Abstract
Sensitivity to the locomotor stimulant effects of methamphetamine is a heritable trait that utilizes neurocircuitry also associated with the rewarding effects of drugs. We used the power of a C57BL/6J x DBA/2J F2 intercross (n = 676) and the precision of a C57BL/6J x DBA/2J F8 advanced intercross line (Aap: B6, D2 – G8; or F8 AIL; n = 552) to identify and narrow quantitative trait loci (QTL) associated with sensitivity to the locomotor stimulant effects of methamphetamine. We used the program QTLRel to simultaneously map QTL in the F2 and F8 AIL mice. We identified six genome-wide significant QTLs associated with locomotor activity at baseline and seven genome-wide significant QTLs associated with methamphetamine induced locomotor activation. The average percent decrease in QTL width between the F2 and the integrated analysis was 65%. Additionally, these QTLs showed a distinct temporal specificity within each session that allowed us to further refine their locations, and identify one QTL with a 1.8-LOD support interval = 1.47 Mb. Next, we utilized publicly available bioinformatics resources to exploit strain-specific sequence data and strain- and region-specific expression data to identify candidate genes. These results illustrate the power of AILs in conjunction with sequence and gene expression data to investigate the genetic underpinnings of behavioral and other traits.
Keywords: addiction, drug abuse, psychostimulant, activity, amphetamine, quantitative trait loci, genetic, GWAS
Introduction
Among humans, there is dramatic individual variability in both the positive and negative subjective effects of numerous drugs, including stimulants (Hart et al. in press). This variability is known to be partially genetic in origin (Nurnberger et al. 1982; Crabbe et al. 1983), and has been associated with subsequent development of drug use disorders (Haertzen et al. 1983; Fergusson et al. 2003; Schuckit & Smith 2011; King et al. 2011).
Mouse models are complementary to human genetic studies and offer unique advantages and opportunities (Parker & Palmer 2011). Numerous classes of drugs increase locomotor activity in mice and this response is mediated through the same neurocircuitry that is implicated in drug reward in animals and drug-induced euphoria in humans (Wise & Bozarth 1987; Di Chiara & Imperato 1988; Koshikawa et al. 1989). Therefore, much attention has been focused on examining genetic factors that influence the locomotor response to various drugs, including methamphetamine (Grisel et al. 1997; Downing et al. 2006; Kamens et al. 2005; Palmer et al. 2005; Bryant et al. 2009a, 2009b). In mice, differences in the sensitivity to the locomotor activating effects of methamphetamine are heritable (Phillips et al. 2008), and predictive of later drug self-administration (Kamens et al. 2005).
Genetic mapping studies in mice have traditionally used recombinant inbred lines (RI), backcrosses (BC), F2 intercrosses (F2), consomic and congenic mice to identify quantitative trait loci (QTLs) for traits like sensitivity to the locomotor stimulating effects of methamphetamine. Due to a lack of recombination, these techniques are only able to identify large genomic regions and are therefore sub-optimal for identifying the genes that underlie QTLs (Peters et al. 2007; Flint 2011; Parker & Palmer 2011). We have recently begun to address this limitation by using populations with greater numbers of accumulated recombinations such as advanced intercross lines (AILs; Cheng et al. 2010; Samocha et al. 2010; Lionikas et al. 2010; Parker et al. 2011). AILs are created by successive generations of pseudo-random mating after the F2 generation. Each additional generation leads to the accumulation of new recombinations, which allows for more precise mapping due to a breakdown in linkage disequilibrium. Because AILs are derived from two inbred founders, they maintain the simplicity of more traditional crosses, possess no rare alleles, and every marker that differs between the parental strains is perfectly informative in terms of identifying which inbred strain the allele is inherited from.
Here, we created an F2 intercross and an F8 AIL derived from C57BL/6J (B6) x DBA/2J (D2) mice (Aap:B6, D2 - G8, hereafter referred to as the F8 AIL). We chose B6 and D2 inbred mice as our progenitor strains in order to take advantage of the vast amount of bioinformatic data that already exists for these specific strains, including genomic sequence and expression QTL (eQTL) data. By combining genome-wide association (GWAS) with complementary bioinformatic resources available for B6 and D2 mice, we utilized sequence data to identify SNPs that may alter proteins directly, and eQTL data to identify putatively causal expression polymorphisms.
