Abstract
We initially studied requirements for 5' and 3' terminal regions (TRs) in flavivirus negative strand synthesis in vitro. Purified West Nile (WNV) and dengue-2 (DV2) RNA polymerases were both active with all-WNV or all-DV2 subgenomic RNAs containing the 5'- and 3'TRs of the respective genomes. However, subgenomic RNAs in which the 5'-noncoding region (5'NCR) or the 5'ORF (nts 100–230) in the 5'TR were substituted by analogous sequences derived from the heterologous genome were modestly to severely defective as templates for either polymerase. We also evaluated the infectivity of substitution-mutant WNV genome-length RNAs. All WNV RNAs containing the DV2 3'SL were unable to replicate. However, WNV RNAs containing substitutions of the 5'NCR, the capsid gene, and/or 3'NCR nt sequences upstream from the WNV 3'SL, by the analogous DV2 nt sequences, were infectious. Combined results suggested that replication was not dependent upon species homology between the 3'SL and NS5.
Keywords: flavivirus RNA replication, RNA cyclization, flavivirus polymerase activity
INTRODUCTION
Dengue and West Nile viruses (WNV) are members of different groups among mosquito-borne flaviviruses, based on serologic and genetic relatedness (Calisher et al, 1989). The flavivirus genome is about 10.5 kb in total length and contains a single long open reading frame (ORF), encoding three viral structural proteins and at least seven nonstructural (NS) proteins. The ORF is flanked by a 5' noncoding region (NCR) which is about 96 to 100 nucleotides (nt) in length and by a 3' NCR which is 400 to 800 nt in length depending on the flavivirus (reviewed in Lindenbach and Rice, 2003). The dengue 3'NCR is among the shortest within the genus, at ~450 nts, while the 3'NCRs of the JE group, including WNV, are ~630 nts in length. Consequently, the WNV 3'NCR contains nt sequences, including but not limited to tandem repeat sequences, CS3 and RCS3, not present in the DV 3'NCRs (reviewed in Markoff, 2003). The 5'- and 3'-terminal 100 nts of the flavivirus genome each form thermodynamically stable stem-loop structures, here referred to as the 5’SL and 3’SL, respectively (Brinton and Dispoto, 1988; Brinton et al, 1986; Grange et al, 1985; Gritsun et al, 1997; Khromykh et al, 2003; Mohan and Padmanabhan, 1991; Proutski et al, 1997; Shi et al, 1996; Wengler and Castle, 1986).
The 3’SL secondary structure is essential for minus strand RNA synthesis and for virus replication in infected mammalian cells (Khromykh et al, 2003; Tilgner and Shi, 2004; You et al, 2001; Yu and Markoff, 2005; Zeng et al, 1998). Within the 3'SL, the long stem segment contains bulges due to mismatching of base pairs in the adjacent strands. The loci of these bulges are not well conserved among flavivirus species. Nonetheless, integrity of specific bulges is required for RNA replication (Tilgner et al, 2005; Yu and Markoff, 2005). This was thought to account for the previous observation that the respective 3'SL nt sequences could not be swapped between the WNV and dengue-2 strain New Guinea C (DV2) genomes. The 3’SL also binds host cellular proteins in vitro (Blackwell and Brinton, 1995; Blackwell and Brinton, 1997; De Nova-Ocampo et al, 2002; Ong et al, 1998; Ta and Vrati, 2000). Finally, the viral nonstructural proteins NS3 and NS5 bind the 3'SL in context with the 3'NCR (Chen et al, 1997). The amino-terminus of NS3 contains a serine protease activity; its carboxy-terminus contains NTPase, RNA helicase, and 5'-RNA triphosphatase activities all thought to be required for RNA replication (for reviews, see Lindenbach, 2003, Padmanabhan, 2006). NS5 contains the RNA-dependent RNA polymerase activity (Ackermann and Padmanabhan, 2001; Tan et al., 1996), as well as 2' O- and N-methyltransferase activities needed for capping nascent viral RNAs (Egloff et al., 2002; Ray et al., 2006). Most if not all the NS proteins are part of the replication complex (Khromykh et al, 2000).
The 5’NCR is required both for RNA synthesis (Ackermann and Padmanabhan, 2001; Cahour et al., 1995; Filomatori et al., 2006; Nomaguchi et al., 2004; You et al., 2001) and for viral RNA translation (Chiu et al, 2005; Holden and Harris, 2004). A recent observation that may account for the requirement for the 5'NCR in RNA synthesis was the demonstration that a specific tri-nucleotide sequence within the conserved stem-loop "A" in the 5'SL is recognized by NS5 as a pre-condition for initiation of negative strand RNA synthesis (Filomatori et al., 2006).
Accumulated evidence suggests that the flavivirus RNA forms a panhandle structure for initiation of negative strand RNA synthesis. RNA elements required for 5'/3' terminal region (TR; here defined as the 5'NCR, 5'ORF, and 3'NCR nt sequences) hydrogen bonding include the 5'CS, an 8-nt sequence within the ORF (nts 137–144 in the WNV genome) that is conserved among mosquito-borne flaviviruses and CS1, a 23- to 25-nt 3'TR element that lies just upstream from the 3'SL in the 3'NCR and contains an 8-nt cyclization sequence (cyc) complementary to the 5'CS (Hahn et al., 1987). Complementarity between the cyc sequence and the 5'CS is required for negative strand RNA synthesis. There is additional complementarity between the 5' and 3' TRs, including but not limited to the 5' and 3' "UAR" nt sequences that lie upstream from the start codon and upstream from the 3'SL, respectively (Ackermann and Padmanabhan, 2001; Alvarez et al., 2005a; Alvarez et al., 2005b; Bredenbeek et al., 2003; Corver et al., 2003; Filomatori et al., 2006; Khromykh et al., 2001; Lo et al., 2003; Nomaguchi et al., 2004; You et al., 2001; Yu and Markoff, 2005; Zeng et al, 1998). Despite the accumulated information, there are still large gaps in our understanding of the specific nature of the interaction of 5' and 3' secondary structures with virus and host proteins to form the replication complex.
To gain additional relevant data, we first examined the ability of RNA synthesis templated by the WNV 3'NCR to be transactivated by added 5'TR nt sequences derived from the WNV and/or the DV2 genome in vitro, using purified WNV NS5 as catalyst. We then created a series of WNV and DV2 "minigenome" RNAs containing 5'NCR, 5'ORF and 3'NCR nt sequences in covalent linkage. Homologous (all-WNV or all-DV2) and WNV/DV2 chimeric minigenome RNAs were assayed for their ability to serve as template for minus strand RNA synthesis, using purified DV2 and WNV NS5 viral polymerase proteins (Ackermann and Padmanabhan, 2001; Nomaguchi et al, 2004). We also engineered WNV infectious RNAs that contained DV2-for-WNV nt substitution mutations of the 5'NCR, 5'ORF, and the 3'NCR or functional elements thereof. These RNAs were analyzed for their replication competence in vivo. The combined data suggested that the requirement for the homologous 3'SL for viral replication is related to specificities of viral NS proteins other than NS5 or to an effect on NS5 specificity conferred by its interaction with other NS proteins or cellular proteins required to form the replication complex. Additional information regarding specific requirements for 5' and 3'TR nt sequence elements in flavivirus RNA replication and parallels between the in vitro and in vivo results were also gained from this work.
RESULTS
5'TR nt sequences required for in vitro RNA synthesis catalyzed by purified NS5 proteins
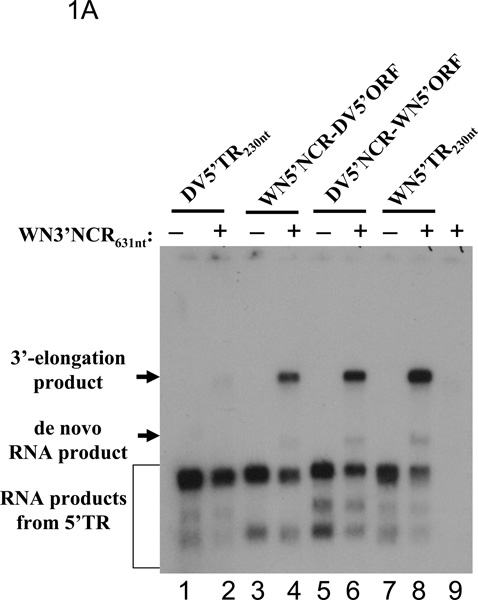
We previously demonstrated that negative strand RNA synthesis templated by the WNV 3'NCR and catalyzed by purified WNV NS5 was dependent upon the presence of WNV 5'terminal nt sequences that included both the 5'NCR and the 5'CS in covalent linkage to the 3'NCR (Nomaguchi et al., 2004). Prior to initiating the present work, we established that RNA synthesis catalyzed by purified WNV NS5 protein could also be activated in trans by WNV 5'terminal region (TR) nt sequences (data not shown), provided that the WNV 5'CS as well as the 5'NCR was included in the transactivating RNA, as had been demonstrated for the DV2 NS5 using analogous DV2 5'TR RNAs (Ackermann and Padmanabhan, 2001; You et al., 2001; You and Padmanabhan, 1999). We then sought to determine whether heterologous or chimeric 5'TR230nt RNAs (i.e., the 5'NCR plus the first 130 nts of the capsid gene ORF) could transactivate in vitro RNA synthesis catalyzed by the WNV NS5, using the WNV 3'NCR631nt RNA as template. Four 5'TR230nt RNAs were generated for this assay. Two control RNAs contained, respectively, the 5' 230nts of the DV2 and WNV genomes. A third RNA contained the DV2 5'NCR (DV2 nts 1–99) covalently linked to the 5'terminus of the WNV ORF (nts 100–230) (DV 5'NCR-WNV 5'ORF RNA), and the fourth contained the WNV 5'NCR (nts 1–99) covalently linked to DV2 nts 100–230 (WNV 5'NCR-DV5' ORF RNA). The 8-nt 5'CS sequence (WNV nts 137–144 or DV2 nts 134–141) per se is conserved between these two genomes (Khromykh et al., 2001). Therefore, any differences between the heterologous or chimeric 5'TRs in trans-activation of RNA synthesis from the WNV 3'NCR, compared to that of the wt WNV 5'TR, were expected to be due to nt sequence heterogeneity within the 5'NCR or within the ORF (nts 100–230), up or downstream from the 5'CS. An effect of these sequences in the corresponding region of the yellow fever genome on RNA replication was previously reported (Corver et al., 2003).
