Abstract
Background
In Africa, relapsing fevers are neglected arthropod-borne infections caused by closely related Borrelia species. They cause mild to deadly undifferentiated fever particularly severe in pregnant women. Lack of a tool to genotype these Borrelia organisms limits knowledge regarding their reservoirs and their epidemiology.
Methodology/Principal Findings
Genome sequence analysis of Borrelia crocidurae, Borrelia duttonii and Borrelia recurrentis yielded 5 intergenic spacers scattered between 10 chromosomal genes that were incorporated into a multispacer sequence typing (MST) approach. Sequencing these spacers directly from human blood specimens previously found to be infected by B. recurrentis (30 specimens), B. duttonii (17 specimens) and B. crocidurae (13 specimens) resolved these 60 strains and the 3 type strains into 13 species-specific spacer types in the presence of negative controls. B. crocidurae comprised of 8 spacer types, B. duttonii of 3 spacer types and B. recurrentis of 2 spacer types.
Conclusions/Significance
Phylogenetic analyses of MST data suggested that B. duttonii, B. crocidurae and B. recurrentis are variants of a unique ancestral Borrelia species. MST proved to be a suitable approach for identifying and genotyping relapsing fever borreliae in Africa. It could be applied to both vectors and clinical specimens.
Author Summary
In Africa, relapsing fevers are caused by four cultured species: Borrelia crocidurae, Borrelia duttonii, Borrelia hispanica and Borrelia recurrentis. These borreliae are transmitted by the bite of Ornithodoros soft ticks except for B. recurrentis which is transmitted by louse Pediculus humanus. They cause potentially undifferentiated fever infection and co-infection with malaria could also occur. The exact prevalence of each Borrelia is unknown and overlaps between B. duttonii and B. crocidurae have been reported. The lack of tools for genotyping these borreliae limits knowledge concerning their epidemiology. We developed multispacer sequence typing (MST) and applied it to blood specimens infected by B. recurrentis (30 specimens), B. duttonii (18 specimens) and B. crocidurae (13 specimens), delineating these 60 strains and the 3 type strains into 13 species-specific spacer types. B. crocidurae strains were classified into 8 spacer types, B. duttonii into 3 spacer types and B. recurrentis into 2 spacer types. These findings provide the proof-of-concept that that MST is a reliable tool for identification and genotyping relapsing fever borreliae in Africa.
Introduction
In Africa, relapsing fevers (RF) are arthropod-borne diseases caused by four cultured species Borrelia crocidurae, Borrelia duttonii, Borrelia hispanica and Borrelia recurrentis [1]. Transmission is by the bite of Ornithodoros soft ticks for the first three species whereas Pediculus humanus louse feces transmit B. recurrentis [2], [3]. In Tanzania, molecular investigations of human and tick specimens further provided evidences for two additional, yet uncultured Borrelia species [1], [4]. Each one of the four cultured Borrelia species is more prevalent in one geographical area of Africa with B. hispanica being reported in Morocco [5], B. crocidurae in Senegal [6], B. duttonii in Tanzania [7] and B. recurrentis in Ethiopia [8]. However, the precise area of distribution of each Borrelia is unknown and may overlap as both B. duttonii and B. crocidurae have been reported in Togo and Tanzania [1], [9].
In these regions of Africa, RF was reported to be the most prevalent bacterial disease, accounting for 8.8% of febrile patients in Togo [9]. In Senegal, average incidence is 11 per 100 person-years [10]. The main clinical symptom of infection is recurrent undifferentiated fever associated with high bacteremia; RF are therefore often diagnosed as malaria and cases of malaria co-infection with have been reported [9], [11], [12]. RF are treatable by antibiotics. Severity ranges from asymptomatic to fatal, particularly if left untreated and can be associated with significant pregnancy loss or peri-natal mortality [13], [14], [15].