Materials and Methods
Animals and housing
All procedures were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC) in accordance with National Institute of Health guidelines for the care and use of laboratory animals. Inbred female B6 and male D2 mice were obtained from Jackson Labs (Bar Harbor, ME) and mated to produce the B6 x D2 F1 mice. Thereafter, mice were pseudo-randomly bred to minimize relatedness and avoid brother-sister mating. The subsequent F2 intercross (319 male, 357 female) was created from 60 different breeding pairs and the F8 AIL (276 male, 276 female) was created from 106 different breeder pairs. The average number of breeder pairs across all seven generations was 58. Colony rooms were maintained on a 12:12 h light-dark cycle (lights on at 0630 h) in same-sex groups of two to five mice with standard lab chow and water available ad libitum. Mice were approximately two months of age at the start of testing (F2 mean age = 76 days, SD = 7.3, range = 60–92; F8 AIL mean age = 61.2 days, SD = 5.8, range = 46–93).
Activity chambers
Locomotor activity was measured using automated Versamax activity chambers (AccuScan, Colombus, OH) as described previously (Bryant et al. 2009a). Briefly, each chamber consisted of a clear acrylic arena (40 × 40 × 30 cm) placed inside a frame containing evenly spaced infrared photobeams from the front to the back and from the left to the right of the arena. Beam breaks were recorded on a computer and converted into distance travelled (cm). Each activity chamber was encased within a sound attenuating PVC/lexan environmental chamber (AccuScan). In each chamber, overhead lighting provided dim illumination (~80 lux) and a fan provided both ventilation and masking of background noise.
Methamphetamine-induced locomotor activity
Testing was conducted over three consecutive days during the light phase, between 0800 and 1700h, as described previously (Bryant et al. 2009a). Mice were transported from the adjacent vivarium and allowed to habituate to the procedure room for 30 minutes in their home cages. On the first and second days of testing, mice were removed from their home cages, weighed, and placed in individual holding cages filled with clean bedding. Mice then received an intraperitoneal (i.p.) injection of physiological saline and were then immediately placed in individual activity chambers where locomotor activity was recorded for 30 minutes. On the third day of testing, mice received an i.p. injection of 2 mg/kg MA and were then immediately placed in the activity chambers to measure locomotor activity for 30 minutes. Methamphetamine response was defined as the total distance travelled on day three during the 30 minute test beginning immediately after drug administration. All systemic injections were administered in a volume of 0.01 ml/g body weight. On all three days, mice were returned to their home cages immediately after the 30 minute test. Activity chambers were cleaned with 10% isopropanol between tests. Mice were returned to the vivarium at the end of each day.
Data analysis
First, independent samples t-tests were calculated to measure generational effects (F2 vs. F8 AIL) on total distance travelled for each day. Next, a one-way ANOVA was used to determine the effects of day on distance travelled in the F2 and in the F8 AIL mice. Finally, two-way ANOVAS were performed to assess the influence of sex and time on distance travelled in the F2 and in the F8 AIL mice on each day. Analyses were conducted in Microsoft Excel 2010 and PASW Statistics 18 (SPSS Inc., Chicago, Illinois).
Genotyping
DNA from the F2 generation was extracted and genotyped by KBiosciences (Hoddesdon, Hertfordshire, UK) using KASPar, a fluorescence-based PCR assay. Markers consisted of 164 evenly spaced, informative SNPs selected from Petkov et al. (2004). In the F8 AIL, DNA was extracted using a salting out protocol and genotyped using the Illumina Mouse Medium Density Linkage Panel (Illumina, San Diego, CA) at the Genomics Core Facility at Northwestern University (http://web.cgm.northwestern.edu/cgm/Core-Facilities/Genomics-Core). The SNP panel consists of 1,449 loci, 1060 of which are polymorphic between B6 and D2 mice.