Trans-activation of RNA synthesis was scored by observation of the relative amounts of both 3'elongation and de novo products. Previous studies demonstrated that the "3'elongation products" represent negative sense extension initiated at the 3'terminus of the 3'NCR which generates a hairpin product molecule that is about twice the length of input templates, whereas "de novo" products represent denatured full-length ds RNAs (Ackermann and Padmanabhan, 2001; Nomaguchi et al, 2004; You et al, 2001). Transactivation was optimal in the presence of the homologous WNV 5'TR230nt RNA (Fig. 1A, lane 8). In contrast, RNA synthesis was barely detectable, when the DV2 5'TR230nt RNA was present in this assay (Fig. 1A, lane 2). Both chimeric 5'TR230nt RNAs exhibited some activity for trans-activating RNA synthesis from the WNV 3'NCR631nt (Fig. 1A, lanes 4 and 6) but less than the WNV 5'TR230nt control. In this regard the DV5'NCR-WNV5'ORF RNA was slightly superior to the WNV5'NCR-DV5'ORF RNA. Thus the in vitro system was sensitive to nt sequence differences between the 5' 230 nts of the WNV genome and those of the DV2 genome for the interaction of exogenous RdRp and the 5' and 3'TRs in mediating RNA synthesis from the WNV 3'NCR, notwithstanding the conservation of the 5'CS and cyc nt sequences. Possibly, the slight superiority of the DV5'NCR-WNV5'ORF RNA, compared to the WNV5'NCR-DV5'ORF RNA as a trans-activator of RNA synthesis, was an indicator that differences in nt sequence between the two ORFs were a determinant of the results. The assay shown in Figure 1A was repeated at least three times and gave similar results each time. Also, visual observations used as a basis for evaluating the relative template activities of these constructs were confirmed by direct quantification of radiolabel in RNA localized to the bands labeled as 3'elongation and de novo products, respectively, in Fig 1A (data not shown).
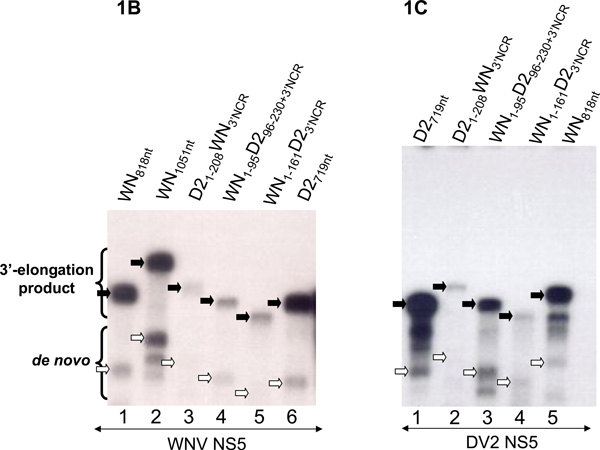
Figure 1. Nt sequence requirements for in vitro RdRP activity using purified WNV and DV2 NS5.
In vitro RdRP assays were performed as previously described (Nomaguchi et al., 2004; You and Padmanabhan, 1999). Radiolabeled RNA product molecules of in vitro RdRp reactions were separated by gel electrophoresis and visualized by autoradiography also as previously described (Ackermann and Padmanabhan, 2001). A. Transactivation of RNA synthesis off the WN3'NCR631nt RNA using heterologous or chimeric 5'TRs. RdRP reactions were carried out in the presence (+) or absence (−) of WN3'NCR631nt RNA. Lanes 1 and 2, reactions contained an RNA bearing the 5'terminal 230 nts of the dengue-2 strain NGC genome (DV5'TR230nt). Lanes 3 and 4, reactions contained a chimeric RNA representing the 5'NCR (nts 1–95) of the WNV genome covalently linked to nts 96–230 of the 5'terminus of the DV ORF (WN5'NCR-DV5'ORF). Lanes 5 and 6, reactions contained a chimeric RNA representing the DV 5'NCR nts 1–95 linked to the WN nts 96–230 (DV5'NCR-WN5'ORF). Lanes 7 and 8, reactions contained WN5'TR230nt RNA. Lane 9, reaction contained only WN3'NCR631nt RNA, as control. B and C: In vitro RdRP assays catalzyed by the WNV NS5 (B) or the DV2 NS5 (C) were done using homologous, heterologous, and chimeric WNV/DV2 minigenome RNAs as templates. WNV1051nt RNA contained the 5'terminal 281 nts of the WNV genome fused to the 3'terminal 770 nts, consisting of the 3'terminal 139 nts of the WN ORF plus the entire 631-nt 3'NCR. WNV818nt RNA contained WNV 5' nts 1–161 fused to the 3'terminal 656 nts of the WNV genome, consisting of the 3'terminal 25 nts of the ORF plus the 631-nt 3'NCR. DV2719nt RNA contained the 5'terminal 227 nts of the DENV2 genome fused to the 3'terminal 41 nt of the ORF plus the entire 451-nt 3'NCR. The construction of chimeric RNAs and conditions for PCR followed by in vitro transcription are described under Materials and Methods. The conditions for standard RdRP assays were previously described (Nomaguchi et al., 2004; You and Padmanabhan, 1999). The locations in the gel of 3'elongation products are indicated by solid arrows. The locations of de novo RNA products are indicated by open arrows.
We then assayed the relative ability of purified DV2 or WNV NS5 polymerases to catalyze RNA synthesis from each of six "minigenome" RNAs that contained homologous, heterologous, or chimeric 5'- and 3'-TRs covalently linked in template RNA molecules. The WNV polymerase recognized both the D2719nt and the WN818nt minigenomes for synthesis of the characteristic product molecules with similar efficiency (Fig. 1B, lanes 1 and 6; Table 1). The DV2 polymerase showed a distinct preference for the homologous wt D2719nt minigenome, but did recognize the WN818nt template, with yields of de novo product less than 3-fold reduced compared to those seen with D2719nt RNA (Fig. 1C, lanes 1 and 5; Table 1). For the WNV polymerase, all of the chimeric mini-genomes were consistently less efficient templates for RNA synthesis than were the two wt ones (Table 1). The D21–208WN3'NCR and WN1–161D23'NCR minigenomes, in that order, were the least efficient templates, regardless of the source of the polymerase (Fig 1B; Fig 1C; Table 1). The result for D21–208WN3'NCR RNA was congruent with that seen in the previous experiment, where 5'TRs were presented in trans (Fig 1A). The construct composed of the WNV 5'NCR (nts 1–95) fused to the DV2 5'ORF (nts 96–230, where the start codon is located at nts 97–99) and the DV2 3'NCR exhibited a vigorous template activity, when the reaction was catalyzed DV2 NS5 and was less efficient as a template for the WNV NS5 (Fig 1B, lane 4; Fig 1C, lane 3; Table 1). This was also analogous to results of the transactivation experiments, where the chimeric 5'TR composed of DV2 nts 1–99 and WNV ORF nts 100–230 was shown to be a relatively efficient trans-activator of RNA synthesis templated by the WNV 3'NCR and catalyzed by the WNV polymerase (Fig 1A) and suggested the signficant positive effect of species homology between the 5'ORF and the 3'NCR. Assays shown in Figs 1B and 1C were repeated at least three times and gave qualitatively identical results each time. Radiolabeled products of indeterminate size, seen particularly in Fig 1C, were interpreted to represent incomplete 3'-elongation or de novo product molecules, respectively, according to their apparent size.
Table 1.
Relative polymerase activity of WNV and DV2 NS5 proteins with indicated templates.
| NS5 | Productb | Yield from indicated templatec | |||||
|---|---|---|---|---|---|---|---|
| WN818nt | D2719nt | Ad | B | C | WN1051nt | ||
| WNa | de novo | 1.0 | 0.96 | 0.41 | 0.58 | 0.37 | 1.9 |
| 3'elongation | 1.0 | 1.22 | 0.31 | 0.25 | 0.12 | 1.06 | |
| D2 | de novo | 0.36 | 1.0 | 0.35 | 0.94 | 0.37 | NTe |
| 3'elongation | 0.72 | 1.0 | 0.12 | 0.49 | 0.06 | NT | |
WN, West Nile; D2, Dengue-2. NS5 proteins were expressed in E. coli and purified as described in Materials and Methods. Reactions were conducted as described elsewhere.
"De novo" RNA molecules represent full-length negative strand RNA copies of the respective RNA templates. "3' elongation" products represent input RNA covalently linked at the 3'terminus to approximately full-length negative stranded copies of the template.
Pixel densities of representative bands on the gel shown as Figs 1C and 1D were determined using the program ImageQuant. Results for the WNV NS5 polymerase were normalized to those obtained with the WN818nt RNA (shown in bold). Results for the DV2 NS5 polymerase were normalized to those obtained with the D2719nt RNA (shown in bold).
A, WN1–161D23'NCR RNA template; B, WN1–95D296–230+3'NCR RNA template; C, D21–208WN3'NCR RNA template.
NT, not tested.