The African RF Borrelia are very closely related species as illustrated by 16S rRNA gene sequence variability ≤1% [2]. Accordingly, a previous comparison of B. duttonii and B. recurrentis genomes indicated that the two organisms formed a unique bacterial species [16]. Such a close genetic and genomic proximity challenged the development of laboratory tools for the accurate discrimination between the African RF Borrelia and genotyping [16]. Sequencing the 16S rRNA and the flagellin genes is unsatisfactory since African RF Borrelia differ by only one base in the flagllin gene sequence and have 16S rRNA gene sequence similarity above 99% [17]. Analysis of the intrergenic spacer (IGS) located between the 16S and 23S rRNA genes only explored the variability between B. duttonii and B. recurrentis [1]. Moreover, IGS sequence overlapped between one B. duttonii phylogenetic group and one B. recurrentis group [1] with a second overlap disclosed with subsequent analyses of further material [7].
We previously observed that multispacer sequence typing (MST), a PCR-sequencing-based method for bacteria genotyping, was efficient in typing otherwise homogenous bacterial species such as the plague agent Yersinia pestis [18] and the typhus agent Rickettsia prowazekii [19]. Ongoing study of the B. crocidurae genome in our laboratory gave us the opportunity to develop MST for African RF Borrelia and to deliver the proof-of-concept that MST is a suitable method for both the species identification and genotyping of RF Borrelia in Africa.
Materials and Methods
Borrelia strains and DNA
B. crocidurae Achema strain, B. recurrentis A1 strain and B. duttonii Ly strain were grown in BSK-H medium (Sigma, Saint Quentin Fallavier, France) supplemented with heat-inactivated 10% rabbit serum (Eurobio, Courtaboeuf, France). B. recurrentis DNA was extracted from 21 blood specimens collected in 1994 in Addis Ababa, Ethiopia Dr. S. J. Cutler (School of Health, Sports and Bioscience, University of East London, London UK). Likewise, B. recurrentis DNA extracted from 9 blood specimens collected in 2011 in Bahir Dah, Highlands of Ethiopia were provided by SC Barker (Parasitology section, School of Chemistry and Molecular Bioscience, University of Queensland, Brisbane, Australia) and KD Bilcha and J Ali (University of Gondar, Ethiopia). In addition, B. duttonii DNA extracted from 17 blood specimens collected in Mvumi, Tanzania were also provided by Dr. S. J. Cutler. B. crocidurae DNA was extracted from 13 blood specimens collected in 2010 in Senegal by C. Sokhna (URMITE, Dakar, Senegal) including 11 specimens from Dielmo and 2 specimens from Ndiop. DNA was extracted from these specimens using QIAamp DNA Blood mini kits (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
Selection of intergenic spacers
The B. crocidurae genome (Genbank accession number CP003426–CP003465) has been sequenced and annotated in our laboratory using pyrosequencing technology on a Roche 454 GS FLX sequencer. The draft genome is comprising of one closed chromosome and scaffolds representing the plasmids. Spacer sequences extracted from B. crocidurae strain Achema, B. recurrentis strain A1 (Genbank accession number CP000993) and B. duttonii strain Ly (Genbank accession number CP000976) genomes using perl script software were compared using ssaha2 software [20]. Spacers were pre-selected for a 300 to 800-bp length. Pre-selected spacers were further analyzed for sequence similarity in order to exclude spacers with <0.1% interspecies sequence similarity. PCR primers were then designed using primer3 software (http://fokker.wi.mit.edu) in order to amplify the entire sequence of each of the selected spacers.