QTL mapping
Genome-wide association analysis was performed in the combined population of the F2 intercross and the F8 AIL using the R package QTLRel (http://cran.r-project.org/web/packages/QTLRel/index.html). This software accounted for the complex relationships (e.g. sibling, half-sibling, cousins) among the F8 AIL mice by using a mixed model as described previously (Cheng et al. 2010). For each analysis, P < 0.05 significance thresholds were estimated using 1000 permutations. Sex was included as an interactive covariate.
Bioinformatic Analyses
The GeneNetwork mapping module (www.genenetwork.org; Wang et al. 2003; Chesler et al. 2004) was used to identify eQTLs in striatum and whole brain mRNA from untreated B6 x D2 F2 mice (accessed on September 15, 2011; OHSU/VA B6D2F2 Striatum M430v2 (Sep05) RMA; OHSU/VA B6D2F2 Brain mRNA M430 (Aug05) RMA; Hitzemann et al. 2004; Hofstetter et al. 2008), as well as nucleus accumbens and prefrontal cortex from saline injected BXD RI mice (accessed on September 15, 2011; VCU BXD NA Sal M430 2.0 (Oct07) RMA; VCU BXD PFC Sal M430 2.0 (Dec06) RMA; Putman & Miles unpublished data) that co-mapped with our behavioral QTLs. We focused on these regions because of their well-known role in drug-induced locomotor activity and reward (Swerdlow et al. 1986; Di Chiara and Imperato 1988); however, examining gene expression in additional brain areas would also be informative. Next, in order to narrow the list of candidate genes within the QTL intervals, we used high density sequence data provided by the Wellcome Trust Sanger Institute (accessed on September 19, 2011; http://www.sanger.ac.uk/cgi-bin/modelorgs/mousegenomes/snps.pl; Keane et al. 2011; Yalcin et al. 2011) to compare genomic regions between B6 and D2 mice. These strains were sequenced to an average of 25x coverage on the Illumina GAII platform (Illumina, San Diego, CA) with a mixture of 54bp, 76bp, and 108bp paired reads. We used this data to search for genes within the QTL intervals that possessed “consequential” polymorphisms between B6 and D2 mice (such as nonsynonymous coding SNPs, stop-gain SNPs, stop-loss SNPs, SNPs resulting in frameshifts and SNPs located in essential splice sites).
Results
Phenotypic Analysis
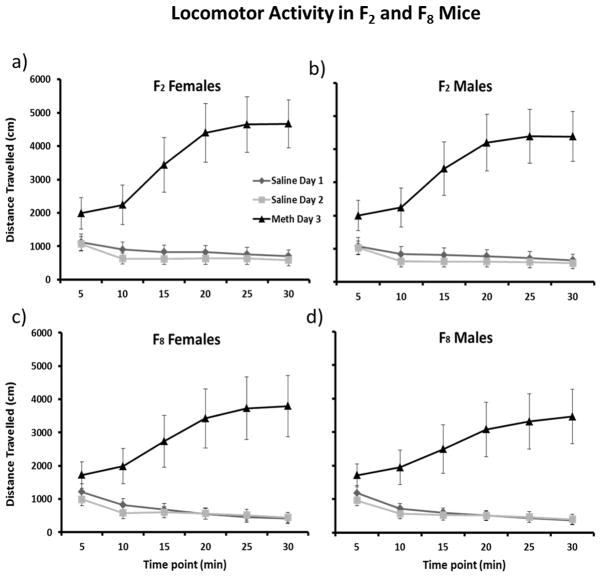
Figure 1 displays the distance travelled across all three sessions in male and female F2 and F8 AIL mice. For both F2 and F8 AIL mice, there was a significant effect of treatment on distance travelled over 30 minutes (in F2: F2, 2006 =2859.02, p < 0.0001; in F8: F2, 1649 = 1499.28, p < 0.0001). Scheffe’s post-hoc analysis revealed that all sessions were significantly different from one another in both the F2 and F8 AIL mice (p < 0.005). The F2 and F8 AIL mice also differed from each other on saline day 1 (F1, 1219 = 169.76; p < 0.0001) and