Requirements for infectivity of wild type and 5' and 3' TR chimeric WNV RNAs
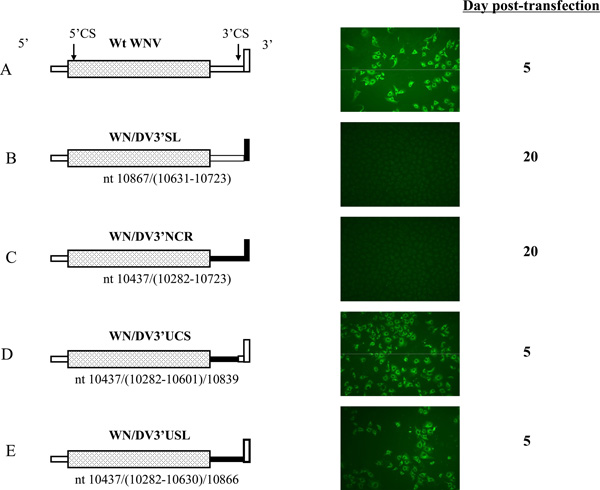
The results described above demonstrated some aspects of the specific interaction between the 3'NCR and the 5'TR that are required for optimal RNA synthesis in vitro, in a system that lacks NS3 and other viral NS and possibly cellular proteins thought to confer specificity (Khromykh et al, 2000). We next sought to examine this issue further in vivo, starting from our previous observation that the 3’SL nt sequence was not interchangeable between the DV2 and WNV genomes (Yu and Markoff, 2005; Zeng et al, 1998). To achieve our goal, several chimeric WNV/DV2 full length RNAs were generated in vitro and tested for replication competence by transfection of Vero cells. We initially created three WNV chimeras with DV2 3’NCR substitutions. In the first, the entire WNV 3’NCR including the 3’SL (WNV nt 10389–10962) was replaced by the corresponding DV2 3’NCR segment (construct C). In the second and third, only the segments of the WNV 3’NCR upstream of CS1 (3’UCS1) and the 3'NCR upstream of the 3'SL (3’USL), respectively, were substituted by the analogous DV2 segments (Fig. 2, constructs D and E, respectively). Replication of viruses produced from transfected wt and mutant RNAs was assayed by an immune fluorescence assay (IFA) specific for expression of WNV antigens in transfected cells. “Viable” viruses gave positive results by IFA within 20 days after transfection, whereas RNAs used to transfect cell monolayers that remained negative during this observation period were considered to bear "lethal" mutations. Chimeric WNV RNA transcripts derived from construct C and a negative control construct containing the 3'SL of DV2 substituted for the WNV 3'SL nt sequence (construct B) produced no WNV-specific antigens (Fig. 2, panels B and C) or virus plaques (data not shown) in Vero cells. In contrast, viruses derived from construct D (Fig. 2, panel D) and E RNAs (Fig. 2, panel D) were similar to wt WNV in kinetics of replication as assessed by IFA (Fig. 2, panel A).
Figure 2.
Replication of WNV viruses containing DV2 3’NCR substitution mutations. The left panel shows schematic representations of the wt and mutant WNV genomes tested. The WNV ORF is represented as a stippled rectangle. WNV 5' and 3'NCR nt sequences are represented by open rectangles, and the DV2 5' and 3'NCR nt sequences are represented by closed rectangles. Nts comprising the 3'SL are indicated by vertically oriented rectangles. Nucleotide numbers in parenthesis indicate nts derived from the DV2 genome that were appended to the WNV genome at the site in the WNV genome indicated by the nt number to the left of the parenthesis. The right panel exhibits the results of IFA on the day indicated. Vero cells were transfected with RNAs of Wt WNV, WNV/DV3’SL, WNV/DV3’NCR, WNV/DV3’UCS, and WN/DV3’USL, as indicated on the left. At 5-day intervals up to 20 days post-transfection, the cells transferred to a coverslip were fixed with acetone, treated with mouse anti-WNV anti-serum, and stained with fluorescein isothiocyanate-conjugated goat-anti-mouse immunoglobulin G.
As shown also by results of an infectious center assay (ICA) and a plaque assay (see below), infectious progeny viruses were produced from cells transfected with construct D and E RNAs. Further, virion RNAs were isolated from infected cell supernatants and sequenced. Results confirmed the stability of the mutations in input RNA in all cases (data not shown). A comparison of the phenotypes of RNA constructs C and E suggested that the lethal phenotype of C was solely due to the substitution of the DV2 3’SL for the WNV 3'SL. Thus DV2 3’NCR nt sequences could functionally replace analogous segments of the WNV genome, despite the divergence in nt sequence and total length of the 3'NCR between the two genomes (reviewed in Markoff, 2003), provided only that the WNV 3'SL was retained within the WNV RNA. These phenotypes were reproducibly observed.
WNV genomes bearing a substitution of the WNV 5'NCR by the DV2 5'NCR
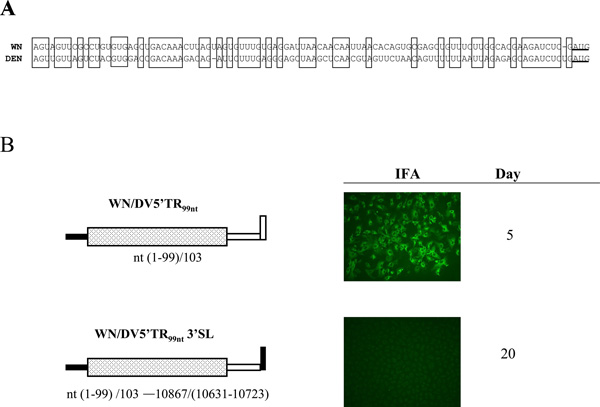
Alignment of the 5’NCR nt sequences in DV2 and WNV RNAs revealed less than 50% identity (Fig. 3A). We therefore sought to determine whether the WNV 5’NCR was a determinant of the specific requirement for the WNV 3’SL for replication of WNV. To address this question, we created two additional chimeric WNV genome RNAs. In the first, the WNV 5'NCR was substituted by the DV2 5’NCR (Fig. 3B; WN/DV5'TR99nt RNA), and in the second, the DV2 5'NCR and the DV2 3'SL replaced the corresponding WNV segments (Fig 3B; WN/DV5'TR99nt3'SL RNA). Once again, virus replication was first assessed by IFA on transfected cells. WN/DV5'TR99nt RNA gave a positive result by day 5 after transfection, similar to wt WNV RNA, and progeny viruses were produced from transfected cells. Further, nt sequence analysis of RNA recovered from transfected cells indicated that WN/DV5'TR99nt RNA was genetically stable. In contrast, substitution of both the 5'NCR and the 3'SL of the WNV genome with the corresponding regions of DV2 in WN/DV5'TR99nt3'SL RNA resulted in a lethal phenotype with no viral antigens detected up to 20 days post-transfection in repeated experiments (Fig. 3B). Thus, the lethal effect of substituting the WNV 3'SL nt sequence with that of the DV2 genome could not be abrogated by substitution of the WNV 5'NCR with the analogous DV2 nt sequence.
Figure 3.
Replication of WNV genomes containing DV2 5’NCR substitution mutations. A. Nucleotide sequence alignment of WNV and DV2 5’NCRs. Conserved nucleotide segments are boxed. There is about 50% sequence identity. The translation start codon AUG is underlined. B. WNV genomes bearing substitutions of the 5’NCR by that of the DV2 genome are depicted, and the results of an IFA to detect virus replication in transfected Vero cells are shown. The experimental details are same as in Figure 2 legend.
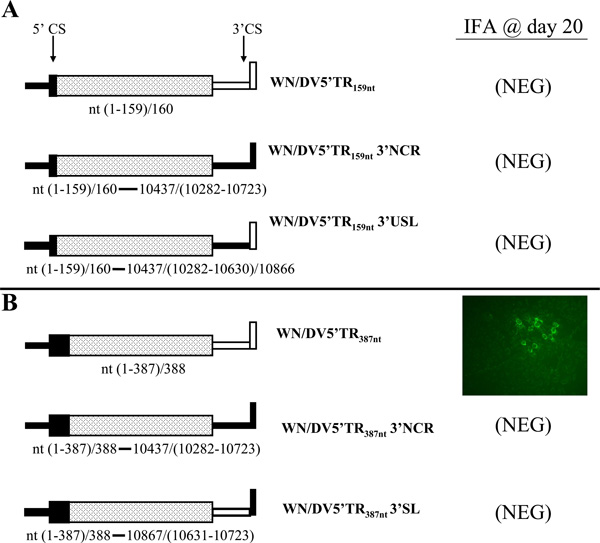
Next, to determine whether the heterologous DV2 capsid nt sequences up or downstream from the 5'CS could affect replication of chimeric WNV/DV2 RNAs, we constructed three mutant WNV infectious DNAs, all of which contained a 5’terminal substitution of WNV nts 1 to 159 by the analogous region from the DV2 genome (Fig 4A). This region includes the entire 5'NCR and the first 21 codons of the capsid gene and therefore encompassed the 5'CS (WNV nts 137–144). This mutation was paired with the wt WNV 3'NCR in WNV/DV5'TR159nt DNA, with the 3'NCR of DV2 in WNV/DV5'TR159nt 3'NCR DNA, or with a chimeric 3'NCR consisting of a substitution of DV2 nt sequences upstream from the WNV 3'SL in WNV/DV5'TR159nt 3'SL DNA. When RNAs synthesized from these mutant DNAs were transfected into Vero cells, none of them showed any signal for replication, as detected by IFA up to 20 days post-transfection (Fig. 4A). No virus release was detected by plaque assays (data not shown). Thus, all three clones exhibited lethal phenotypes.
Figure 4.
Replication of WNV genomes containing heterologous or homologous DV2 5’ and 3’CS elements. A. WNV/DV2 chimeric genome RNAs containing the DV2 5’NCR and nts 100–159 of the 5'ORF, which includes the 5' cyclization sequence (CS), minus (WN/DV5’TR159nt RNA) or plus the entire DV2 3'NCR (WN/DV5’TR159nt3’NCR RNA) or the DV2 3'NCR upstream from the WNV 3'SL (WN/DV5’TR159nt3’USL RNA) are depicted on the left. These RNAs were transfected into vero cells, and WN virus replication was monitored by an IFA conducted at 5-day intervals up to day 20 post-tranfection, on an aliquot of cells that had been transferred to a coverslip. Negative results on day 20 post-transfection of the RNAs is indicated on the right. B. WNV/DV2 chimeric genomes analogous to those depicted in part A were constructed and are depicted on the left, except that in each case the entire ORF of the DV2 capsid gene segment (nts 100–387) was substituted for the ORF of the WNV capsid. Results of the IFA (described in part A and in the legend to Fig 2) on day 20 post-transfection of the indicated RNAs are shown on the right.