Multispacer sequencing typing
Five microliters of Borrelia DNA and 10 pmol of each primer (Eurogentec, Seraing, Belgium) were added to the PCR mixture, containing 0.4 U Phusion DNA Polymerase (Finnzymes, Espoo, Finland), 4 µl of 5× Phusion HF Buffer (Finnzymes) and 0.4 µl of 10 mM dNTPs. The volume was adjusted to 24 µL by adding distilled water. Thermal cycling was performed on a 2720 DNA thermal cycler (Applied Biosystems, Courtaboeuf, France) with an initial 30-sec cycle at 98°C followed by 35 cycles consisting of 10 seconds at 98°C, 30 seconds at 58°C and 1 minute at 72°C, followed by a 10-min final extension step at 72°C. To rule out amplicon carry-over, nucleotide-free water negative control was used throughout the steps of the protocol. PCR products were purified prior to sequencing by using the Nucleo-Fast 96 PCR Kit (Macherey-Nagel, Hoerdt, France). Three microliters of the resulting DNA were added to each primer mixture comprised of 10 pmol of each primer, 4 µL water and 3 µL BigDye Terminator reaction mix (Applied Biosystems). Sequencing thermal cycling was performed on a Applied Biosystems DNA thermal cycler with an initial 5-min cycle at 96°C followed by 25 cycles consisting of 30 seconds at 96°C, 20 seconds at 55°C, and 4 minutes at 60°C, followed by a 7-min final extension step at 15°C. Sequencing products were purified using sephadex plates (Sigma-Aldrich, Saint Quentin Fallavier, France) and sequencing electrophoresis was performed on a 3130 Genetic Analyzer (Applied Biosystems).
Sequence analysis
The nucleotide sequences were edited using ChromasPro software (www.technelysium.com.au/chromas.html). Similarities between spacers were determined after multiple alignments using the MULTALIN software [21]. MST discrimination power was calculated using the Hunter-Gaston Index [22]:
where D is the numerical index of discrimination, N is the total number of isolates in the sample population, s is the total number of different types, and nj is the number of isolates belonging to the jth type.
The five spacer sequences analyzed herein were concatenated and neighbor-joining phylogenetic tree was reconstructed using the maximum likelihood method in PhyML 3.0 [23]. Each particular sequence of a given spacer was assigned to a spacer type (ST) number.
Ethics statement
This study was approved by the IFR48 Ethic Committee. All patients provided informed written consent.
Results
Spacer selection
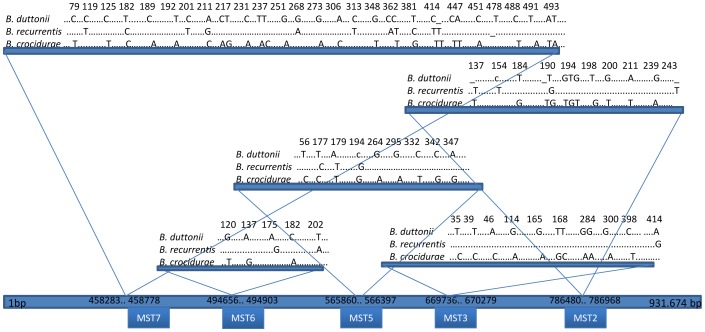
Chromosome sequence alignment of the three Borrelia reference genomes studied herein revealed that 23 intergenic spacers that were common to all three species. Of these, five spacers fulfilled our selection criteria and were named MST2, MST3, MST5, MST6 and MST7. Use of the PCR primers listed in Table 1 to amplify each of the five spacers produced amplicons ranged from 333-bp to 738-bp and sequence reads ranging from 246-bp to 543-bp (Table 1).
Table 1. List of primers and genes flanking five intergenic spacers herein studied in relapsing fever Borrelia.
| Spacers | Start End | Spacer flanking genes (5------3) | Primers | PCR product size (bp) | Spacer size (bp) |
| MST2 | 786480.. 786968 | penicillin-binding protein//uncharacterized conserved protein | F:TTTTTGCTAAAATTAACCCTTTTCAR:CTCATTTTAATTTCCTTACCCCTA | 578 | 487 |
| MST3 | 669736.. 670279 | N-acetylmuramoyl-L-alanine amidase, putative//vacuolar X-prolyl dipeptidyl aminopeptidase I | F:GCAGGTGGCTGTTAACCACTR:ATGTGGGGAATGCACTCTTT | 687 | 543 |
| MST5 | 565860.. 566397 | translation elongation factor G//uncharacterized conserved protein | F:CCTGAGTCGATATGGGCACTR:CAACCTGACATATCTTACTCAATTCAT | 653 | 536 |
| MST6 | 494656.. 494903 | tRNA-ser//DNA polymerase III subunits gamma and tau | F:GGGTTCGAATCCCATTTTCTR:CTCTGGGACGCCTCTTAATG | 333 | 246 |
| MST7 | 458283.. 458778 | 16S ribosomal RNA//hypothetical protein | F:TTCGCCACTGAATGTATTGCR:TGCCAATGTTCTTGTTGGTC | 738 | 494 |
Start and end of spacer are according to B. duttonii genome.