methamphetamine day 3 (F1, 1218 = 6.04; p = 0.014), but not saline day 2 (Figure 1). Additionally, F2 and F8 AIL mice displayed sex and time differences for all three sessions. In the F2 mice, a two-way ANOVA reported varying effects of sex and time-point on distance travelled for saline day 1 (sex: F1, 676 = 56.2; p = 0.0007, time: F5, 676 =375.3; p < 0.0001), saline day 2 (sex: F1, 676 =18.6; p = 0.008, time: F5, 676 = 674.1; p < 0.0001) and methamphetamine day 3 (sex: F1, 676 = 5.0; p = 0.08, time: : F5, 676 = 273.2; p < 0.0001). A two-way ANOVA also reported significant effects of sex and time-point on distance travelled in the F8 AIL mice for saline day 1 (sex: F2, 552 = 17.2; p = 0.009, time: F5, 552 = 336.4; p < 0.0001), saline day 2 (sex: F2, 552 = 30.9; p = 0.003, time: F5, 552 = 335.9; p < 0.0001) and methamphetamine day 3 (sex: F2, 552 = 11.4; p = 0.02, time: F5, 552 = 95.7; p < 0.0001). While the disparity between F2 and F8 AIL populations may be due to the segregation of alleles associated with high locomotor activity during the creation of the F8 AIL mice, we suspect it is more likely a result of handling effects of different testers across the 2 year period between the F2 and F8 generations. The F2 mice were an average of about 15 days older than the F8 AIL mice when tested, we have not seen much impact of similar differences in age on these behaviors in the past. As a result of these differences, both the F2 and the F8 AIL data were converted to z-scores prior to genome-wide analysis.
Figure 1.
Distance travelled on Saline Day1 (◆), Saline Day 2 (■) and Meth Day 3 (▲) in the F2 female (a) and male (b) mice; as well as in F8 female (c) and male (d) mice. For both the F2 and F8 mice, distance travelled on day 1 was slightly greater than on day 2 (p < 0.005) and distance traveled on day 3 was dramatically greater than on either day 1 or 2 (p < 0.0001). The F2 and F8 generation also differed from each other on day 1 (p < 0.0001) and day 3 (p = 0.014), but not day 2. Additionally, F2 and F8 AIL mice displayed sex and time differences for all three sessions. In the F2 mice, a two-way ANOVA reported varying effects of sex and time-point on distance travelled for saline day 1 (sex: F1, 676 = 56.2; p = 0.0007, time: F5, 676 =375.3; p < 0.0001), saline day 2 (sex: F1, 676 =18.6; p = 0.008, time: F5, 676 = 674.1; p < 0.0001) and methamphetamine day 3 (sex: F1, 676 = 5.0; p = 0.08, time: : F5, 676 = 273.2; p < 0.0001). A two-way ANOVA also reported significant effects of sex and time-point on distance travelled in the F8 AIL mice for saline day 1 (sex: F2, 552 = 17.2; p = 0.009, time: F5, 552 = 336.4; p < 0.0001), saline day 2 (sex: F2, 552 = 30.9; p = 0.003, time: F5, 552 = 335.9; p < 0.0001) and methamphetamine day 3 (sex: F2, 552 = 11.4; p = 0.02, time: F5, 552 = 95.7; p < 0.0001).
