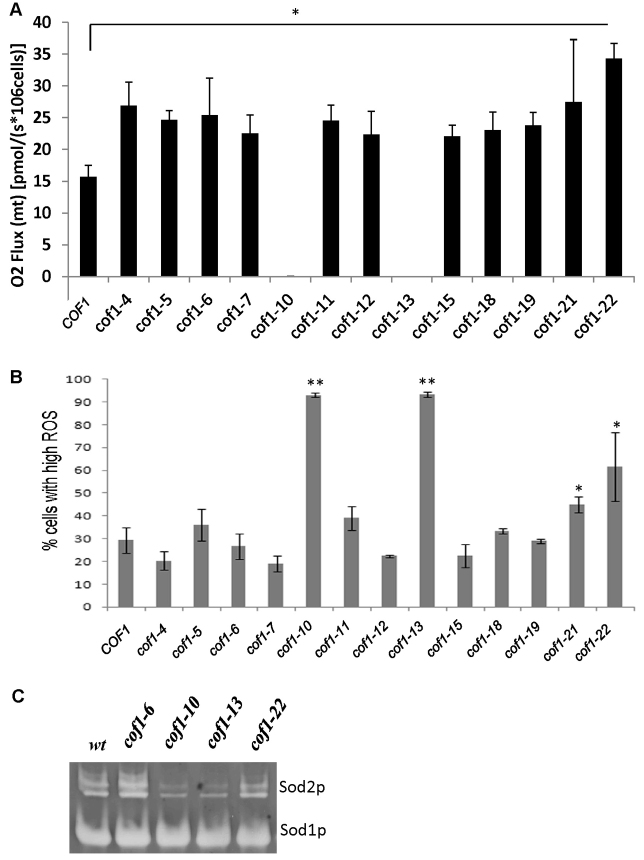
To examine the role of cofilin in regulating mitochondrial function in more detail, we performed high-resolution respirometry (Fig. 2A). Using this technique, changes in oxygen consumption at the cytochrome c oxidase (COX) complex (Complex IV), can be accurately measured within living cells, and is a direct measure of electron transport chain (ETC) activity. Initially, we assessed the effects of cofilin surface charge alteration upon routine respiration (Fig. 2A). In line with their inability to grow on glycerol-containing media, cof1-10 and cof1-13 strains failed to respire (Fig. 2A). However, cof1-5 and cof1-22 strains, which displayed reduced growth on glycerol and displayed sensitivity to H2O2 showed a significant increase in routine respiration (Fig. 2A). In fact, with the exception of strains expressing cof1-10 and cof1-13, all of the cofilin mutant alleles tested exhibited significantly elevated levels of routine respiration when compared with the wild type (Fig. 2A).
Fig. 2.
Cofilin plays a role in the regulation of respiration and ROS production. Cells were grown in triplicate for 24 hours to diauxic shift in YPD. (A) Mitochondrial respiration was measured in a known number of whole cells and (B) ROS accumulation was assessed using the indicator dye H2DCF-DA by flow cytometry. (C) Superoxide dismutase activity present in protein extracts from wild type, cof1-6, cof1-10, cof1-13 and cof1-22 (Sod1p and Sod2p) strains grown for 24 hours to diauxic shift in YPD were assessed after separation on separated in a native gel. Statistical significance with reference to wild type was assessed using a Student's t-test, *P<0.05, **P<0.01.
Changes in respiratory activity can be associated with measurable differences in the production of reactive oxygen species (ROS), a natural by-product of aerobic growth. We therefore assessed the effects of cofilin surface charge alteration on ROS levels, using the reporter dye H2DCF-DA (Fig. 2B). Four strains demonstrated significantly higher numbers of ROS-producing cells when compared with the wild type. Two of these high-ROS-producing strains were those expressing the cof1-10 and cof1-13 alleles, which we had previously determined not to have detectable respiratory function and which were sensitive to H2O2 (Fig. 2A; Fig. 1D). Strains expressing the cof1-21 and cof1-22 alleles also showed elevated numbers of ROS-producing cells (Fig. 2B). Interestingly, despite exhibiting elevated levels of respiratory activity, we did not observe increased ROS production in all other strains tested (Fig. 2B).
ROS accumulation can occur as a result of altered ETC activity as well as changes in the expression or activity of detoxification enzymes. We therefore tested whether the activity of the antioxidant enzymes Sod1p and Sod2p were affected in cofilin surface charge mutant strains using an in gel in vitro assay (Fig. 2C). Although Sod1p activity appeared to remain constant in all strains, the level of Sod2p was observed to change. Sod2p activity appeared elevated in the high-respiring cof1-6 strain, which maintains wild-type ROS levels (Fig. 2B; Fig. 3C), but reduced in the high-ROS-producing cof1-10 and cof1-13 mutant strains (Fig. 2C). Interestingly, Sod2p activity was not increased in the cof1-22 strain, which exhibits elevated respiration and higher ROS levels (Fig. 2C).
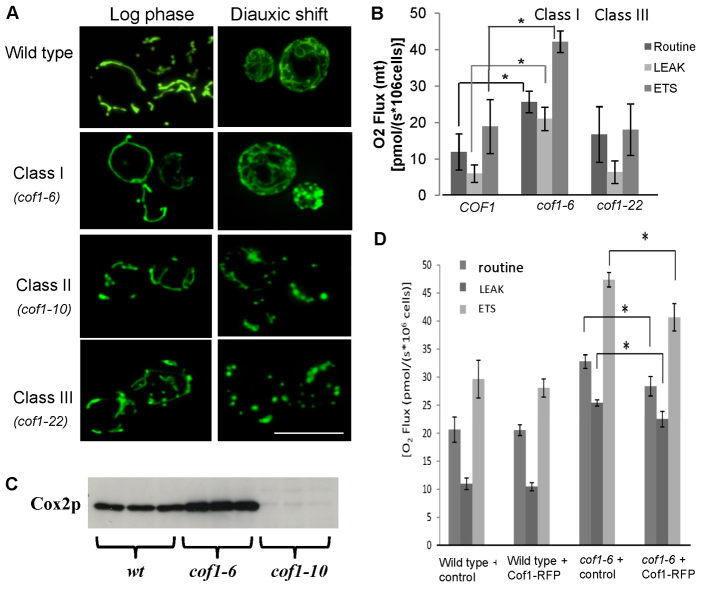
Fig. 3.
Analysis of the effects of cofilin mutation on mitochondrial morphology and respiratory performance. Mutants with similar phenotypes were grouped into Class I (cof1-4, cof1-6, cof1-7, cof1-11, cof1-12, cof1-15, cof1-18, cof1-19 and cof1-21), Class II (cof1-10 and cof1-13) and Class III (cof1-5 and cof1-22). (A) Routine, LEAK (by addition of TET, 150 μM) and ETS (by addition of FCCP, 12 μM) respiration were measured in triplicate wild-type, Class I and Class III cofilin mutants grown for 24 hours to diauxic shift in YPD. Mean values ± s.d. are presented. (B) Protein was extracted from a known number of cells and Cox2p levels were assessed in wild-type, cof1-6 and cof1-10 cells grown for 24 hours to diauxic shift in YPD. (C) An image of the Coomassie-stained gel used in this western blotting experiment is provided in supplementary material Fig. S3. A plasmid expressing a functional Cof1–RFP fusion protein was introduced into wild-type or cof1-6 expressing cells and grown for 24 hours to diauxic shift before routine, LEAK and ETS respiration levels were assessed as above. (D) Statistical significance was assessed using a Student's t-test, *P<0.05. Scale bar: 10 μm.