Interspecies analysis
Pairwise comparison of the five spacers (Table 2 and figure 1) revealed they had species-specific sequence with interspecies sequence differences relying on single nucleotide polymorphism in 36 (90%) cases, deletion in 3 (7.5%) cases and insertion in 1 (2.5%) case. Comparing B. duttonii MST7 with B. crocidurae and B. recurrentis MST7 yielded 93% and 97% similarity, respectively, whilst comparing B. crocidurae MST7 with B. recurrentis MST7 showed 93% similarity. The other four spacers yielded pairwise sequence similarity of 97–99% (Table 2). Sequences for each allele of each spacer have been deposited in GenBank under accession number (JQ398815: JQ398841) as well as in our local data base (http://www.ifr48.com).
Table 2. Pairwise comparison of each spacer of B. duttonii, B. recurrentis and B. crocidurae.
| Species spacers | B. duttonii | B. recurrentis | B. crocidurae | B. duttonii | B. recurrentis | B. crocidurae | B. duttonii | B. recurrentis | B. crocidurae | B. duttonii | B. recurrentis | B. crocidurae | B. duttonii | B. recurrentis | B. crocidurae |
| MST2 | MST3 | MST5 | MST6 | MST7 | |||||||||||
| B. duttonii | 99–100 | 99 | 99 | 100 | 99 | 98 | 98–100 | 97–100 | 98–99 | 98–100 | 99 | 98 | 100 | 97 | 93–94 |
| B. recurrentis | 99–100 | 99 | 100 | 98 | 100 | 99 | 100 | 99 | 100 | 93 | |||||
| B. crocidurae | 99 | 99–100 | 99–100 | 99–100 | 98–100 | 99–100 |
Bold characters indicate range of similarity within the species.
Figure 1. Distribution of spacers among the chromosome of B. duttonii and main differences within each spacer.
Intra-species analysis
While the concatenation of the five spacers yielded a discrimination index of 0.825,1, this index was of 0.7814 for MST2, 0.6896 for MST6, 0.6749 for MST5, 0.6623 for MST7, and 0.6579 for MST3. Concatenation of the five spacers yielded 8 STs named ST6–ST13 for the 13 B. crocidurae samples and the B. crocidurae Achema type strain (Table 3; Figure 2). 3 STs named ST1–ST3 for the 18 B. duttonii samples and the B. duttonii Ly type strain and 2 STs named ST4–ST5 for the 30 B. recurrentis samples and the B. recurrentis A1 type strain. MST2 sequencing classified latter samples into ST-4 (11 samples) and ST-5 (19 samples) due to the insertion of a G at position 190. The genotype ST-5 represented 47.6% (10 out 21 samples) detected in 1994 and all the nine samples detected in 2011.
Table 3. List of spacer types (ST) found in this study.