QTL Mapping
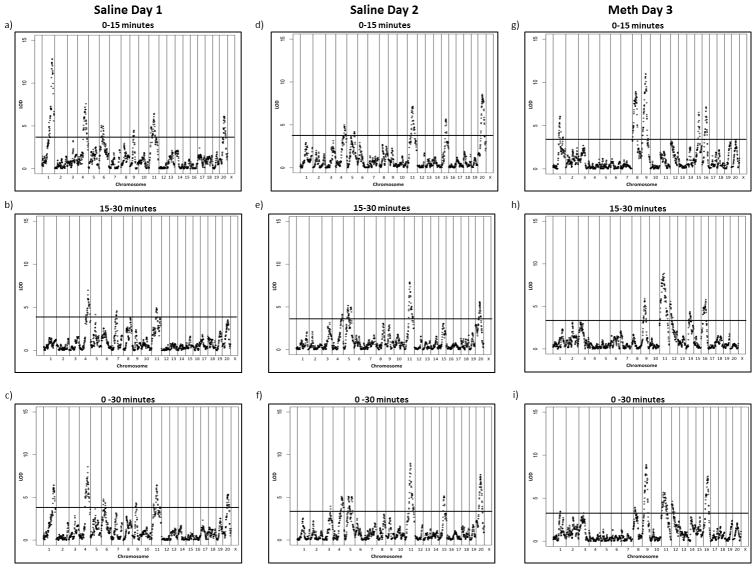
We performed genome-wide analysis on the integrated F2 and F8 AIL populations for distance travelled during the first 15 minutes (0–15 min) as well as distance travelled from 15–30 minutes, and distance travelled during the entire 30 minute testing period (0–30 min) across all treatments (Figure 2). However, we focused our subsequent analyses on the 0–15 min and 0–30 min time periods given the fact that previous studies in our lab have identified QTLs for methamphetamine-induced locomotor activity specific to these time-points (Palmer et al. 2005; Bryant et al. 2009a, 2009b; Cheng et al. 2010; Sokoloff et al. 2011). Some of the QTLs we identified were present only in the first 15 minutes whereas others were only present for the total 30 minute time period. For example, we identified the same six QTLs associated with distance travelled on day 1 from 0–15 minutes and 0–30 minutes (on chromosomes 1, 4, 6, 9, 11 and X; Figure 2a & 2c). For day 2, we identified five QTLs associated with distance travelled from 0–15 minutes (on chromosomes 4, 5, 11, 15, and X; Figure 2d). All of these QTLs were also significant for the 0–30 minute time period, in addition to a QTL on chromosome 3 (Figure 2f). However, on day 3 we identified five QTLs associated with distance travelled from 0–15 minutes (on chromosomes 1, 8, 9, 15, and 16; Figure 2g). Only four of these QTLs were significant for distanced travelled from 0–30 minutes (on chromosomes 1, 8, 9 and 16), and three additional QTLs were identified (on chromosomes 3, 11 and 12; Figure 2i). Using 1000 permutations, significance thresholds were determined to range from 3.84–4.04 LOD. We also mapped QTLs for the difference between activity on day 3 and day 2, which we and others have sometimes used as a way to distinguish between differences that are specific to drug treatment vs. those that are secondary to differences in basal locomotor activity and occur even in the absence of drug exposure. The QTLs identified were similar to those of day 3 (data not shown). We were most interested in locomotor activity in a novel environment and methamphetamine induced locomotor activity; thus, the remainder of our analyses focused on the day 1 and day 3 results. Because the use of F2 and F8 AIL mice is most interpretable when there is concurrence between the QTL locations in both generations, we further constrained our analyses to QTLs that were evident (though not necessarily significant) in both the F2 and the F8 cohorts. This left 3 QTLs for activity on day 1 (in a novel environment - Act1, Act4, and ActX), and 3 QTLs for activity on day 3 (methamphetamine response - Meth9, Meth15, and Meth16).
Figure 2.
Integrated genome-wide results for distance travelled on (a) saline day 1 for 0–15 minutes (P < 0.05 significance threshold LOD = 3.93), (b) saline day 1 for 15–30 minutes (P < 0.05 significance threshold LOD = 4.00), (c) saline day 1 for 0–30 minutes (P < 0.05 significance threshold LOD = 4.03), (d) saline day 2 for 0–15 minutes (P < 0.05 significance threshold LOD = 4.04),(e) saline day 2 for 15–30 minutes (P < 0.05 significance threshold LOD = 3.92), (f) saline day 2 for 0–30 minutes (P < 0.05 significance threshold LOD = 3.87), (g) meth day 3 for 0–15 minutes (P < 0.05 significance threshold LOD = 3.98), (h) meth day 3 for 15–30 minutes (P < 0.05 significance threshold LOD = 4.01), and (i) meth day 3 for 0–30 minutes (P < 0.05 significance threshold LOD = 3.84).