Mutations in cofilin influence mitochondrial morphology, function and biogenesis
Changes in mitochondrial morphology are often associated with altered function. We therefore characterised the effects of cofilin mutation upon mitochondrial morphology (Fig. 3A). As we observed identical phenotypes for particular cofilin mutant strains, we grouped them as either wild type, Class I (cof1-4, cof1-6, cof1-7, cof1-11, cof1-12, cof1-15, cof1-18, cof1-19 and cof1-21), Class II (cof1-10 and cof1-13) or Class III (cof1-5 and cof1-22). Mitochondria in all strains, with the exception of Class III mutants, were indistinguishable from the wild type during log-phase growth, appearing as a tubular network that extended throughout mother and daughter cells (Fig. 3A). Cells from the Class III mutant strains, cof1-5 and cof1-22, which are known to have impaired actin regulatory function (Lappalainen et al., 1997), showed a partially fragmented morphology during log phase (Fig. 3A). During diauxic shift, cells cultured in glucose-containing media undergo mitochondrial biogenesis as they shift to a respiratory mode and use the ethanol produced during fermentative growth. The elaboration of mitochondrial membranes during this period of metabolic change can be observed in wild-type cells (Fig. 3A). In the Class I mutants, mitochondrial biogenesis occurred as in wild-type cells, despite their apparent elevated respiratory activity (a representative image of class I mutants is shown in Fig. 3A). In Class II (cof1-10 and cof1-13) and Class III mutant (cof1-5 and cof1-22) strains, mitochondria did not proliferate during the diauxic shift period of growth, and appeared fragmented and enlarged (representative images of Class II and III mutants are shown in Fig. 3A).
To investigate the effects of cofilin mutants upon mitochondria further we performed a more detailed analysis of electron transport chain (ETC) function using high-resolution respirometry. To do this, we applied a number of cell-permeable ETC inhibitors in sequence to wild-type and cofilin mutant strains within the respirometer sample chamber and assessed their effects on oxygen consumption. After measurement of routine respiration, the ATP synthase and Complex V inhibitor triethyltin bromide (TET) (Cain and Griffiths, 1977) was introduced at a concentration of 150 μM to determine LEAK respiration (Fig. 3B). LEAK represents non-phosphorylating respiration that occurs as a result of proton leakage back across the inner mitochondrial membrane. A Tet-induced block of the proton pumping function of Complex V results in an accumulation of protons within the intermembrane space that inhibits electron flow. Any ETC activity measured therefore results from the leak of protons across the inner mitochondrial membrane itself. Cells were then exposed to 12 μM FCCP, a protonophore that dissipates the mitochondrial membrane potential. Because the proton gradient established across the inner membrane is a major control point of the ETC, its removal allows the ETC to run at its maximum capacity (denoted as ETS in Fig. 3B). A typical wild-type (healthy) respiratory profile should show that LEAK oxygen consumption is significantly reduced compared with that of routine respiration, with the ETS respiration significantly higher (Fig. 3B).
Analysis of Class I cofilin mutants revealed that the elevated level of routine respiration was accompanied by a very large increase in ETS (data for cof1-6 respirometry, as an example of Class I mutants, is shown in Fig. 3B, whereas the results for all other strains, except Class II, can be found in supplementary material Fig. S1). In addition, the level of LEAK respiration of Class I mutants was found to be similar to the routine level of respiration (Fig. 3B; supplementary material Fig. S1). Class III mutants displayed a very different profile, with the routine and maximal (ETS) respiratory levels being the same (Fig. 3B). The ratio of routine/LEAK respiration (R/L), gives an indication as to how much control is being exerted by the proton-pumping activity of Complex V. Our analysis suggests that a trend exists whereby Class I cofilin mutant strains exhibit a reduction in the R/L ratio, whereas Class III mutants exhibit an increase, when compared with the wild type (supplementary material Fig. S2). These data imply that the mitochondrial inner membrane of Class I cofilin mutants does not rely on Complex V activity as a major point of control of the ETC and protons might move across the inner membrane by another means.
Our data suggest that mutations in cofilin lead to altered respiratory output. We therefore examined whether mitochondrial (Cox2p) COX component protein levels were influenced by these mutations. In all mutants where respiration was elevated, we found that Cox2 levels were increased (Fig. 3C shows cof1-6 as an example). By contrast, the levels of COX complex components in respiratory incompetent cof1-10 and cof1-13 mutant strains were greatly reduced (Fig. 3C shows cof1-10 as an example).
To verify that elevated levels of respiration observed in Fig. 4B were attributable to expression of the mutant cof1-6 allele, we introduced a plasmid expressing a wild-type Cof1–RFP fusion protein that has previously been demonstrated to be functional in vivo (Lin et al., 2010). Our rationale was that we should observe a reduction in the level of the elevated respiration observed in cof1-6-expressing cells, when Cof1–RFP is co-expressed, as a result of competition for cellular processes between wild-type and mutant cofilin proteins. Importantly, we found that expression of Cof1–RFP in wild-type cells did not alter mitochondrial performance as assessed by high-resolution respirometry (Fig. 3D). However, we did observe a significant and reproducible reduction in respiration in cof1-6-expressing cells that co-expressed Cof1–RFP, when compared with levels in controls (Fig. 3D).
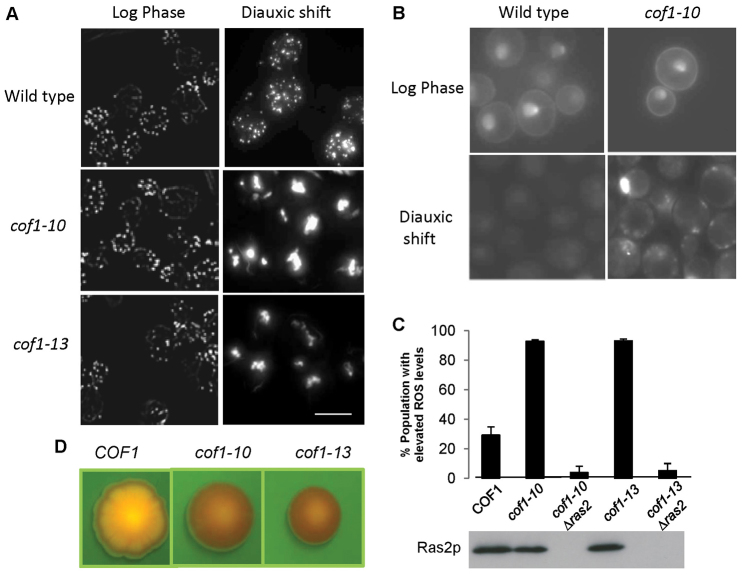
Fig. 4.