The lethal phenotype of WNV/DV5'TR159nt RNA was most likely due either to dysfunction of the chimeric capsid protein encoded by the latter genome or to a requirement for nts within the WNV ORF up- or downstream from the 5'CS for 5'/3' end interaction. To test these alternative hypotheses, we constructed three more mutant WNV infectious cDNAs in which the entire coding sequence of the WNV capsid was replaced by that of the DV2 capsid (Fig. 4B). The WNV mutant RNA containing the DV2 5'NCR and the entire DV2 capsid gene segment (WNV/DV5'TR387nt) was able to replicate but gave a positive result by IFA only at day 20 post-transfection (Fig 4B, top panel). The lag in viral replication as determined by IFA was consistent with the results from other analyses based on the kinetics of plaque formation and plaque morphology (see below). Sequencing of progeny WNV/DV5'TR387nt virion RNA revealed that the expected DV2 sequences from nts 1 to 387 were retained. However, we detected three point mutations, two within the prM gene segment and one within the NS5 gene segment (Table 2). Whether these point mutations were due to PCR-introduced errors or represent necessary compensatory mutations was not directly examined. Thus, the chimeric structure of the capsid protein was at least partly a cause of the lethal phenotype of WN/DV5’TR159nt RNA and the related mutant RNAs derived from it (Fig 4A), although replacement of the entire WNV capsid gene segment in WN/DV5’TR159nt RNA with that of DV2, in the context of WN/DV5’TR387nt RNA, did not completely "repair" the defect. In contrast, an analogous mutant RNA in which the entire WNV 3'NCR was replaced by that of DV2 (WN/DV5’TR387nt 3’NCR RNA) and one in which only the WNV 3'SL was replaced by that of DV2 (WN/DV5’TR387nt 3’SL RNA) were completely unable to replicate (Fig 4B; bottom two panels). These results again demonstrated the prime importance of the homologous 3'SL nt sequence in WNV RNA replication, regardless of the origin of the 5'TR or upstream 3'NCR nt sequences.
Table 2.
Second site mutations in WN/DV5’TR387nt RNA.
| Nucleotide No.a |
AA No.b | Mutation | AA change | Gene segment |
|---|---|---|---|---|
| 522 | 142 | C to U | Silent | prM |
| 611 | 172 | U to C | Ile to Thr | prM |
| 8763 | 2889 | C to A | Silent | NS5 |
Based on nt sequence of WNV strain 956 genome (GenBank M12994).
Amino acid (AA) number referenced to the start codon for the single long ORF in the WNV genome.
Specific infectivity of mutant WNV RNAs and growth kinetics and plaque morphology of viable viruses
We estimated the specific infectivity of viable mutant WNV RNAs by an infectious center assay for plaque formation on vero cell monolayers (Table 3). Results suggested that wt WNV, WN/DV5'TR99nt, and WN/DV3'USL RNAs were comparably infectious, whereas WN/DV5'TR387nt RNA was at least 100-fold less infectious than any of the others. This could indicate that the WN/DV5'TR387nt genome was relatively defective for RNA replication or alternatively that the structurally chimeric virus encoded by WN/DV5'TR387nt RNA was defective either in virion morphogenesis or in the capacity of chimeric particles to infect adjacent cells in the monolayer. The finding that transfected cells did not become positive for WNV antigens until late times after transfection suggests that in fact the defect is at the level of RNA synthesis, since we would have expected WNV antigens to have been detectable in transfected cells at early times post-transfection, if the defect was at the level of virion morphogenesis.
Table 3.
Results of infectious center assays for wild type and mutant RNAs in Vero cells.
| Virus RNA | Specific infectivitya (pfu/ug of RNA) |
|---|---|
| Wt WNV | 36000 |
| WN/DV5’TR99nt | 34000 |
| WN/DV3’USL | 23000 |
| WN/DV5’TR387nt | 160 |
Vero cells were transfected with the indicated viral RNAs. Then transfected cells were serially diluted, plated on 6-well plates, and overlaid with agarose. Plates were incubated for 5–8 days, then cells were fixed and stained to visualize plaques. Average results of two separate experiments are shown.
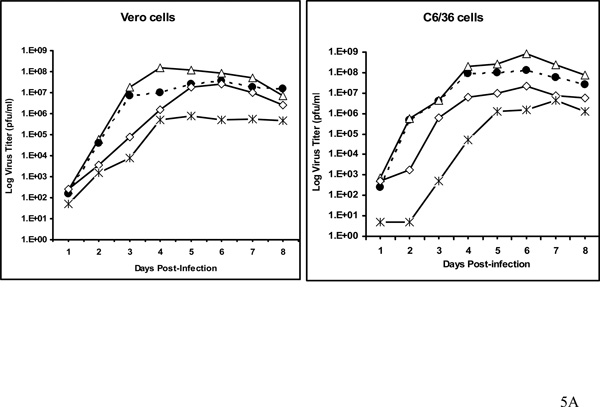
As another measure of virus replication competence, the growth rates of wt and viable mutants in Vero and C6/36 cells were determined at a multiplicity of infection of ~0.01 in each case, using plaque-titered stocks. After infection, plaque assays were done daily for the virions secreted into the medium, for 8 days. The peak titer for wt WNV in Vero cells was about 1.5 × 108 pfu/ml, achieved at day 4 post-infection (Fig. 5A), whereas, the titer of WNV/DV5'TR99nt virus was about 10-fold lower than wt at day 4. However, by day 6 this virus reached a peak titer of 3.5 × 107 pfu/ml. Similarly, the titer of WNV/DV3'USL virus was almost 100-fold lower than that of wt WNV at day 4 after infection, but it ultimately reached a peak titer of about 2.5 × 107 pfu/ml (Fig. 5A). In contrast, WNV/DV5'TR387nt virus reached a titer comparable to that of WNV/DV3'USL virus at day 4, but the peak titer never exceeded 1 × 106 pfu/ml in Vero cells. Thus if the spontaneous mutations detected in the WNV/DV5TR387nt genome (Table 2) were compensatory, they were not sufficient to restore replication competence to the level of wt virus.
Figure 5.
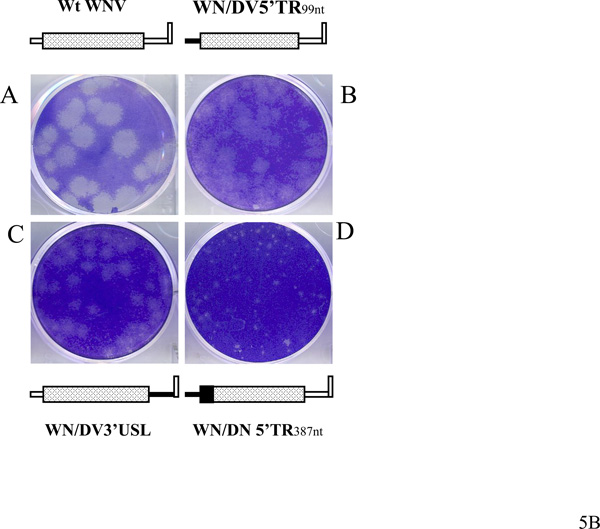
A. Growth of viable WN/D2 chimeric viruses in Vero and C6/36 cells. Plaque titers were determined for pools of viruses derived from transfected Vero cells. These viruses were used to infect confluent monolayers of Vero and C6/36 cells at a multiplicity of infection of 0.01. Viruses in the supernatant of infected cell monolayers were plaque titered on Vero cells on the days shown post-infection. Wt WNV, open triangles; WN/DV5’TR99nt virus, closed circles; WN/DV3’USL virus, open diamonds; WN/DN5’TR387nt virus, data points indicated by "x". B. Plaque size differences among chimeric mutant viruses as compared to wt WNV. Vero cells were infected with stock viruses as described under Materials and Methods, and plaques were stained with crystal violet at day 5 post-infection, except for plaques formed by WNV/DV5’TR387nt virus, which were stained at day 8 post-infection.
The peak titer for wt WNV in C6/36 cells was about 8.75 × 108 pfu/ml, achieved on day 6 post-infection (Fig. 5A). WNV/DV5'TR99nt and WNV/DV3'USL viruses attained slightly lower peak titers of 1.25 × 108 and 2.25 × 107 pfu/ml, respectively. The replication of WNV/DV5'TR387nt virus was retarded at early times post-infection, but the kinetics paralleled those of wt virus between days 3 and 5. However, the peak titer only reached 4.25 × 106 pfu/ml at day 7, almost 200-fold lower than that of the wt virus. Results in C6/36 cells roughly paralleled those seen in Vero cells, and differences among the growth curves in general paralleled the differences in the respective specific infectivities of the RNAs, as demonstrated in the infectious center assay.
Viruses recovered from infectious supernatants were used for a determination of plaque size on day 5 post-infection of confluent monolayers of vero cells. The plaque size of WNV/DV5'TR99nt and WNV/DV3'USL viruses (construct D in Fig. 2) ranged from 2 to 3.5 mm in diameter, smaller than that of wild type virus (about 4 to 6 mm). In contrast, WNV/DV5'TR387nt virus produced minute plaques at day 5 post-infection (data not shown), and plaques were still smaller than 1.5 mm diameter at day 8 post-infection (Fig. 5B). These results, taken together, suggested that both the DV2 5’NCR and the DV2 3’NCR upstream from the 3'SL were quite functional in the context of WNV replication. However, substitution of the WNV capsid protein gene by the corresponding DV2 gene, in the context of the relevant chimeric genomes, reduced the efficiency of WNV replication by all measurements.
Analysis of interaction between 5’ and 3’ terminal regions using MFold
The nt sequences of the 5’NCRs and 5'ORFs in the DV2 and WNV genomes are divergent, as noted above. The 3'NCRs of the respective genomes are also highly divergent except for the CS1 nt sequence already mentioned and CS2, a 23-nt sequence that is strongly conserved between the WNV and DV2 genomes (reviewed in Markoff, 2003). In order to assess the effects of the substitution of WNV sequences with corresponding regions of the DV2 genome on 5'/3' base pairing, we used both the MFOLD algorithm (v3.2) for RNA secondary structure analysis (Zuker et al, 1991) and the program RNAdraw (Matzura and Wennborg, 1996). Results were the same for either method of predicting secondary structure. To avoid potential generation of alternate secondary structures arising from different lengths of RNA and complications in interpretation of data, we analyzed equal lengths of wild type and chimeric RNAs. All mini-genome RNAs entered into this analysis contained a total of 438 nts, 195 nts derived from the respective 5'TRs plus 243 nts derived from the respective 3'NCRs.