Time-dependent nature of QTLs
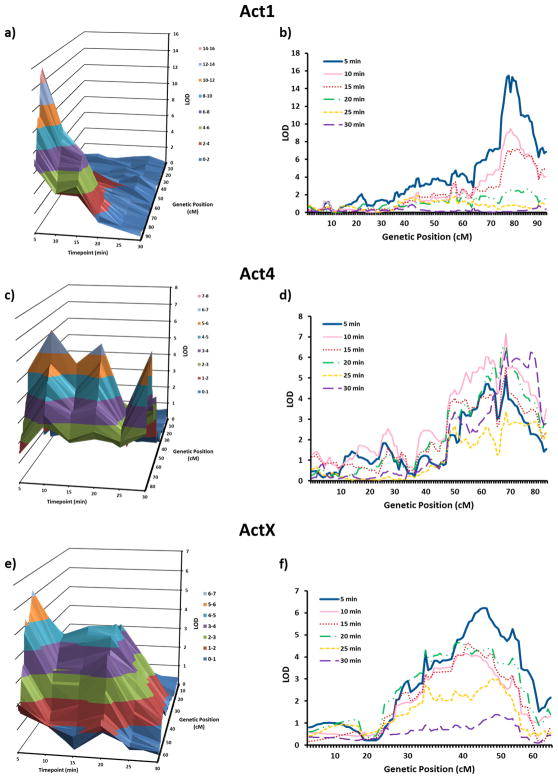
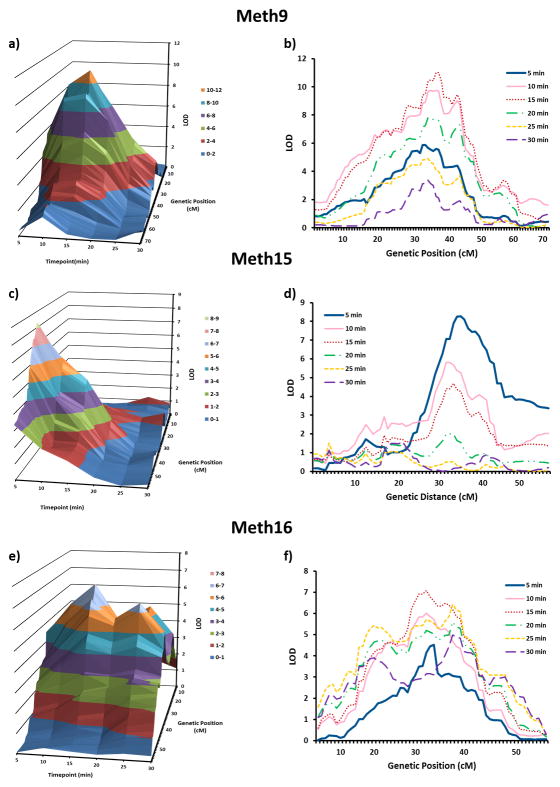
Next, we split the 30 minute testing period into 6 consecutive five-minute bins in order to determine if a particular time point was driving each QTL. In both Act1 (Figure 3a, 3b) and ActX (Figure 3e, 3f), the 0–5 minute time bin was largely responsible for the QTLs. In contrast, Act4 (Figure 3c, 3d) reached its peak during the 5–10 minute bin. For methamphetamine response, Meth15 (Figure 4c, 4d) peaked during the 0–5 minute bin and Meth9 (Figure 4a, 4b) and Meth16 (Figure 4e, 4f) both showed the strongest effect at the 10–15 minute bin. The 1.8-LOD intervals for these six QTLs ranged from 1.5 to 50.0 Mb, with a median width of 15.6 Mb. Table 1 displays the LOD scores, peak SNP, Mb location and width for each QTL interval for the peak time-point.
Figure 3.
Time-dependent nature of baseline locomotor activity QTLs. a) 3D plot of the integrated QTL results for the Act1 QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis) b) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the Act1 QTL. c) 3D plot of the integrated QTL results for the Act4 QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis). d) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the Act4 QTL. e) 3D plot of the integrated QTL results for the ActX QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis). f) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the ActX QTL.
Figure 4.
Time-dependent nature of methamphetamine-induced locomotor activity QTLs. a) 3D plot of the integrated QTL results for the Meth9 QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis) b) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the Meth9 QTL. c) 3D plot of the integrated QTL results for the Meth15 QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis). d) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the Meth15 QTL. e) 3D plot of the integrated QTL results for the Meth16 QTL across time-point (depth axis), genetic distance (horizontal axis) and LOD score (vertical axis). f) plot of 6 LOD curves, separated by time-point (0–5, 5–10, 10–15, 15–20, 20–25, 25–30) for the Meth16 QTL.