Expression of cof1-10 and cof1-13 alleles stabilises F-actin and triggers Ras hyperactivation and ROS production. F-actin was stained using Rhodamine–phalloidin in fixed wild-type, cof1-10 and cof1-13 cells grown to either log or diauxic shift in YPD. (A) The localisation of active Ras was assessed in vivo in wild-type and cof1-10 cells grown to either log or diauxic shift in YPD. (B) ROS levels were assessed in wild type (COF1), cof1-10, cof1-10Δras2, cof1-13 and cof1-10Δras2 grown for 24 hours in YPD to diauxic shift using H2DFC-DA. (C) Western blot was carried out to confirm the deletion of RAS2 in cof1-10 and cof1-13 strains. (D) Cell death was assessed in wild-type, cof1-10 and cof1-13 colonies by growing on YPD agar plates containing 10 μg/ml Phloxine B and visualised using a GFP filter set under a dissecting microscope. Because Phloxine B is actively pumped out of healthy cells, its accumulation marks regions within the colony that possess dead or dying cells. Scale bars: 10 μm.
Our previous work has shown that elevated Ras activity can lead to higher respiratory rates by the elevation of cAMP–PKA activity (Leadsham and Gourlay, 2010). To determine whether the elevation of respiration observed occurred as a result of elevated cAMP levels, we overexpressed the high-affinity phosphodiesterase PDE2, which hydrolyses cAMP and reduces intracellular levels of this secondary messenger (Wilson et al., 1993). Overexpression of PDE2 did not reduce the respiratory rate of cof1-6-expressing cells but in fact caused a significant increase (supplementary material Fig. S4). This suggests that elevated cAMP–PKA signalling levels are not the cause of increased respiratory levels observed in cofilin surface-charge mutants.
Expression of cof1-10 and cof1-13 alleles stabilises F-actin and triggers Ras hyperactivation and ROS production
Previous work suggests that the expression of cof1-10 and cof1-13 alleles has no effect upon growth or the regulation of the actin cytoskeleton (Lappalainen et al., 1997). In line with this view, we could find no differences between the cytoskeletal phenotype observed in wild-type, cof1-10 or cof1-13 cells during log growth in glucose-containing medium (Fig. 4A). During diauxic shift, F-actin can be observed as numerous punctuate foci in wild-type cells; however, the F-actin cytoskeleton in cof1-10 and cof1-13 cells accumulated into large F-actin aggregations during this phase of growth (Fig. 4A). Our previous work has identified the premature aggregation of F-actin as a trigger for the hyperactivation of the small GTPase Ras, which leads to an elevation of cAMP–PKA activity and to the induction of dysfunctional ROS-producing mitochondria that drive apoptosis (Gourlay and Ayscough, 2006). To test whether this was also the case in cof1-10 and cof1-13 strains, we used a probe that fuses three consecutive active Ras binding domains from the human Raf1 protein to GFP. Our previous work has shown that this probe binds to and highlights areas of concentrated active Ras within living yeast cells (Leadsham et al., 2009). In log-phase cells, active Ras could be found at the plasma membrane and within the nuclei of wild-type and cof1-10 cells (Fig. 4B). Similar results were also found in cof1-13 cells at this stage of growth. During diauxic shift, the active Ras probe was found as a weak and diffuse signal because Ras becomes de-activated during nutrient depletion (Fig. 4B). However, in strains expressing cof1-10 (Fig. 4B) and cof1-13, Ras proteins remained active within intracellular foci (Fig. 4B). To confirm that Ras pathway activity is the source of ROS within cof1-10 and cof1-13 cells, we deleted RAS2 in these strains and confirmed the loss of expression by western blot (Fig. 4C). The accumulation of ROS was then examined using the indicator dye H2DCF-DA (Fig. 4C). We observed that the deletion of Ras2 almost completely prevented the accumulation of ROS within cof1-10- and cof1-13-expressing populations (Fig. 4C). ROS accumulation has been shown to lead to the stabilisation of F-actin (Farah et al., 2011) and might account for the aggregation phenotype observed. However, we found that cof1-10Δras2 and cof1-13Δras2 cells still accumulated F-actin aggregates (supplementary material Fig. S5). The failure to control Ras activation has been shown to result in elevated levels of cell death (Leadsham et al., 2009); this was confirmed by growing wild-type, cof1-10 and cof1-13 colonies on medium containing the cell death marker Phloxine B (Fig. 4D). Phloxine B dye accumulates within dead and dying cells and is highly fluorescent, allowing visualization within colonies under UV illumination. Although wild-type colonies developed a striking pattern of Phloxine B accumulation, indicating the controlled zonation of cell death, cof1-10- and cof1-13-expressing colonies showed heavy dye accumulation over the entire colony (Fig. 4D). This finding is consistent with the observed failure to control Ras signalling and suggests that widespread cell death occurs within colonies formed from cells expressing cof1-10 or cof1-13.
Transcriptome analysis reveals a role for cofilin in the regulation of multi-drug resistance and fatty acid β-oxidation
Our data suggested that certain cofilin surface charge alterations can influence metabolic status. We therefore sought to determine the transcriptional alterations associated with the elevated respiratory rate observed in Class I cofilin mutants. To this end, we compared the transcriptomes of wild-type and cof1-6 (D18A, K20A)-expressing cells grown in YPD medium for 24 hours with the diauxic phase of growth. Using this approach we identified 69 genes as being significantly upregulated (log2 fold change ≥1, P≤0.05) and 23 downregulated (log2 fold change ≥1, P≤0.05). The genes assigned as upregulated or downregulated are documented in supplementary material Table S2 and Table S3. Surprisingly, because cof1-6 cells displayed elevated levels of respiration, we did not observe the upregulation of genes that are associated with ECT function (supplementary material Table S3). These data suggest that the effects of cof1-6 on respiration are not achieved by engaging mitochondrial biogenesis by the upregulation of nuclear-encoded genes. To address this, we used a well-known reporter for the activity of the mitochondrial biogenesis transcription factor HAP4 (Lascaris et al., 2004). The reporter construct assesses levels of a lacZ reporter that is driven by the CYC1 promoter, which contains multiple HAP4 binding sites (Mösch et al., 1999). In line with the microarray data, we found that levels of HAP4 activity were not elevated in cof1-6 cells when compared with the wild type, and in fact showed a decrease in the level of lacZ transcription (Fig. 5A).
Fig. 5.