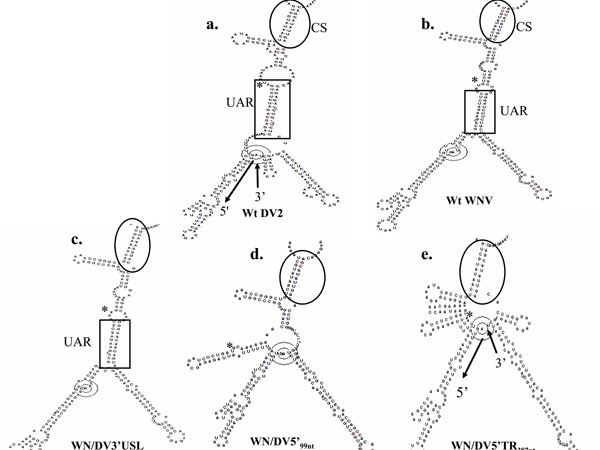
The results predict that a 5’/3’ terminal association occurs in viable mutant RNAs, resulting in secondary structures at least loosely comparable to those predicted for the respective wt RNAs (Fig 6). Comparison of wt DV2 and WNV minigenome RNA secondary structures (Figs. 6a and b, respectively) with those of mutant RNAs with heterologous 5’- or 3’-end sequences, for example, WNV/DV3'USL (Fig 6c), WNV/DV5'TR99nt RNA (Fig 6d) and WNV/DV5'TR387nt RNA (Fig 6e), revealed that base pairing between the 5'CS and the "cyc" nt sequence was predicted to occur in all cases. In addition, wt DV2, wt WNV, and WNV/DV3'USL RNAs (Fig. 6) formed a 12- to 15-bp duplex between 5' and 3' nt sequences previously defined as "Upstream AUG Regions" (UARs) (Alvarez et al., 2005a). UAR base-pairing was also predicted to be retained in mutant RNAs WNV/DV5'TR159nt3'NCR, WN/DV5’TR159nt3’SL, and WNV/DV5'99nt/DV3'SL (data not shown), yet none of these latter mutant RNAs were viable (Figs 3A and 4B), possibly owing to the chimerization of the capsid gene or to the dominant influence of the 3'SL nt sequence in determining viability. In contrast, there were no UAR base pairings predicted in some viable mutant RNAs, including WNV/DV5'TR99nt (Fig 6d) and WNV/DV5'TR387nt RNAs (Fig 6e). This was a possible indication that the UAR base pairings are dispensible for replication in the presence of the homologous 3'SL nt sequence. The predicted secondary structure of WN/DV5'TR387nt minigenome was aberrant in several other ways compared to those of all other viable RNAs, all of which replicated with much greater efficiency. Perhaps this was a factor in the relative lack of infectivity of this construct.
Figure 6.
Predicted RNA Secondary structures resulting from 5’ and 3’end interactions. The potential 5' and 3'end interactions were predicted using both RNAdraw (Matzura and Wennborg, 1996) and MFOLD v3.2 (Zuker et al, 1991) algorithms. a. Wt DV2 RNA; b. WNV RNA; c. WN/DV3’USL RNA; d. WN/DV5’99nt RNA; e. WN/DV5’TR387nt RNA. WN/DV5'99nt RNA contained an AAT to CAA mutation of nts 100–102, which was introduced in association with generating the chimera. The 5’/3’ CS base-pair interactions are indicated by ovals. Additional hydrogen bonding due to an interaction of the 5' and 3' UAR nt sequences (in the DV2 genome, 5’ nts 82–96 and 3’ nts 10642–10656) are boxed. The ATG codon is labeled with an asterisk. Arrows in 6a indicate 5’ and 3’ directions.
DISCUSSION
We used a previously established in vitro assay for polymerase activity of purified WNV and DV2 NS5 proteins (Ackermann and Padmanabhan, 2001; Nomaguchi et al., 2004; You et al., 2001; You and Padmanabhan, 1999) to determine their ability to catalyze synthesis of de novo RNAs off the 3'terminus of DV2, WNV, and chimeric RNAs containing nt sequences derived from the 5'TRs and the 3'NCRs of the two genomes. To correlate the in vitro results with virus physiology, we then determined the infectivity of genome-length WNV RNAs containing strategic substitutions of DV2 nt sequences for WNV nt sequences comprising the 5'TR and/or the 3'NCR. We had previously shown in two separate studies that 3'SL nt sequences en bloc could not be swapped in either direction between the DV2 and WNV genomes without abrogating infectivity of the parent genome (Yu and Markoff, 2005; Zeng et al, 1998). A goal of the present work was to determine whether further substitution of 5'TR and 3'NCR WNV nt segments known to be required for RNA replication by analogous DV2 nt segments, in the context of the nonviable WNV genome in which the WNV 3'SL was substituted by DV2 3'SL nt sequences (WN/DV3'SL RNA; Fig 2), could restore replication competence. The effects of these additional substitution mutations on WNV replication were also assessed by introducing them into a fully wt WNV genome.
As for other RNA viruses (Blumenthal and Carmichael, 1979; Herold and Andino, 2001; Hewlett et al, 1977; Hsu et al, 1973; Hu et al, 2007; Ooms et al., 2007), circularization of the genome appears to be a pre-requisite for replication of flavivirus RNAs (Alvarez et al, 2005b; Filomatori et al, 2006). The conserved 5' and 3' cyclization motifs in flavivirus genomes were initially shown to be required for negative strand RNA synthesis by in vitro methods (Ackermann and Padmanabhan, 2001; Nomaguchi et al, 2004; You et al., 2001; You and Padmanabhan, 1999). A physical interaction between 5' and 3'terminal nt sequences dependent on complementarity between the two cyclization motifs was demonstrated by psoralen-UV-cross-linking (You et al., 2001), and recently by atomic force microscopy (Alvarez et al., 2005b). The requirement for complementarity of the cyclization motifs in RNA replication was also established using flaviviral replicon RNAs (Alvarez et al., 2005a; Corver et al., 2003; Khromykh et al., 2001; Lo et al., 2003) and mutant genomes derived from infectious DNAs (Alvarez et al., 2005b; Bredenbeek et al., 2003).
Given the conservation of the 5'CS and its 8-nt complement within CS1 in the WNV and DV2 genomes, we assumed that differences we observed in activity of the relevant mutant RNAs in vitro and replication efficiency of analogous mutant genomes in vivo were determined by nt sequence differences between the two genomes that occur up and downstream from the formally recognized cyclization segments, either by an effect on 5'/3' hydrogen bonding or by the possible effects of nt substitutions on binding of viral or cellular proteins necessary for replication. For example, the 5'CS is part of a larger conserved nt sequence element that was originally predicted to participate in 5'/3' hydrogen bonding with the entirety of CS1 in the 3'NCR (Hahn et al., 1987). In support of that hypothesis, Corver et al (2003) demonstrated that nts 147 through 165 (within the capsid gene ORF) were required for fully efficient replication of a replicon derived from the Yellow Fever strain 17D (YF) genome. Since the 5'terminus of the 8-nt 5'CS maps to nt 156 in the YF genome (Khromykh et al., 2001), the domain mapped by Corver et al (2003) closely corresponds to the one originally described by Hahn et al (1987) in that it included the 5'CS plus an additional 9 nts upstream and 2 nts downstream and is complementary to CS1 in the YF genome. CS1 within the WNV and DV2 genomes are slightly divergent from each other; they differ in nt sequence at 2 positions, and in addition CS1 of the DV2 genome is one nt shorter than that of the WNV genome (reviewed in Markoff, 2003). These small differences in nt sequence may in fact account for some of our observations of specificity in the in vitro system (see below).
Hydrogen bonding between the 5' and 3'UARs was deemed to be an additional important factor for RNA circularization and replication competence (Alvarez et al., 2005a). However, the 5' and 3' UAR nt sequences are also conserved between the WNV and DV2 genomes. Thus the failure of chimeric minigenomes to serve efficiently as template for in vitro RNA synthesis or of chimeric genome RNAs to replicate in transfected cells was unlikely per se to be due to a lack of complementarity between 5' and 3' UARs, although alteration of the nt sequences up or downstream from the UARs might conceivably interfere with their hydrogen bonding. In fact, using MFold, the 5' and 3'UARs in two viable mutant RNAs, WNV/DV5'TR99nt and WNV/DV5'TR387nt, were not predicted to base pair. Since the analysis was based on limited sequence data and is further limited by the predictive accuracy of MFold per se, we would hesitate to draw any firm conclusions from this finding. However, it should be noted that the 3'UAR is composed of nts that alternatively base pair with each other to form the bottom part of the long stem in the 3'SL (Alvarez et al., 2005b). Thus the consequence of hydrogen bonding between the UARs is a foreshortening of the length of the long stem in the 3'SL. Results of the MFold analysis are indicative that the "classical" 3'SL structure (Brinton et al, 1986; Hahn et al., 1987; Rice et al., 1985; Wengler and Castle, 1986) is thermodynamically favored over hydrogen bonding between the UARs in the context of the two mutant RNAs used for the MFold analysis.
According to the proposed model (Filomatori et al., 2006), NS5 polymerase initially recognizes and binds the SLA. Circularization of the genome (and possibly the action of other proteins) presumably brings the 3'terminus into a close spatial relationship with SLA, permitting the replicase to commence negative strand RNA synthesis. The SLA nt sequence is also conserved between the WNV and DV2 5'NCR nt sequences, despite their roughly 50% overall nt sequence similarity. Conservation of the SLA nt sequence and of the 5'UAR nt sequence may account in part for our finding that the 5'NCR of the DV2 genome can substitute for the 5'NCR of the WNV genome in the context of the WNV RNA with no significant loss of viability.
Some known characteristics of the requirement for the 3'SL in negative strand RNA synthesis were briefly discussed in the Introduction. In contrast to the formal cyclization sequences, the 5' and 3'UARs, and SLA, the nt sequences of the WNV and DV2 3'SLs are highly divergent, and the loci of bulges in the long stems of these structures that are required for RNA replication are different for the two different 3'SLs. This appeared to account for the finding that the WNV and DV2 3'SLs en bloc are not interchangeable between the two genome RNAs (Yu and Markoff, 2005; Zeng et al, 1998). Finally, the 3'NCRs of the DV2 and WNV genomes differ significantly in total length (451 nts vs 631 nts, respectively). Both contain additional mosquito-borne flavivirus-conserved features (such as CS1, CS2, and RCS2; reviewed in Markoff, 2003) upstream from the 3'SL, and there are data to implicate some of the conserved stem-loop structures upstream from the 3'SL as functionally important in RNA synthesis (for example see Men et al, 1996). However, the WN genome contains a pair of tandem repeat nt sequences (CS3 and RCS3) not found in the DV2 3'NCR. Our results suggest that CS3 and RCS3 are not needed for WNV replication in Vero or C6/36 cells.