| Species | Strains | ST | MST2 | MST3 | MST5 | MST6 | MST7 |
| B. duttonii | Bd 9, 11,17 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bd 1, 2, 3, 5, 6, 8, 12, 13,16, 19,20, 22, Ly | 2 | 2 | 1 | 2 | 2 | 1 | |
| Bd15, 18 | 3 | 2 | 1 | 3 | 2 | 1 | |
| B. recurrentis | Br 1,2,3,4,5,6,7,8,9,10,11 | 4 | 3 | 2 | 4 | 3 | 2 |
| Br12, 13, 14, 15, 16, 17, 18, 19, 20,21,22,23,24,25,26,27,28,29,30,A1 | 5 | 4 | 2 | 4 | 3 | 2 | |
| B. crocidurae | B.cr18, B.cr89, B.cr88 B.cr85 | 6 | 5 | 3 | 5 | 7 | 4 |
| B.cr34 | 7 | 5 | 4 | 5 | 7 | 5 | |
| B.cr30 | 8 | 6 | 4 | 5 | 4 | 4 | |
| B.cr35 | 9 | 5 | 4 | 5 | 5 | 3 | |
| B.cr936 | 10 | 5 | 3 | 5 | 6 | 5 | |
| B.cr81 B.cr57 B.cr40, B.cr23 | 11 | 5 | 4 | 5 | 6 | 5 | |
| B.cr66 | 12 | 5 | 4 | 5 | 7 | 4 | |
| Achema | 13 | 7 | 5 | 6 | 8 | 6 |
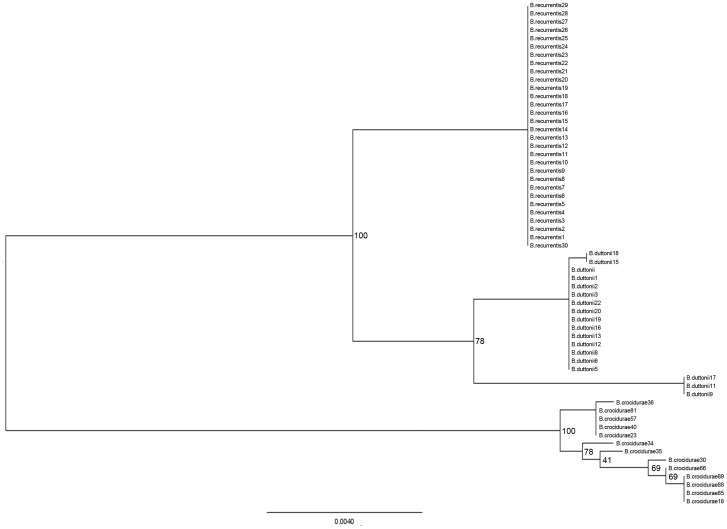
Figure 2. Maximum-likelihood tree based on five intergenic spacers sequences for 61 Borrelia strains.
To examine the confidence of ML tree, 100 bootstrap replicates were used.
MST-based phylogenetic analysis of RF Borrelia
The phylogenetic tree constructed after concatenation of the five intergenic spacer sequences separated the RF Borrelia into three clades, each clade containing only one Borrelia species (figure 2). A first clade comprised of all the 30 B. recurrentis isolates; a second clade comprised of three groups representing the three B. duttonii spacer types and a last clade comprised of 7 B. crocidurae spacer types.
Discussion
PCR-derived data reported herein were interpreted as authentic as the negative controls used in every PCR-based experiment remained negative, all the PCR products were sequenced and experiments yielded reproducible sequences. We therefore established the proof-of-concept that MST could be used for species identification and genotyping of 3 out of 4 cultured RF borreliae (B. hispanica was not available for this study) in Africa. MST combines the sensitivity of PCR with unambiguous, portable data yielded by sequencing. Indeed, all the sequences determined are freely available in GenBank and in our local database website at ifr48.com. Therefore, any laboratory with a capacity in PCR-sequencing could easily confirm and compare their data with that reported herein to further increase the knowledge of RF Borrelia species and genotypes circulating in African countries.
In the present study, five intergenic spacers were selected from the alignment of B. crocidurae, B. duttonii and B. recurrentis reference genomes, representing approximately ∼0.2% of the total genome length. The spacers were scattered across the chromosome thus representative of the whole genome. Such a multi-target approach offers distinct advantages over the one single locus methods previously used, such as the 16S–23S IGS for typing that may be less representative of the whole genome. Based on this spacer sequencing, a total of 61 RF strains could be separated into 12 STs. Interestingly, we observed that isolates grouped into three clades corresponding to the three Borrelia organisms under study. Indeed, MST yielded no overlap between B. duttonii and B. recurrentis organisms contrary to that observed when using IGS typing [1], [7]. We observed that sequencing MST7 spacer alone accurately discriminated between B. duttonii and B. recurrentis with 3% sequence divergence, a result not previously achieved. Therefore, sequencing MST7 spacer alone could be used for the molecular identification of RF Borrelia in Africa at the species level, but not for genotyping which requires sequencing the four other spacers in addition to MST7.