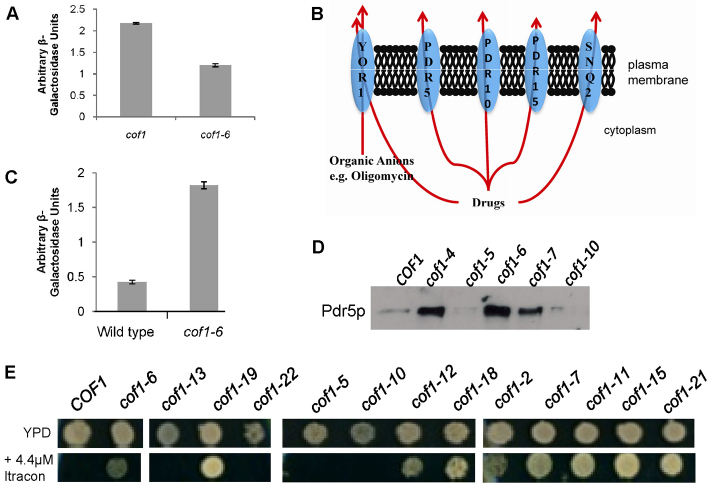
Transcriptome analysis reveals a role for cofilin in the regulation of multi-drug resistance and fatty acid β-oxidation. (A) Activity of the HAP4 transcription factor was estimated in wild-type and cof1-6 cells using a lacZ reporter gene and CYC1 promoter construct. (B) Following a comparative transcriptomic analysis, we identified a number of plasma membrane transporters that were significantly upregulated in cof1-6 when compared with wild-type cells. (C) To examine the regulation of PDR5 transcription in wild-type and cof1-6 cells, we used a construct containing a lacZ reporter upstream of the PDR5 promoter and conducted a β-galactosidase assay. (D) We also examined Pdr5p protein levels by western blotting in wild-type (COF1), cof1-4, cof1-5, cof1-6, cof1-7 and cof1-10 strains using an antibody against Pdr5p. (E) Growth of wild-type (COF1) and cofilin mutant strains was assessed on YPD or YPD + 4.4 μM itraconazole containing agar plates.
We did, however, note the differential expression of a number of plasma-membrane-localised transporters when comparing cof1-6 with the wild type (supplementary material Table S3). Of particular interest were the upregulation of a cluster of ATP-binding cassette (ABC) transporters (YOR1, PDR5, PDR10, PDR15 and SNQ2), which are known to be required for the acquisition of drug resistance (Fig. 5B). To confirm the array data, we examined the levels of PDR5 expression in cof1-6 and wild-type cells using a PDR5 promoter lacZ fusion reporter construct (Hallstrom and Moye-Rowley, 2000). In line with the microarray data, we were able to detect a significant increase in the transcription of lacZ from the PDR5 promoter in cof1-6 cells when compared with the wild type (Fig. 5C). We also confirmed that Pdr5p protein levels were significantly elevated in cof1-6 cells by western blotting (Fig. 5D). Interestingly, Pdr5p levels were also significantly increased in all of the Class I cofilin mutant strains (examples in cof1-4, cof1-6 and cof1-7 are shown in Fig. 5D), but not in class II (cof1-10) and III (cof1-5) mutant strains (Fig. 5D). To investigate the effects of cofilin mutation on drug sensitivity, we spotted wild-type and cofilin mutant strains onto YPD medium containing itraconazole, an antifungal that is a known ABC transporter substrate. Wild-type cells could not tolerate addition of itraconazole at a concentration of 4.4 μM (Fig. 5E). However, all mutants that we had previously grouped as Class I (high respiring) grew well on medium containing 4.4 μM Itraconazole (Fig. 5E). By contrast, Class II cofilin mutants (non-respiring) were more sensitive than the wild type to itraconazole (unpublished observations).
We also observed that the complete set of genes required for the peroxisomal process of fatty acid β-oxidation were upregulated in cof1-6 cells when compared with the wild-type strain (supplementary material Fig. S6 and Table S3). In support of cofilin surface charge alteration leading to the upregulation of fatty acid β-oxidation, we found that all Class I cofilin mutant strains were able to use oleic acid more readily than the wild-type strain could (unpublished observations). Interestingly, Class III mutant strains grew more slowly than the wild type when grown on oleic acid as the sole carbon source (unpublished observations). Additional confirmation of the microarray data quality was sought by introducing a peroxisome-targeted GFP-expressing plasmid under the control of the CTA1 (peroxisomal catalase) promoter into cof1-6 and wild-type cells (supplementary material Fig. S7). Using this approach, we found that the level of GFP–PTS1 fluorescence was considerably higher in cof1-6 cells than was observed in the wild type (supplementary material Fig. S7).
Cofilin-mediated azole resistance is dependent on the ABC transporter and requires the transcription factor PDR1
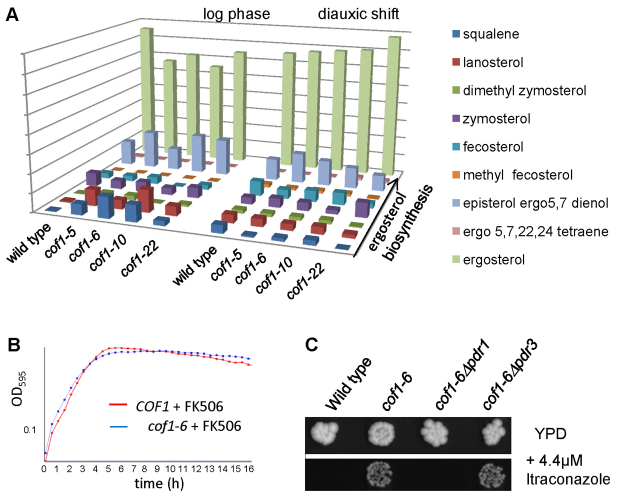
Our experiments suggest that changes to the surface charge of cofilin that elevate respiration also impart resistance to azole antifungals. The biosynthesis of ergosterol, a membrane sterol that is unique to fungi, is thought to be a primary target of azole antifungals (Vanden Bossche et al., 1987) and mutations that alter the flux of intermediates can lead to drug resistance. We therefore examined the levels of ergosterol biosynthesis intermediates in representatives of Class I, II and III cofilin mutants. An analysis of a number of biological replicates was carried out and typical data sets from log and post diauxic shift cultures are presented in Fig. 6A. During log-phase growth, we observed that cof1-5, cof1-6 (Class I) and cof1-10 (Class II) mutants did accumulate early pathway intermediates and exhibited reduced levels of ergosterol (Fig. 6A). However, at later stages of growth, there were no discernible differences (Fig. 6A), indicating that ergosterol biosynthesis is not grossly affected by mutations to cofilin. In light of this result, we examined whether the upregulation of ABC transporter activity was responsible for imparting drug resistance in cofilin mutant strains. Overexpression of PDR5 is sufficient to impart azole resistance in S. cerevisiae (Leppert et al., 1990). FK506 is a macrocyclic lactone and immunosuppressive agent that has been shown to specifically inhibit Pdr5p activity (Egner et al., 1998). We found that both cof1-6 and wild-type cells were inhibited equally by itraconazole when FK506 was present, indicating a direct role for Pdr5p in mediating drug resistance in cofilin mutants strains (Fig. 6B). PDR5 expression is regulated by the transcription factors PDR1 and PDR3. These transcriptional regulators control overlapping gene sets and are a major control point in the regulation of drug resistance (Shahi and Moye-Rowley, 2009). We deleted PDR1 and PDR3 individually in cof1-6 cells and examined the effects on sensitivity to itraconazole (Fig. 6C). Interestingly, although PDR1 deletion abolished the drug resistance observed in cof1-6 cells, the loss of PDR3 did not.
Fig. 6.