In the in vitro system, the DV5'TR230nt RNA was not able to stimulate highly efficient synthesis of 3'-elongation product RNA off the WNV 3'NCR, catalyzed by the WNV NS5, whether the 5'TR was provided in trans or in cis. Conversely, both the WNV and the DV2 NS5 were relatively inactive when presented with constructs in which the 5'TR and 3'NCR were heterologous (in D21–208WN3'NCR and WN1–161D23'NCR minigenome RNAs). These in vitro results paralleled those observed after transfection of a WNV mutant RNA that contained a DV2-derived 5'TR, including the entirety of the DV2 capsid gene ORF (WN/DV5'TR387nt RNA). The mutation was not ultimately lethal, but evidence of virus replication by IFA was not obtained until day 20 post-transfection, and the RNA had a significantly lower specific infectivity compared to wt and other viable mutant RNAs. Data indicated that inefficient replication of the WN/DV5'TR387nt mutant virus was due to a defect at the level of RNA synthesis possibly resulting from relative lack of complementarity between nts in the DV2 capsid gene segment with nt sequences in the WNV 3'NCR. This was consistent with the previously cited work with the YF replicon (Corver et al., 2003) where replication efficiency was sensitive to even a 2-nt deletion of the YF capsid gene nt sequence complementary to CS1 and was completely abrogated by a 3-nt substitution mutation that reduced 5'/3' complementarity within this segment.
One major difference between the DV2 and WNV NS5 RdRP activities in the in vitro system was seen with WN1–95D296–230+3'NCR minigenome RNA as template. The DV2 polymerase was consistently more active with this template than was the WNV polymerase. It is tempting to ascribe this difference in activity to the species specificity of the 3'SL that was demonstrated in the previous (Yu and Markoff, 2005; Zeng et al, 1998) and present in vivo studies. However, this specificity was not otherwise observed in vitro; the WNV polymerase did not discriminate between minigenomes containing all-DV2 and all-WNV 5' and 3'NCR nt sequences, and the DV2 polymerase exhibited significant (although reduced compared to control) activity in the presence of the WNV818nt template. Therefore, we speculate that the superior activity of the DV2 NS5 compared to the WNV NS5 with the WN1–95D296–230+3'NCR template could have been due in part to an interaction between the homologous DV2 5'ORF nts included in the construct (i.e., nts 96–230) and the DV2 3'NCR that favored the DV2 polymerase. In light of these results, the observed phenotypes of the D21–208WN3'NCR and WN1–161D23'NCR minigenome RNAs in vitro and of WN/DV5'TR387nt RNA in vivo, and the cited work with the YF replicon (Corver et al, 2003), it appears that replication efficiency is extremely sensitive to nt sequences, especially those within the capsid gene ORF, that may lie up or downstream from the commonly described conserved segments needed for genome circularization.
Other new data presented here show that no segment of the 5'TR or 3'NCR upstream from the 3'SL can "rescue" a nonviable mutant WN genome bearing the DV2 3'SL; all mutant genome RNAs in which the 3'SL was heterologous to the non-structural proteins were unable to replicate. Conversely, no other DV2 nt segment within the 5'TR and 3'NCR was per se lethal to replication of substitution mutant WNV RNAs. (Specifically, the DV2 5'NCR, capsid ORF, and the 3'NCR upstream from the 3'SL were individually tested in the context of substitution mutant WNV genomes, and all RNAs yielded viable viruses.) These observations favor the hypothesis that viral NS proteins other than NS5 (rather than cellular proteins) are the principle determinant of the specificity of the 3'SL. Taken together with our previous work demonstrating that the loci of bulges in the long stem of the DV2 and WNV 3'SLs are determinants of their specific requirements for negative strand RNA synthesis (Yu and Markoff, 2005; Zeng et al, 1998), we would now therefore posit that binding of viral NS proteins other than NS5 to such bulges in the long stem mediates this specificity. Cellular proteins also play an as yet undefined role in these processes, possibly a la the role of PABP in the circularization of poliovirus RNA (Herold and Andino, 2001). One cellular protein likely to be involved in WNV replication is eEF1A (Davis et al., 2007).
MATERIALS AND METHODS
Construction of homologous and chimeric minigenome DNA templates
The construction of DNA plasmids encoding the D2719nt and WN1051nt minigenome RNAs (pSY-2719nt and pSP64-WNV1051nt DNAs, respectively) has been described elsewhere (Nomaguchi et al., 2004; You and Padmanabhan, 1999; You and Padmanabhan, 2001). In summary, pSY-2719nt DNA contained the 227-nt 5'terminus of the dengue-2 virus (DV2) strain NGC genome in covalent linkage to a 3'terminal fragment of the genome containing the last 41 nts of the NS5 ORF (including the stop codon) plus the entire 451-nt 3'NCR. pSP64-WNV1051nt DNA contained the 5'terminal 281 nts of the West Nile virus (WNV) strain Eg101 genome fused to the 3'terminal 770 nts, i.e., the last 139 nts of the NS5 ORF plus the 631-nt 3'NCR. To construct the pSP64-WNV818nt plasmid DNA encoding WN818nt RNA, the pSP64-WNV1051nt plasmid (3µg) was sequentially incubated in 50µl reaction volume with BsmI, which cut the DNA at nt 161 within the 5'terminal nt sequences, and BbsI, which cut the DNA at nt 385 of the construct, 666 nts upstream from the 3'terminus of the genome, for 3h at 37°C and 3h at 65°C. The DNA was purified by QIAquick purification kit (Qiagen), blunt-ended with Klenow DNA polymerase (5U/50µL volume), and incubated with T4 DNA ligase. The ligation product was cloned using standard protocols. This procedure removed nucleotides between BsmI and BbsI sites to yield the pSP64-WNV818nt plasmid.
The recombinant plasmid template for D21–208WN3'NCR RNA was created by digestion of pSP64-WNV1051nt and DV2 pSY-2 plasmids (3µg each) with EcoRI (24U) (which cut at a single site in the vector upstream from the T7 promoter) for 2h at 37°C, followed by digestion with BsmI (20U) (which cut the WNV DNA at 161 and the DV2 DNA at nt 208, within the respective 5'terminal nt sequences in these fragments) at 65°C for 2h. The plasmids were purified on a QIAquick column after each digestion. The larger product of SP64-WNV1051n plasmid DNA digestion was treated with calf intestinal alkaline phosphatase (CIAP) at 37°C for one hour, purified by agarose gel electrophoresis, and extracted using a QIAquick gel extraction kit. The DV2-specific insert released from the pSY-2 plasmid by EcoRI and BsmI digestion was cloned between the EcoRI and BsmI sites of the pSP64-WNV1051nt plasmid. The product molecule was then sequentially digested to remove redundant WNV nts between the BsmI site and the BbsI site, blunt-ended with Klenow fragment, re-ligated with T4 DNA ligase, and cloned to yield D21–208WN3'NCR DNA. The WN1–161D23'NCR plasmid DNA was constructed in an analogous manner, by digesting pSP64-WNV1051nt DNA and the DV2 pSY-2 plasmid with EcoRI and BsmI. The resulting recombinant vector DNA fragments containing the 5'terminal 161 nts of the WNV genome and the 3'NCR of the DV2 genome, respectively, were gel purified, ligated, and cloned to yield the desired construct. To construct the WN1–95D296–230+3'NCR DNA, the recombinant plasmids SP64-WNV818nt and DV2 pSY-2 were doubly digested with the restriction endonucleases EcoRI and BglII, releasing fragments bearing nts 1–95 of the WNV and DV2 genomes, respectively. The WNV-specific fragment was then cloned into DV2 pSY-2 plasmid DNA from which the DV2-specific EcoRI-BglII fragment had been removed. All constructs were verified by nt sequence analysis.
Generation of full length WNV-DV2 chimeric cDNAs
A recombinant plasmid DNA containing a full-length copy of the WNV strain 956 genome, pSP6WN/Xba (strain 956, GenBank M12294), was reported previously (Yamshchikov et al., 2001). The full-length WNV DNA insert was initially sub-cloned into a pRS424 yeast shuttle vector (http://seq.yeastgenome.org/vectordb/vector_descrip/PRS424.html) at ClaI and NotI sites, resulting in recombinant pRSWN DNA. To construct either 5’- or 3’-terminal WNV/DV2 chimeric mutants, the 5’terminal ~2 kb of the WNV DNA was released from pRSWN DNA by digestion with restriction endonucleases KpnI (pRS424 1894) and SacI (WNV nt 2199) and inserted into the pBluscript vector (Stratagene, La Jolla, CA) at SacI-KpnI sites, to yield pBWN2K. Then a ~4 kb fragment of the DNA containing the 3'terminus of the WNV genome was released from the pSP6WN/Xba plasmid by digestion with SphI (WNVnt 6777) and XbaI (WNVnt 10962) and cloned into the pGEM vector (Promega, Madison, WI) at the same sites to yield pGWN4K.
The pGWN4K DNA was used to make substitution mutations in the WNV 3’NCR. To create pGWN4K/DV.3’UCS (a substitution of WNV 3'NCR nt sequences upstream from CS1 by analogous DV2 nt sequences), A pair of primers were used to synthesize a PCR copy of the DV2 "3’UCS" nt sequence. A DV2 10282 Bgl II sense primer, AACTAGATCTAACATGAAACAAGGCTAGAAGRCAGG, containing the DV2 nts 10282 to-10290 and a 5'terminal BglII recognition sequence (underlined), and a DV2 10601 AgeI anti-sense primer, AAATACCGGTCTCCTCTAACCTCTAGCCTTCCAG, containing DV2 nts 10601-10574 and a 5'terminal AgeI site (underlined). Wild type DV2 DNA (Polo et al., 1997) was used as a template. The entire pGWN4K DNA sequence was also PCR-amplified using a sense primer with a 5'terminal AgeI site, AAATACCGGTGCCAAAAACACCAAAAGAAACAGCATATT (WNVnt 10833–10863) and an antisense primer with a 5'terminal BglII site, AATTAGATCTCATTCTCAATAAATCCTATTTACACTAA (WNVnt 10437-10410). The DV2 3’UCS and the linear pGWN4K PCR products were treated with BglII and AgI, ligated using T4 DNA ligase, and transformed into E.coli to produce pGWN4K/DV.3’UCS DNA.