Further analysis indicated that each one of the three Borrelia species under study was comprised of several spacer-types. B. recurrentis was the least diverse Borrelia comprising of only two very closely related groups. This finding supports the previous genomic analysis that concluded that B. recurrentis was a subset of B. duttonii [16]. In our study also, there was an inverse correlation between the RF Borrelia MST diversity and the reported mortality rate for these RF Borrelia [8], [15].
Despite the fact that we tested a small set of B. crocidurae, nevertheless we found a high diversity index in this species since 13 B. crocidurae samples collected in Senegal yielded 7 MST types and the B. crocidurae Achema type strain collected in Mauritania yielded an additional MST type. This first genotyping method for B. crocidurae is therefore very promising to probe its geographic repartition as well as potential association of B. crocidurae genotypes with vectors. Indeed, four genogroups could be identified in O. sonrai ticks collected in Senegal and Mauritania [24]. In this study, B. crocidurae flagellin sequence was found identical among the four O. sonrai tick groups but the B. crocidurae infection rate significantly differed among the four tick groups; MST may help studying such discrepancy and may reveal previously unknown relationships between B. crocidurae genotypes and O. sonrai genotypes. Moreover, a recent study indicated that B. crocidurae may be transmitted by soft tick Ornithodoros erraticus in Tunisia, challenging O. sonrai as the only B. crocidurae vector in West Africa [25]. MST is new laboratory tool to question whether the unexpected higher diversity in B. crocidurae than in B. duttonii and B. recurrentis is linked to a more complex cycle involving several mammals and ticks species.
Present data indicate that MST is offering a new sequencing-based technique for further exploring the identification and genotypes of RF Borrelia in vectors and clinical specimens collected in Africa.
Footnotes
The authors have declared that no competing interests exist.
The authors acknowledge the financial support of ANR 2008 BORETIC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Scott J, Wright D, Cutler S. Typing African relapsing fever spirochetes. Emerg Infect Dis. 2005;11:1722–1729. doi: 10.3201/eid1111.050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ras NM, Lascola B, Postic D, Cutler SJ, Rodhain F, et al. Phylogenesis of relapsing fever Borrelia spp. Int J Syst Bacteriol. 1996;46:859–865. doi: 10.1099/00207713-46-4-859. [DOI] [PubMed] [Google Scholar]
- 3.Houhamdi L, Raoult D. Excretion of living Borrelia recurrentis in feces of Infected Human Body Lice. J Infect Dis. 2005;191:1898–1906. doi: 10.1086/429920. [DOI] [PubMed] [Google Scholar]
- 4.Kisinza WN, McCall PJ, Mitani H, Talbert A, Fukunaga M. A newly identified tick-borne Borrelia species and relapsing fever in Tanzania. Lancet. 2003;18:1283–1284. doi: 10.1016/s0140-6736(03)14609-0. [DOI] [PubMed] [Google Scholar]
- 5.Sarih M, Garnier M, Boudebouch N, Bouattour A, Rihani A, et al. Borrelia hispanica relapsing fever, Morocco. Emerg Infect Dis. 2009;15:1626–1629. doi: 10.3201/eid1510.090403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, et al. Tick-borne relapsing fever borreliosis, rural Senegal. Emerg Infect Dis. 2011;17:883–885. doi: 10.3201/eid1705.100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutler SJ, Bonilla EM, Singh RJ. Population structure of East African relapsing fever Borrelia spp. Emerg Infect Dis. 2010;16:1076–1080. doi: 10.3201/eid1607.091085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgnolo G, Denku B, Chiabrera F, Hailu B. Louse-borne relapsing fever in Ethiopian children: a clinical study. Ann Trop Paediatr. 1993;13:165–171. doi: 10.1080/02724936.1993.11747641. [DOI] [PubMed] [Google Scholar]