Cofilin-mediated azole resistance is dependent on the ABC transporter and requires the transcription factor PDR1. (A) The biosynthesis of ergosterol was assessed in log and post diauxic shift wild-type, cof1-5, cof1-6, cof1-10 and cof1-22 cells by measuring pathway intermediates using a Gas-chromatography/Mass spectrometry approach as described in the Materials and Methods. (B) The effects of adding the Pdr5p inhibitor FK506 (10 μg/ml) upon the sensitivity of cof1-6 cells to itraconazole (4.4 μM) was observed in liquid medium. (C) The influence of the transcription factors PDR1 and PDR3 upon resistance to itraconazole (4.4 μM) was investigated by comparing growth of wild-type, cof1-6, cof1-6Δpdr1 and cof1-6Δpdr3 cells.
Mitochondrial function is required for cofilin-mediated multi-drug resistance
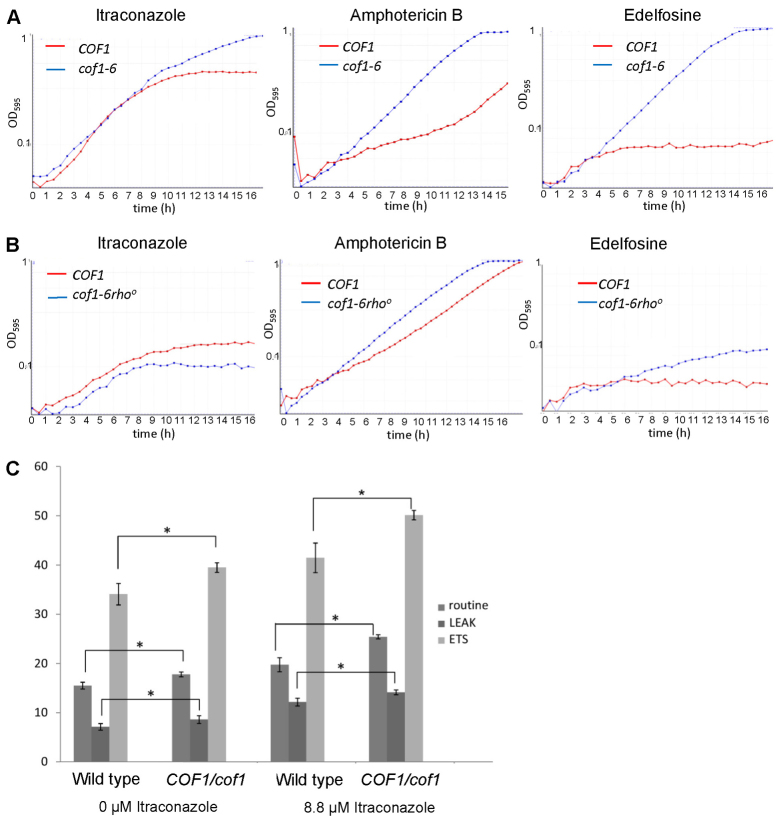
We next examined whether the alteration of cofilin surface charge imparted resistance to a broader range of antifungal drugs with different modes of action. The effects of itraconazole, amphotericin B and edelfosine exposure on growth of wild-type and cof1-6 cells in liquid culture were examined. Amphotericin B is generally thought to disrupt membrane integrity by association with ergosterol (Baginski and Czub, 2009). Edelfosine (1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine) is a lysophospholipid that is known to affect the composition of lipid rafts (Zaremberg et al., 2005). Cells expressing cof1-6 were found to be resistant to itraconazole, amphotericin B and edelfosine when compared with wild-type cells in liquid medium growth-rate assays (Fig. 7A). We then tested whether mitochondrial function is required for the resistance associated with the cof1-6 mutation by generation of a strain that lacks mitochondrial DNA (cof1-6 rho0) rendering it incapable of respiration. We found that the cof1-6-associated resistance to itraconazole and edelfosine was lost completely, whereas growth in amphotericin B was greatly reduced when mitochondrial function was impaired in this way (Fig. 7B). These data suggest mitochondrial function is required for the MDR response observed on cofilin mutant strains.
Fig. 7.
Cofilin-mediated multi-drug resistance requires mitochondrial function. (A,B) Growth of wild-type, cof1-6 cells and cof1-6rho0 cells in YPD liquid medium containing the antifungal compounds itraconazole (4.4 μM), amphotericin B (0.3 μg/ml) or edelfosine (10 μg/ml). (C) Routine, LEAK (by addition of TET, 150 μM) and ETS (by addition of FCCP, 12 μM) respiration were measured in triplicate wild-type and COF1,cof1 diploid cells grown for 24 hours to diauxic shift in YPD in the presence or absence of 8.8 μM itraconazole. Average values and s.d. are presented. Statistical significance with reference to wild type was assessed using a Student's t-test, *P<0.05.
To more directly assess whether mitochondrial function is engaged in response to anti-fungal exposure and whether cofilin is involved in this response we examined mitochondrial performance in wild-type and COF1,cof1 diploid cells, in which cofilin levels are reduced by approximately 25% (unpublished observations), grown in the presence or absence of itraconazole (Fig. 7C). Our analysis identified that routine, LEAK and ETS respiration are elevated in wild-type cells when exposed to itraconazole, suggesting that mitochondrial biogenesis is part of a physiological response to drug exposure. However, we also noted that whereas untreated COF1,cof1 cells displayed a small increase in respiratory potential, their exposure to itraconazole led to an exaggerated response when compared with the wild type (Fig. 7C). We also found that COF1,cof1 cells exhibited a reproducible increase in ROS production during diauxic shift (supplementary material Fig. S8) and a small increase in sensitivity to itraconazole (supplementary material Fig. S9). These data suggest a role for cofilin in mediating mitochondrial function in response to itraconazole exposure.
Discussion
Structural and mutagenesis studies have indicated that particular residues are crucial for interactions of cofilin with actin (Lappalainen et al., 1997). A number of residues on helix 3 are important for cofilin–actin interactions in yeast (Lappalainen et al., 1997). Arg96 and Lys98 on helix 3 play crucial roles in actin binding and dynamics (Lappalainen et al., 1997), whereas additional residues important for cofilin–F-actin interactions in yeast include those within the β5 strand (Arg80, Lys82), the loop between β6 and α4 (Asp123, Glu126), the α4 helix (Glu134, Arg135, Arg138) and within the disordered N-termini (Lappalainen et al., 1997). Our analysis of F-actin arrangement reveals that additional residues mutated in the cof1-10 (K42 and E43) and cof1-13 (E59 and D61) strains are important in maintaining a dynamic actin cytoskeleton during nutritional stress. In strains expressing the cof1-10 and cof1-13 alleles we found that the F-actin cytoskeleton becomes aggregated as cells start to become nutritionally depleted and enter the diauxic shift phase of growth. We found that the stabilisation of F-actin in cof1-10 and cof1-13 cells led to the hyperactivation of Ras, mitochondrial fragmentation and the build up of ROS. Oxidation of particular cysteine residues can also lead to actin stabilisation (Farah et al., 2011), raising the possibility that actin aggregates observed in cof1-10 and cof1-13 cells may occur as a result of ROS production. However, the deletion of RAS2 in cof1-10 and cof1-13 strains, which prevented a build-up of ROS, did not relieve the aggregation of F-actin. Because the mutations associated with cof1-10 and cof1-13 strains do not affect the actin cytoskeleton during log-phase growth, we propose that different modes of cofilin-based regulation of F-actin exist in vivo.