To create pGWN4K/DV.3’NCR DNA, a DNA fragment containing the DV2 3’NCR was PCR-synthesized using the DV2.10282Bgl II sense primer (see above) and an antisense primer (DV2.3'Xba) containing a 5'terminal Xba I site and nts complementary to DV2 nts 10723-10696 (CATATCTAGAGAACCTGTTGATTCAACAGCACCATTC). The PCR fragments were digested with BglII and XbaI, and ligated to pGWN4K/DV.3’UCS DNA that had been linearized with the same restriction endonucleases in a fragment swap, resulting in pGWN4K/DV.3’NCR DNA.
To create pGWN4K/DV.3'USL DNA, the plasmid pGWN4K/DV.3'UCS was PCR-amplified with a pair of primers (a sense primer CCTGGGATAGACTAGGGGATCTTCTGC, WNV nt 10868–10894), and a 5'phosphorylated antisense primer containing the complement of the DV2 3'CS nt sequence, CGTCAATATGCTGTTTTTTGTTTTGGGGGGGTCTCCTCTAACCTCTAGTCCTTCCAGTG, DV2 nt 10631-10573). The PCR products were self-ligated at blunt ends and used to transform E.coli to produce pGWN4K/DV.3'USL. Next, all three mutant recombinant plasmid DNAs, pGWN4K/DV.3’UCS, pGWN4K/DV.3’USL, and pGWN4K/DV.3’NCR were digested with SphI (which cuts WNV DNA at nt 6774) and XbaI (which cuts WNV DNA at nt 10962). The SphI-XbaI fragments containing 3’NCR substitution mutations were then inserted into pSP6WN/Xba DNA that had been linearized with SphI and XbaI, to produce WN/DV3’UCS, WN/DV3’USL, and WN/DV3’NCR cDNAs, respectively.
Next, we created the WNV/DV2 chimera in which the DV2 5’NCR replaced the corresponding region in the WNV genome, except for a Ser to Gln mutation at the second amino acid (aa) in the ORF, as follows. DV2 5’ NCR fragment (nt 1–99) was obtained by PCR using the DV2 strain NGC infectious DNA (Polo et al., 1997) as the template, a forward primer containing a ClaI restriction site upstream of SP6 promoter sequences, TAACATCGATTAGAATTTAGGTGACACTATAG, and a 5'phosphorylated reverse primer containing DV2 nts 99-73, CATCAGAGATCTGCUCTCTAATTAAAAAAC. The pBWN2K plasmid DNA was also PCR-amplified with a pair of primers (an antisense Cla primer, GCTTATCGATACCGTCGACCTCGAGGGGGG, complementary to pRS424 nts 1939-1914, and a sense primer containing WNV nts 103–130 with the second code mutation, CAAAAGAAACCAGGAGGGCCCGGTAAAACC). Both the DV2 5’NCR and the pBWN2K plasmid DNA PCR products were treated with ClaI, ligated, and transformed into E.coli to produce the chimeric recombinant DNA plasmid, pBWN2K/DN5'TR99nt. Then the pBWN2K/DN5'TR99nt DNA was digested with ClaI and MluI (WNV nt 918), and the excised WNV DNA fragment of about 1 kb, containing the DV2 5’NCR, was inserted into pSP6WNV/XbaI and WNV/DV3'SL DNAs (Yu and Markoff, 2005) that had been linearized by digestion at the same unique ClaI and MluI sites, resulting in WNV/DV5'TR99nt and WNV/DV5'TR99nt3'SL DNAs, respectively (Fig. 3B, top and bottom).
A similar cloning strategy was used to construct WNV/DV5'TR159nt and WNV/DN5'TR387nt chimeric cDNAs. For WNV/DV5'TR159nt, which contained the DV2 5’NCR plus the N-terminal first 21 codons of the DV2 ORF substituted for the analogous WNV nt sequences in the context of the WNV DNA backbone, DV2 5’TR159nt fragment was PCR-synthesized with DV2 DNA as template and with a pair of primers (a sense Cla primer as shown above, and a 5'phosphorylated antisense primer complementary to DV2 nts 159-133, GTTTCTCTCGCGTTTCAGCATATTGAAG). The pBWN2K vector was PCR-amplified to produce a linear product molecule from which WNV nts 1–159 had been deleted, using an antisense Cla primer and a sense primer complementary to WNV nt160–184, CCCCGCGGATTGTCCTTGATAGGAC). Both PCR products were digested with ClaI, ligated, and transformed into E.coli cells to produce pBWN2K/DN5'TR159nt. The 5’-chimeric fragment was then cleaved from pBWN2K/DN5'TR159nt recombinant plasmid DNA with ClaI and MluI, and inserted into pSP6WN/Xba DNA, after that DNA was also doubly digested, to produce WNV/DV5'TR159nt DNA. To construct chimeric WN/DV5'TR159nt DNAs containing substitution mutations of the WNV 3'NCR by the DV2 3'NCR or of WNV 3'NCR nt sequences upstream from the 3'SL by analogous DV2 nt sequences (3'USL), 5’-end one kbp fragments were cleaved from the pBWN2K/DN5'TR159nt DNA with ClaI and MluI and were inserted into WN/DV3’NCR and WNV/DV3’USL (described above) at the same sites to produce WNV/DV5'TR159nt3’NCR and WNV/DV5’TR159nt3’USL cDNAs, respectively.
To construct the WN/DV5'TR387nt DNA, the DV2 5’-TR387nt fragment was PCR-amplified using DV2 DNA as template and a pair of primers (a sense Cla primer as shown above, and a 5'phosphorylated antisense primer complementary to DV2 nts 387-361 (CCTGTTCAAGATGTTCAGCATCCTTCC). The pBWN2K vector was also PCR-amplified to produce a linear product that lacked nts 1–387 of the WNV genome, using a pair of primers (an antisense Cla primer, and a sense primer containing WNV nts 388–416, CGGAGCACAAAACAAAAGAAAAGAGGAGG). Both PCR products were digested with ClaI, ligated, and used for transformation of E.coli to produce pBWN2K/DV5'TR387nt. The chimeric fragment containing DV2 capsid gene nt sequences were then cleaved from pBWN2K/DV5'TR387nt with ClaI and MluI and inserted into pSP6WN/Xba, WN/DV3'SL, and WN/DV3’NCR DNAs at the same sites to obtain WN/DV5'TR387nt 3'SL DNA and WN/DV5'TR387nt 3'NCR DNA, respectively.
All cloned plasmids were analyzed by restriction endonuclease digests (New England BioLabs, Beverly, MA) and by partial sequencing using Big Dye Terminator kit and an ABI model 377 DNA sequencer (Applied Biosystems, Foster City, Calif.). All plasmids were purified with Miniprep kit (QIAGEN, Valencia, Calif.) and then used for synthesis of their RNA counterparts.
Preparation of RNAs containing 5'TR and the WNV 3'NCR nt sequences
RNAs containing sequences from the 5' and 3'TRs of the DV2 and WNV genomes, as well as the DV2/WNV chimeric RNAs, were synthesized in vitro from the corresponding PCR amplified DNA fragments. The DNAs were obtained as follows. The PCR fragment encoding DV2 nts 1–230 was obtained as described previously (You and Padmanabhan, 1999). The PCR fragment containing WNV nts 1–230 downstream from the T7 promoter was also obtained as described previously (Nomaguchi et al., 2004), with a WNV DNA template. The PCR fragment containing the chimeric 5'TR RNA, WN5'NCR-DV5'ORF (WN nts 1–95 fused to DV2 nts 96–230) was obtained using WN1–95D296–230+3'NCR DNA as template (see above). The EcoRI primer described above was used as forward primer, and the reverse primer was complementary to DV2 nts 233-217. The PCR fragment encoding the 5'TR RNA, DV5'NCR-WN5'ORF (DV2 nts 1–95 fused to WNV nts 96–230) was obtained in the converse manner using a minigenome DNA containing D21–95WN96–230+3'NCR nt sequences (created using the respective wt template DNAs and techniques based on the conserved Bgl II recognition sequence in both genomes, as described above for derivation of WN1–95D296–230+3'NCR DNA) and a reverse primer complementary to WN nts 230-207. Derivation of the RNA containing the 631-nt sequence of the WNV strain Eg101 genome was previously described (Nomaguchi et al., 2004).
All PCR reactions were performed with Vent polymerase (New England Biolabs). PCR products were purified using the QIAquick gel extraction kit (Qiagen) prior to use in the in vitro transcription reaction, as described previously (Nomaguchi et al., 2004). To generate RNAs used in transactivation assays, the PCR products derived as per above, containing T7 RNA polymerase promoters, were used for in vitro transcription. Transcripts were purified as previously described (You and Padmanabhan, 1999) and used in the in vitro RdRP assays (see below).
In vitro RdRP Assays using purified WNV and DV2NS5
DV2 and WNV NS5 were expressed in E. coli and purified as described previously (Ackermann and Padmanabhan, 2001; Nomaguchi et al., 2004). Purified NS5 was stored in a buffer containing 40% glycerol at 20°C prior to its use in RdRP assays. RNA templates were prepared by in vitro transcription on linearized DNA templates containing the region to be transcribed. In vitro transcription was carried out using the Ampliscribe T7 Transcription kit (Epicentre Technologies). The standard reaction mixture (50 µl) contained 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 0.1 mM dithiothritol, template RNA (0.3 µg), 500 µM (each) ATP, GTP, and UTP, 10 µM unlabeled CTP, and 10 µCi of [α-32P]CTP along with 270 ng (2.6 pmol) of purified NS5. The reaction was incubated for 60min at 30°C. For trans-activation assays, the 5'TR RNA was labeled with [32P] in an in vitro transcription reaction The reaction was terminated by extraction with acid chloroform/phenol, followed by ethanol precipitation in the presence of 50 µg of E. coli tRNA as a carrier. The unincorporated nucleoside triphosphates were removed by using a Biorad P30 column. The in vitro RdRP products were analyzed by formaldehyde-agarose (2%) gel electrophoresis and autoradiography.