Table 1.
QTLs selected for fine-mapping. Table includes peak time-point, LOD at peak SNP, 1.8 LOD support interval, and the width of the support interval.
| Saline Day 1 Total Distance Travelled QTLs | Chr | Peak Time-point | LOD | Peak SNP | 1.8 LOD start Mb | 1.8 LOD end Mb | Peak Mb Position | Width (Mb) | Genes with Coding SNPs |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0–5 min | 15.4 | rs8245216 | 172.357 | 173.832 | 173.174 | 1.475 | 20 | |
| 4 | 5–10 min | 7.1 | rs13478002 | 133.275 | 141.023 | 136.412 | 7.749 | 64 | |
| X | 0–5 min | 6.2 | gnfX.086.039 | 81.842 | 131.953 | 99.159 | 50.111 | 12 | |
| Meth Day 3 Total Distance Travelled QTLs | Chr | Peak Time-point | LOD | Peak SNP | 1.8 LOD start Mb | 1.8 LOD end Mb | Peak Mb Position | Width (Mb) | Genes with Coding SNPs |
| 9 | 10–15 min | 10.7 | rs3655717 | 57.888 | 72.491 | 68.204 | 14.603 | 24 | |
| 15 | 0–5 min | 9.3 | rs13482642 | 68.959 | 85.617 | 75.523 | 16.659 | 52 | |
| 16 | 10–15 min | 7.6 | rs4186744 | 42.090 | 70.632 | 54.759 | 28.542 | 42 |
Bioinformatic Analyses
Numerous eQTLs across multiple brain regions were identified that co-mapped with these six behavioral QTLs (Supplemental Table 1). The majority of these eQTLs were cis-eQTLs, meaning that the eQTL was coincident with the location of the gene. Additionally, we confirmed the results of previous studies in our lab with the identification of casein kinase 1, epsilon (Csnk1e; Palmer et al. 2005; Bryant et al. 2009a, 2009b) and identified other genes that had been implicated in amphetamine sensitivity (epha receptor A3, Epha3, Sieber et al. 2004; epha receptor A6, Epha6, Sieber et al. 2004; galanin receptor 3, Galr3, Kuteeva et al. 2005). We then examined our 1.8-LOD support interval for the presence of SNPS that had the potential to directly alter proteins (ie.nonsynonymous coding, stop-gain, stop lost, framshift, splice sites; Supplemental Table 2).
Discussion
We performed genome-wide mapping of QTL affecting locomotor activity in a novel environment, as well as QTL associated with methamphetamine induced locomotor activity in an F2 and F8 AIL population of mice. We identified a total of six QTLs (on chromosomes 1, 4, 6, 9, 11, and X) associated with locomotor activity in a novel environment and eight QTLs (on chromosomes 1, 3, 8, 9, 11, 12, 15, and 16) associated with methamphetamine induced locomotor activation. Four of the QTLs associated with methamphetamine sensitivity (Meth9, Meth11, Meth12, and Meth15) replicated the results of previous studies in STSL derived from B6 x D2 F2 mice (chromosomes 9, 11, 12, 15; Palmer et al. 2005), and one (Meth11) overlapped with a QTL region identified in a LG/J x SM/J F34 AIL (chromosome 11; Cheng et al. 2010). In addition, five agreed with results from a B6 x AJ consomic panel (chromosomes 8, 9, 11, 12 and 16; Bryant et al. 2009b), and one agreed with results from BXD RI strains (chromosome 15; Grisel et al. 1997). Importantly, our AIL provided greater resolution and narrower support intervals as compared to the STSL, CSS, and the BXD RI panel. Additionally, some of the QTLs we identified for methamphetamine sensitivity (Meth1, Meth11, Meth12) also overlapped with QTLs underlying ethanol (chromosomes 1, 11; Bennett et al. 2006; Downing et al. 2006), opioid (chromosome 11; Bryant et al. 2009b) and etomidate (chromosome 12; Downing et al. 2003) sensitivity, suggesting that the genes underlying these QTL regions may not be drug specific.