Little attention has focused on the conserved regions of cofilin that lie outside of the recognised actin binding sites. Our data suggest that conserved charged residues on cofilin, which are not directly implicated in the regulation of the actin cytoskeleton, have important functions within the cell. The mutation of residues over a broad non-actin-binding surface led to a significant increase in respiration by a mechanism that did not rely on engaging mitochondrial biogenesis at the level of the nuclear genome. We also observed that these mutations elevated the process of peroxisomal fatty acid β-oxidation. The production of acetyl CoA from the β-oxidation of fatty acids within the peroxisome is essential for mitochondrial function. One possibility is that cofilin acts to upregulate this peroxisomal function to support the upregulation of respiratory activity (Fig. 8). How cofilin surface charge alterations lead to the elevation of respiratory function remains elusive. One possibility is that, as in mammalian cells, cofilin is able to interact with the outer surface of mitochondria in response to stress, possibly by modulating actin dynamics at the organelle outer membrane. In mammalian cells, the oxidation of all four cysteine residues is required for translocation of cofilin to the mitochondrial surface (Klamt et al., 2009). However, because yeast cofilin possesses a single cysteine residue, it is unclear whether oxidative stress would lead to mitochondrial translocation in the same manner. Interestingly, we found that mutation of the single yeast cysteine (C62) to alanine led to the elevation of mitochondrial function. Because this cysteine is conserved in all cofilins, and does not appear to be essential for protein stability or growth, it might be that this residue plays an important signalling or regulatory function. Our preliminary data also suggest that a stress-specific interaction between cofilin–actin and the voltage-dependent ion channel exists in yeast in vivo (unpublished observations). Further evidence that cofilin can alter mitochondrial integrity comes from the high-resolution respirometry data presented in this study. Mutations in cofilin that elevated respiratory levels were also found to lead to an increase in the complex-V-independent movement of protons from the inner membrane space to the matrix. In addition, mitochondrial fragmentation that is probably coupled to the regulation of actin dynamics was observed in Class II (cof1-10 and cof1-13) and Class III (cof1-5 and cof1-22 mutant strains).
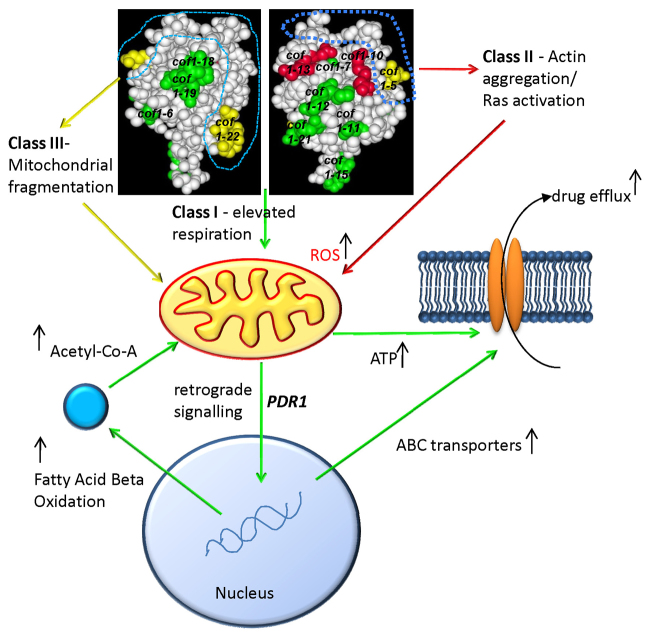
Fig. 8.
Cofilin function links the control of mitochondrial function to multi-drug resistance. Mutations that lie within the known actin binding face (Class III mutations), which are labelled in yellow, lead to fragmented mitochondria. Mutations of residues (Class II mutations) that lie outside of the defined actin-interacting face, demarcated by the blue dotted line, were found to stabilise F-actin during nutrient depletion. Actin aggregation in these mutants triggered the inappropriate activation of Ras, which in turn leads to the accumulation of ROS and cell death. The loss of charge in regions not associated with actin regulation results in an apparently Ras–cAMP–PKA-independent post-transcriptional upregulation of mitochondrial function. The altered mitochondrial function leads to a multi-drug resistance response that is dependent upon PDR1 and to the upregulation of peroxisomal fatty acid β-oxidation, which might support elevated levels of respiration. We also postulate that elevated mitochondrial function might provide the ATP required to fuel drug pump activity.
The binding of cofilin to PtdIns(4,5)P2 is also known to mediate cofilin–actin interactions. Some of the residues implicated in PtdIns(4,5)P2 binding (Ojala et al., 2001), K105/D106 (cof1-18) and R109/R110 (cof1-19) were included in our study and we found that these mutants displayed elevated levels of respiration. The residues implicated in binding to PtdIns(4,5)P2 are distributed widely over the positively charged surface of cofilin, and it has been suggested that this facilitates binding to multiple phospholipid molecules (Zhao et al., 2010). Our data show that the loss of surface charge over a wide area of the cofilin molecule leads to the acquisition of identical mitochondrial and drug resistance phenotypes. One possibility is therefore that the influence of cofilin upon mitochondrial function is mediated by PtdIns(4,5)P2 interaction. PtdIns(4,5)P2 can be found within many cellular membranes, including those of mitochondria, where it was shown to be important for the maintenance of fission–fusion control and mitophagy (Rosivatz and Woscholski, 2011). It is conceivable therefore that cofilin–PtdIns(4,5)P2 interaction might occur at multiple locations, and provide a mechanism through which the regulation of actin dynamics can be compartmentalised in response to environmental change.
Mitochondrial function has been linked previously to the regulation of a multi-drug resistance (Hallstrom and Moye-Rowley, 2000). However, studies to date have implicated severe mitochondrial dysfunction as a trigger of retrograde signalling that can upregulate the transcription of drug transporters and elicit multi-drug resistance in a PDR3-dependent manner. Our study differs from these reports because cofilin mutations were found to trigger a retrograde response that upregulated drug pump activity in a PDR1-dependent manner from apparently healthy mitochondria. We also found that the resistance to a number of drugs with apparently different modes of action required mitochondrial function. It might be the case therefore that multiple mitochondrial states exist that can trigger the retrograde control of drug pump transcription. These observations might also have significance within the field of cancer biology, because tumour cells are frequently reported to exhibit altered mitochondrial function, and to rapidly acquire multi-drug resistance within chemotherapy regimes. In line with this possibility, recent studies have highlighted that the human cofilin, CFL2 can be used as an effective prognostic marker for non-small cell lung cancer and drug resistance (Castro et al., 2010; Müller et al., 2011). It will be interesting to investigate whether cofilin plays a widespread role in the regulation of drug resistance through its ability to regulate mitochondrial function. Another possibility that cannot be ruled out at present is that altered cofilin function might elicit transcriptional changes that lead to drug resistance because it has recently been shown that cofilin-1 can directly facilitate Pol II transcription through its interaction with actin (Obrdlik and Percipalle, 2011).