RNA transfection and indirect immunofluorescence assays (IFAs)
Plasmid DNA (1 µg) linearized by digestion with the XbaI was used as the template for RNA transcription catalyzed by SP6 RNA polymerase (Promega, Madison, Wis.), as previously described (Polo et al., 1997; Yu and Markoff, 2005; Zeng et al, 1998). Wt and mutant WNV RNAs derived by in vitro transcription were transfected into Vero cells to determine the replication phenotypes of mutant WNV viruses. IFA was performed to examine expression of viral protein during replication. Detailed procedures for RNA transfection of Vero cells and IFA were previously described (Polo et al., 1997; Yu and Markoff, 2005; Zeng et al, 1998).
Infectious center assay, plaque assay, and virus growth curve
Approximately 106 Vero cells in a volume of 300 µl of PBS were transfected by electroporation as previously described (Yu and Markoff, 2005), using 0.5 µg of RNA derived by transcription of cloned Wt or mutant recombinant plasmid DNAs. Briefly, serial 10-fold dilutions of transfected Vero cells were performed with MEM containing 2 × 105 fresh Vero cells, and were seeded onto paired wells of six-well tissue culture plates. Plates were incubated for 4 h at 37°C in 1x MEM. The media were removed and the cell monolayers were overlaid with 1 % agarose (BioWhittaker, Rockville, MD) containing 1x Earle's balanced salts (Sigma, Saint Louis, MO), 1 mM sodium pyruvate (Gibco-Invitrogen, Carlsbad, CA), 1x NEAA (BioWhittaker, Rockville, MD), 1x vitamins (Gibco-Invitrogen, CA), and 10% fetal calf serum (Atlanta Biologicals, Lawrenceville, GA). Plates were incubated for 5–8 days at 37°C under 5% CO2. Cells were then fixed with 7% formaldehyde for 1 h at room temperature. The agarose layer was then removed, and cell monolayers were stained with 1% crystal violet to visualize plaques which were counted. The RNA specific infectivity was expressed as the number of pfu (plaque-forming units) per microgram of RNA.
For titration of virus infectivity by plaque assays, each of the supernatants derived from transfected or infected Vero cells was harvested when about 100% of the cells were positive for viral antigens. Monolayers of Vero cells were infected with 10-fold serial dilutions of stock viruses at 37°C for 1–2 hour in 1x MEM. The monolayers were overlaid with agarose, and after 5–8 days of incubation at 37°C under 5% CO2, were stained with 1% crystal violet as described above.
To determine virus growth kinetics, Wt WNV virus and each of the viable mutant viruses released from Vero cells were used to infect both Vero cells in six-well plates and C6/36 cells in T-25 flasks, at a multiplicity of infection (MOI) of 0.01. Then, 300 µl of supernatant from infected cells was harvested daily for virus titration. The volume of supernatant removed each day was replaced with fresh medium. Virus titers for each cell line were determined by plaque assay on Vero cells as described above.
Sequencing of RNAs isolated from viable mutant viruses
Details of the sequencing of mutant virus RNA prepared from recovered progeny viruses were previously described (Yu and Markoff, 2005).
Computer analysis of predicted secondary structures of wt and mutant RNA
A 438-nt segment (195 nt from 5’-end and 243 nt from 3’-end) of wt or mutant RNAs were analyzed for the predicted secondary structures for potential 5’-3’ interaction using the MFOLD algorithm (Zuker et al, 1991) and the program "RNAdraw”, an integrated program for RNA secondary structure calculation and analysis on a Microsoft Windows platform (Matzura and Wennborg, 1996).
ACKNOWLEDGMENTS
We thank Barry Falgout for many helpful discussions. We also thank Dr. Vladimir Yamshchikov for the generous gift of WNV infectious DNA. This study was partially supported by an NIH grant AI32078 (to RP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3'UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339(2):200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005b;79(11):6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA. BHK cell proteins that bind to the 3' stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69(9):5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3' stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71(9):6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Carmichael GG. RNA replication: function and structure of Qβ replicase. Annu. Rev. Biochem. 1979;48:525–532. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol. 2003;84(Pt 5):1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5'-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Fernandez AV, Dispoto JH. The 3'-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153(1):113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. Growth-restricted dengue virus mutants containing deletions in the 5' noncoding region of the RNA genome. Virology. 1995;207(1):68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70(Pt 1):37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Kuo MD, Chien LJ, Hsu SL, Wang YM, Lin JH. RNA-protein interactions: involvement of NS3, NS5, and 3' noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71(5):3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WW, Kinney RM, Dreher TW. Control of translation by the 5'- and 3'-terminal regions of the dengue virus genome. J Virol. 2005;79(13):8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol. 2003;77(3):2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis WG, Blackwell JL, Shi PY, Brinton MA. Interaction between the Cellular Protein eEF1A and the 3'-Terminal Stem-Loop of West Nile Virus Genomic RNA Facilitates Viral Minus-Strand RNA Synthesis. J Virol. 2007;81(18):10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. Translation elongation facto-1alpha, La, and PTB interact with the 3' untranslated region of dengue 4 virus RNA. Virology. 2002;295(2):337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20(16):2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T, Bouloy M, Girard M. Stable secondary structures at the 3'-end of the genome of yellow fever virus (17 D vaccine strain) FEBS Lett. 1985;188(1):159–163. doi: 10.1016/0014-5793(85)80895-4. [DOI] [PubMed] [Google Scholar]
- Gritsun TS, Venugopal K, Zanotto PM, Mikhailov MV, Sall AA, Holmes EC, Polkinghorne I, Frolova TV, Pogodina VV, Lashkevich VA, Gould EA. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5' and 3'-UTRs. Virus Res. 1997;49(1):27–39. doi: 10.1016/s0168-1702(97)01451-2. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3' untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7(3):581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett MJ, Petterson RF, Baltimore D. Circular forms of Uukenimi virion RNA: an electron microscopic study. J. Virol. 1977;75:5009–5017. doi: 10.1128/jvi.21.3.1085-1093.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden KL, Harris E. Enhancement of dengue virus translation: role of the 3' untranslated region and the terminal 3' stem-loop domain. Virology. 2004;329(1):119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Hsu MT, Kung HJ, Davidson N. Secondary structure of tRNA. Cold Spring Harbor Symp Quant Biol. 1973;38:943–950. doi: 10.1101/sqb.1974.038.01.096. [DOI] [PubMed] [Google Scholar]
- Hu B, Pillai-Nair N, Hemenway C. Long-distance RNA-RNA interactions between terminal elements and the same subset of internal elements on the potato virus X genome mediate minus- and plus-strand RNA synthesis. Rna. 2007;13(2):267–280. doi: 10.1261/rna.243607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Kondratieva N, Sgro JY, Palmenberg A, Westaway EG. Significance in replication of the terminal nucleotides of the flavivirus genome. J Virol. 2003;77(19):10623–10629. doi: 10.1128/JVI.77.19.10623-10629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75(14):6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Westaway EG. cis- and trans-acting elements in flavivirus RNA replication. J Virol. 2000;74(7):3253–3263. doi: 10.1128/jvi.74.7.3253-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3' untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L. 5'- and 3'-noncoding regions in flavivirus RNA. Adv Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput Appl Biosci. 1996;12(3):247–249. doi: 10.1093/bioinformatics/12.3.247. [DOI] [PubMed] [Google Scholar]
- Men R, Bray M, Clark D, Chanock RM, Lai CJ. Dengue type 4 virus mutants containing deletions in the 3'noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan PM, Padmanabhan R. Detection of stable secondary structure at the 3' terminus of dengue virus type 2 RNA. Gene. 1991;108(2):185–191. doi: 10.1016/0378-1119(91)90433-c. [DOI] [PubMed] [Google Scholar]
- Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem. 2004;279(13):12141–12151. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- Ong CC, Lam SK, AbuBakar S. Cellular proteins bind to the 3' and 5' untranslated regions of dengue 2 virus genome. Malays J Pathol. 1998;20(1):11–17. [PubMed] [Google Scholar]
- Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35(15):5253–5261. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J Virol. 1997;71(7):5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski V, Gaunt MW, Gould EA, Holmes EC. Secondary structure of the 3'-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J Gen Virol. 1997;78(Pt 7):1543–1549. doi: 10.1099/0022-1317-78-7-1543. [DOI] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5'-cap structure is formed by sequential guanine N-7 and ribose 2'-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Shi PY, Brinton MA, Veal JM, Zhong YY, Wilson WD. Evidence for the existence of a pseudoknot structure at the 3' terminus of the flavivirus genomic RNA. Biochemistry. 1996;35(13):4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Ta M, Vrati S. Mov34 protein from mouse brain interacts with the 3' noncoding region of Japanese encephalitis virus. J Virol. 2000;74(11):5108–5115. doi: 10.1128/jvi.74.11.5108-5115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216(2):317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- Tilgner M, Deas TS, Shi PY. The flavivirus-conserved penta-nucleotide in the 3' stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology. 2005;331(2):375–386. doi: 10.1016/j.virol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Tilgner M, Shi PY. Structure and function of the 3' terminal six nucleotides of the west nile virus genome in viral replication. J Virol. 2004;78(15):8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Castle E. Analysis of structural properties which possibly are characteristic for the 3'-terminal sequence of the genome RNA of flaviviruses. J Gen Virol. 1986;67(Pt 6):1183–1188. doi: 10.1099/0022-1317-67-6-1183. [DOI] [PubMed] [Google Scholar]
- Yamshchikov VF, Wengler G, Perelygin AA, Brinton MA, Compans RW. An infectious clone of the West Nile flavivirus. Virology. 2001;281(2):294–304. doi: 10.1006/viro.2000.0795. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5'- and 3'- terminal regions that influence RNA structure. J Biol Chem. 2001;276(19):15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3'-end of exogenous viral RNA templates requires 5'- and 3'-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274(47):33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Yu L, Markoff L. The topology of bulges in the long stem of the flavivirus 3' stem-loop is a major determinant of RNA replication competence. J Virol. 2005;79(4):2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3'- SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72(9):7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Jaeger JA, Turner DH. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991;19(10):2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]