- 9.Nordstrand A, Bunikis I, Larsson C, Tsogbe K, Schwan TG, et al. Tickborne relapsing fever diagnosis obscured by malaria, Togo. Emerg Infect Dis. 2007;13:117–123. doi: 10.3201/eid1301.060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vial L, Diatta G, Tall A, Ba el H, Bouganali H, et al. Incidence of tick-borne relapsing fever in west Africa: longitudinal study. Lancet. 2006;368:37–43. doi: 10.1016/S0140-6736(06)68968-X. [DOI] [PubMed] [Google Scholar]
- 11.Ramos JM, Reyes F, Tesfamariam A, Malmierca E. Louse-borne relapsing fever and malaria co-infection in Ethiopia. Trop Doct. 2007;37:121–122. doi: 10.1177/004947550703700229. [DOI] [PubMed] [Google Scholar]
- 12.Miron D, Olshinsky A, Assy N, Zuker M, Efrat M, et al. Plasmodium and Borrelia co-infection. J Travel Med. 2004;11:115–116. doi: 10.2310/7060.2004.17073. [DOI] [PubMed] [Google Scholar]
- 13.Jongen VH, van Roosmalen J, Tiems J, Van Holten J, Wetsteyn JC. Tick-borne relapsing fever and pregnancy outcome in rural Tanzania. Acta Obstet Gynecol Scand. 1997:834–839. doi: 10.3109/00016349709024361. [DOI] [PubMed] [Google Scholar]
- 14.Dupont HT, La Scola B, Williams R, Raoult D. A focus of tick-borne relapsing fever in southern Zaire. Clin Infect Dis. 1997:139–144. doi: 10.1086/514496. [DOI] [PubMed] [Google Scholar]
- 15.Ramos J, Malmierca E, Reyes F, Wolde W, Galata A, et al. Characteristics of louse-borne relapsing fever in Ethiopian children and adults. Ann Trop Med Parasitol. 2004;98:191–196. doi: 10.1179/000349804225003136. [DOI] [PubMed] [Google Scholar]
- 16.Lescot M, Audic S, Robert C, Nguyen TT, Blanc G, et al. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 2008;12:e1000185. doi: 10.1371/journal.pgen.1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toledo A, Anda P, Escudero R, Larsson C, Bergstrom S, et al. Phylogenetic analysis of a virulent Borrelia species isolated from patients with relapsing fever. J Clin Microbiol. 2010;48:2484–2489. doi: 10.1128/JCM.00541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drancourt M, Roux V, Dang LV, Lam THCD, Chenal-Francisque V, et al. Genotyping, Orientalis-like Yersinia pestis, and plague pandemics. Emerg Infect Dis. 2004;10:1585–1592. doi: 10.3201/eid1009.030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Fournier PE, Ogata H, Raoult D. Multispacer typing of Rickettsia prowazekii enabling epidemiological studies of epidemic typhus. J Clin Microbiol. 2005;43:4708–4712. doi: 10.1128/JCM.43.9.4708-4712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 24.Vial L, Durand P, Arnathau C, Halos L, Diatta G, et al. Molecular divergences of the Ornithodoros sonrai soft tick species, a vector of human relapsing fever in West Africa. Microbes Infect. 2006;8:2605–2611. doi: 10.1016/j.micinf.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Bouattour A, Garnier M, M'Ghirbi Y, Sarih M, Gern L, et al. Borrelia crocidurae infection of Ornithodoros erraticus (Lucas, 1849) ticks in Tunisia. Vector Borne Zoonotic Dis. 2010;10:825–830. doi: 10.1089/vbz.2009.0151. [DOI] [PubMed] [Google Scholar]