In summary, this study provides evidence for new roles for cofilin within the cell. We have shown that cofilin plays a role in the regulation of mitochondrial function beyond that previously described, and that this interaction can, in turn, influence the downstream signalling events. We hypothesise that cofilin has a role in coupling the environment to both cytoskeletal and mitochondrial functions, and that this biosensory role has been conserved and adapted to provide additional flexibility within eukaryotic cells.
Materials and Methods
Yeast strains, plasmids, media and growth conditions
Yeast strains used in this study are listed in supplementary material Table S1. Unless stated otherwise, cells were grown in a rotary shaker at 30°C in liquid YP medium (1% yeast extract, 2% bactopeptone) containing either 2% glucose (YPD) or 3% glycerol (YPG). For drug-sensitivity growth assays, strains were grown overnight in YPD and re-inoculated at an OD600 of 0.1 in fresh YPD medium containing itraconazole (4.4 μM), edelfosine (10 μg/ml) or amphotericin B (0.3 μg/ml). Cells were assayed for growth in 24-well plates using double orbital shaking in a FluoStar Optima plate reader (BMG Labtech). CYC1-lacZ and PDR5-lacZ reporter plasmids used to assess HAP4 activity (Chevtzoff et al., 2010) and PDR5 transcription (Hallstrom and Moye-Rowley, 2000) have been described previously. The GFP–3×RBD reporter plasmid used to assess the activity of Ras within living yeast cell was also previously described (Leadsham et al., 2009). Mitochondrial morphology was visualised using the plasmids pVTU100U-mtGFP and pYX122-mtGFP (Westermann and Neupert, 2000) by fluorescence microscopy. ROS accumulation was assessed using the indicator dye H2DCD-DA as previously described (Leadsham et al., 2009). Primers used for the deletion of RAS2, PDR1 and PDR3 are listed in supplementary material Table S4.
High-resolution respirometry
Intact cell respiration of cells grown in YPD for 24 hours to diauxic shift was carried out as previously described (Leadsham et al., 2009).
Immunoblotting
Cell number was accurately determined using a haemocytometer and total protein extracted using an optimised protein extraction method suitable for quantitative proteomics (von der Haar, 2007). Samples were separated by SDS-PAGE and transferred to PVDF membranes before probing with primary antibodies at the final concentrations, anti-Cox2 (Mitoscience; 1:1000) and anti-Pdr5 (a gift from Karl Kuchler, Department of Molecular Genetics, University and Biocenter of Vienna, Austria; 1:20,000).
Fluorescence microscopy
Rhodamine–phalloidin and DAPI staining were performed as previously described for F-actin (Gourlay et al., 2004). Cells were viewed with an Olympus IX-81 fluorescence microscope with a 150 W Xenon mercury lamp and an Olympus 150× Plan NeoFluor oil-immersion objective. Images were captured using a Hammamatsu ORCA AG digital camera using Olympus Cell R software.
Analysis of cell death in yeast colonies
Between 10 and 20 cells were plated onto 9 cm YPAD agar plates containing 10 μM Phloxine B (Sigma). Colonies were grown for 5 days before being visualised using a Leica M2FLIII microscope and documented with a Leica DC300F colour camera.
β-galactosidase assay
1×107 cells were harvested from a culture grown for 24 hours to diauxic shift in YPD medium. β-galactosidase assay was carried out as previously described (Kippert, 1995).
Microarray analysis
Triplicate samples of wild-type and cof1-6 cells were grown in YPD medium for 24 hours to diauxic shift. Total RNA from 5×107cells, broken by mechanical glass bead lysis directly into denaturation buffer, was extracted using a Qiagen RNeasy mini kit as per the manufacturer's instructions. The data obtained from Affymetrix Yeast Genome 2.0 array chips was transformed and normalised before further analysis using RMA (Robust Multichip Average). Statistical significance of the changes in expression levels between sample types was examined using the limma package. Differentially expressed genes were ranked by P value, the adjusted P-value cut-off value used was 0.05. Downstream analysis of gene ontology was performed using the GO Slim Mapper tool (http://db.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl).
Analysis of ergosterol biosynthesis
Yeast cells (50–100 ml) were grown to log phase or early stationary phase. Cells were washed with H2O before being resuspended in 3 ml KOH in ethanol (25% w/v), and incubated under reflux conditions for 1 hour at 87°C. The non-saponifiable lipid fraction was extracted with the addition of 3 ml n-heptane. Samples were dried under nitrogen gas and kept at −20°C for storage until used. The non-saponifiable lipid fraction was analysed with a HP5890 series II equipped with a Hewlett Packard CHEMSTATION software package and a fused silica DB5-MS capillary column (15 m ×0.25 mm ×0.25 μm). Samples were run in the splitless mode and the temperature program was 3 minutes at 195°C, then ramped at 5.5°C/minute until the final temperature of 300°C was reached and held for 10 minutes. Nitrogen was used as a carrier gas at a linear velocity of 30 cm/second. Sterols were identified by retention times and where required GC/MS. GC/MS analyses were performed with a HP5890 GC coupled to a HP5972 mass selective detector. The GC separations were done on a fused silica column (DB-5 15 m ×0.32 mm ×0.25 μm film thickness), programmed from 40°C to 300°C (40°C for 1 minute, 30°C/minute to 300°C and then held for 4 minutes). Helium was the carrier gas with a linear velocity of 50 cm/second in the splitless mode. Mass spectra were generated in the electron impact mode at electron energy of 70 eV and an ion source temperature of 150°C. The instrument was programmed to scan from 40 to 700 a.m.u. at 1 second intervals. Representative experiments from log phase and early stationary phase are presented.
Superoxide dismutase assay
Cells were grown for 24 hours to diauxic shift in YPD. 2×107 cells were lysed using glass beads in 0.5 ml lysis buffer [10 mM NaPO4, pH 7.8, 5 mM EDTA, 0.1% Triton X-100, 50 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and Complete EDTA-free Protease Inhibitor cocktail (Roche)]. The assay was carried out as previously described (Jakubowski et al., 2000).
Acknowledgements
Thanks to Tobias von der Haar for critical reading of this manuscript and for his helpful comments. We are also grateful to Hans Ulrich-Mösch (Marburg) and Phillip Wittmann (Medical University of Vienna) for reagents.
Footnotes
Funding
This work was sponsored by a Medical Research Council (MRC) career development fellowship to C.W.G. [grant number 78573]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.099390/-/DC1
References
- Agrawal P. B., Greenleaf R. S., Tomczak K. K., Lehtokari V. L., Wallgren-Pettersson C., Wallefeld W., Laing N. G., Darras B. T., Maciver S. K., Dormitzer P. R., et al. (2007). Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am. J. Hum. Genet. 80, 162-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginski M., Czub J. (2009). Amphotericin B and its new derivatives - mode of action. Curr. Drug Metab. 10, 459-469 [DOI] [PubMed] [Google Scholar]
- Bernstein B. W., Chen H., Boyle J. A., Bamburg J. R. (2006). Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am. J. Physiol. Cell Physiol. 291, C828-C839 [DOI] [PubMed] [Google Scholar]
- Cain K., Griffiths D. E. (1977). Studies of energy-linked reactions. Localization of the site of action of trialkyltin in yeast mitochondria. Biochem. J. 162, 575-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M. A., Dal-Pizzol F., Zdanov S., Soares M., Müller C. B., Lopes F. M., Zanotto-Filho A., da Cruz Fernandes M., Moreira J. C., Shacter E., et al. (2010). CFL1 expression levels as a prognostic and drug resistance marker in nonsmall cell lung cancer. Cancer 116, 3645-3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevtzoff C., Yoboue E. D., Galinier A., Casteilla L., Daignan-Fornier B., Rigoulet M., Devin A. (2010). Reactive oxygen species-mediated regulation of mitochondrial biogenesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285, 1733-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua B. T., Volbracht C., Tan K. O., Li R., Yu V. C., Li P. (2003). Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 5, 1083-1089 [DOI] [PubMed] [Google Scholar]
- Dominguez R., Holmes K. C. (2011). Actin structure and function. Annu. Rev. Biophys. 40, 169-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R., Rosenthal F. E., Kralli A., Sanglard D., Kuchler K. (1998). Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell 9, 523-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M. E., Sirotkin V., Haarer B., Kakhniashvili D., Amberg D. C. (2011). Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton (Hoboken) 68, 340-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R. (2006). Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 6487-6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Carpp L. N., Timpson P., Winder S. J., Ayscough K. R. (2004). A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 164, 803-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom T. C., Moye-Rowley W. S. (2000). Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 37347-37356 [DOI] [PubMed] [Google Scholar]
- Hawkins M., Pope B., Maciver S. K., Weeds A. G. (1993). Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 32, 9985-9993 [DOI] [PubMed] [Google Scholar]
- Hayden S. M., Miller P. S., Brauweiler A., Bamburg J. R. (1993). Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32, 9994-10004 [DOI] [PubMed] [Google Scholar]
- Jakubowski W., Biliński T., Bartosz G. (2000). Oxidative stress during aging of stationary cultures of the yeast Saccharomyces cerevisiae. Free Radic. Biol. Med. 28, 659-664 [DOI] [PubMed] [Google Scholar]
- Kippert F. (1995). A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128, 201-206 [DOI] [PubMed] [Google Scholar]
- Klamt F., Zdanov S., Levine R. L., Pariser A., Zhang Y., Zhang B., Yu L. R., Veenstra T. D., Shacter E. (2009). Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol. 11, 1241-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D. G. (1997). Cofilin promotes rapid actin filament turnover in vivo. Nature 388, 78-82 [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Fedorov E. V., Fedorov A. A., Almo S. C., Drubin D. G. (1997). Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 16, 5520-5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris R., Piwowarski J., van der Spek H., Teixeira de Mattos J., Grivell L., Blom J. (2004). Overexpression of HAP4 in glucose-derepressed yeast cells reveals respiratory control of glucose-regulated genes. Microbiology 150, 929-934 [DOI] [PubMed] [Google Scholar]
- Leadsham J. E., Gourlay C. W. (2010). cAMP/PKA signaling balances respiratory activity with mitochondria dependent apoptosis via transcriptional regulation. BMC Cell Biol. 11, 92-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadsham J. E., Miller K., Ayscough K. R., Colombo S., Martegani E., Sudbery P., Gourlay C. W. (2009). Whi2p links nutritional sensing to actin-dependent Ras-cAMP-PKA regulation and apoptosis in yeast. J. Cell Sci. 122, 706-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppert G., McDevitt R., Falco S. C., Van Dyk T. K., Ficke M. B., Golin J. (1990). Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics 125, 13-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. C., Galletta B. J., Sept D., Cooper J. A. (2010). Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J. Cell Sci. 123, 1329-1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver S. K., Pope B. J., Whytock S., Weeds A. G. (1998). The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256, 388-397 [DOI] [PubMed] [Google Scholar]
- Minamide L. S., Striegl A. M., Boyle J. A., Meberg P. J., Bamburg J. R. (2000). Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2, 628-636 [DOI] [PubMed] [Google Scholar]
- Mösch H. U., Kübler E., Krappmann S., Fink G. R., Braus G. H. (1999). Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10, 1325-1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C. B., de Barros R. L., Castro M. A., Lopes F. M., Meurer R. T., Roehe A., Mazzini G., Ulbrich-Kulczynski J. M., Dal-Pizzol F., Fernandes M. C., et al. (2011). Validation of cofilin-1 as a biomarker in non-small cell lung cancer: application of quantitative method in a retrospective cohort. J. Cancer Res. Clin. Oncol. 137, 1309-1316 [DOI] [PubMed] [Google Scholar]
- Nadella K. S., Saji M., Jacob N. K., Pavel E., Ringel M. D., Kirschner L. S. (2009). Regulation of actin function by protein kinase A-mediated phosphorylation of Limk1. EMBO Rep. 10, 599-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik A., Percipalle P. (2011). The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus 2, 72-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala P. J., Paavilainen V., Lappalainen P. (2001). Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry 40, 15562-15569 [DOI] [PubMed] [Google Scholar]
- Pope B. J., Gonsior S. M., Yeoh S., McGough A., Weeds A. G. (2000). Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 298, 649-661 [DOI] [PubMed] [Google Scholar]
- Posadas I., Pérez-Martínez F. C., Guerra J., Sánchez-Verdú P., Ceña V. (2012). Cofilin activation mediates Bax translocation to mitochondria during excitotoxic neuronal death. J. Neurochem. 120, 515-527 [DOI] [PubMed] [Google Scholar]
- Rosivatz E., Woscholski R. (2011). Removal or masking of phosphatidylinositol(4,5)bisphosphate from the outer mitochondrial membrane causes mitochondrial fragmentation. Cell. Signal. 23, 478-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P., Moye-Rowley W. S. (2009). Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 1794, 852-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649-667 [DOI] [PubMed] [Google Scholar]
- Vanden Bossche H., Willemsens G., Marichal P. (1987). Anti-Candida drugs--the biochemical basis for their activity. Crit. Rev. Microbiol. 15, 57-72 [DOI] [PubMed] [Google Scholar]
- von der Haar T. (2007). Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE 2, e1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., Neupert W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427 [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Renault G., Jacquet M., Tatchell K. (1993). The pde2 gene of Saccharomyces cerevisiae is allelic to rca1 and encodes a phosphodiesterase which protects the cell from extracellular cAMP. FEBS Lett. 325, 191-195 [DOI] [PubMed] [Google Scholar]
- Zaremberg V., Gajate C., Cacharro L. M., Mollinedo F., McMaster C. R. (2005). Cytotoxicity of an anti-cancer lysophospholipid through selective modification of lipid raft composition. J. Biol. Chem. 280, 38047-38058 [DOI] [PubMed] [Google Scholar]
- Zhao H., Hakala M., Lappalainen P. (2010). ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys. J. 98, 2327-2336 [DOI] [PMC free article] [PubMed] [Google Scholar]