Interestingly, one of the QTLs we identified, which was associated with locomotor activity in a novel environment (Act1, 1.8-LOD interval = 172.36 Mb to 173.83 Mb), mapped to the proximal region of known QTL hotspot called Qrr1. Qrr1 extends from 172.5 Mb to 177.5 Mb on chromosome 1, and is highly enriched in QTL that control neural and behavioral phenotypes including basal locomotor behavior, escape latency, emotionality, ethanol-induced locomotor activity, and responses to caffeine, pentobarbital and haloperidol (Mozhui et al. 2008). Qrr1 contains 164 known genes and is thought to contain a highly complex gene expression regulatory interval composed of multiple loci modulating the expression of functionally similar sets of genes. In addition to Act1, a chromosome 1 QTL associated with methamphetamine sensitivity (Meth1, 1.8-LOD interval = 111.83 Mb to 184.52 Mb) also maps to Qrr1. Because Qrr1 consists of multiple regions (each associated with the expression of distinct subsets of genes and QTLs), it is possible that Act1 and Meth1 represent either the same or distinct loci.
Traditionally, F2 intercrosses are used to identify QTLs underlying phenotypic variation, and fine-mapping is carried out as second step, often in congenic strains. Efforts at subsequent dissection and gene identification are often impeded by the existence of multiple causative loci of small effect located in the same chromosomal region (Mott et al. 2000; Shao et al. 2010). An AIL is an improvement over these traditional methods because of the additional recombinations it accumulates over successive generations. The accumulated recombinations allow identification and fine-mapping to be merged into a single step, which can often discriminate between loci that are due to single versus multiple alleles (Parker et al. 2011). The integration of the F2 and F8 AIL population combines the detection power of the F2 with the precision of the F8 AIL. In the integrated analysis, we reduced the 1.8-LOD support intervals by approximately 65% over the F2 analysis alone (Supplemental Figure 1). In several instances significant QTLs identified in the F2 population were not supported by the F8 AIL data. These regions are difficult to interpret, as they may be caused by either a false positive result in the F2 mice (this is very unlikely when the LOD score vastly exceeds the threshold for significance), or a false negative in the F8 AIL mice, which has less power than a similarly sized F2 (resulting from the reduced association between genotypes at markers). Alternatively, lack of a significant peak in the F8 AIL may be due to the presence of multiple loci of small effect located in the same chromosomal region, which segregate as a unit in the F2 but segregate independently in the F8 AIL. Because of the ambiguity of QTLs not replicated in the F8 AIL, we chose to focus our fine-mapping efforts on regions where the F2 and F8 AIL QTLs were in agreement.
Most studies of drug response traits identify QTLs based on summary measures that collapse out the within-subjects factor time. This approach implies that the QTLs are expected to have a uniform effect over the testing period. In order to better determine if a QTL was driven by a particular time point, we split the 30 minute testing period into six bins of five-minutes each. This indicated a temporal nature to our QTLs, although a formal test examining the QTL-by-time interaction would be necessary to definitively state that differences across time-bins are statistically significant. We plotted these results in three dimensions (time x position x LOD score). QTLs for initial locomotor activity in a novel environment as well as QTLs for methamphetamine-induced locomotor activity displayed peak LOD scores in the first half of the testing period. By considering the time-course in greater detail, we were able to observe that in some situations, the peak LOD scores were primarily driven by a particular time-point, as was the case with Act1, ActX, Meth9, and Meth15. In other instances, the LOD scores for the QTLs were consistently high across all time-points; this was the case for Act4 and Meth16.
To further narrow our QTLs and to attempt to identify the underlying genes, we used a series of bioinformatic approaches. First, we identified eQTLs that co-mapped with our QTLs. eQTLs are believed to underlie many QTLs for more complex traits (Nicolae et al. 2010; Li & Deng 2010). We used an existing database (www.GeneNetwork.org) of eQTLs from whole brain and striatum of untreated B6 x D2 F2 mice and from the nucleus accumbens and prefrontal cortex of saline-injected BXD RI mice. In many cases this identified a smaller number of genes that co-mapped within the 1.8-LOD intervals of our QTLs (Supplemental Table 1). Some of the genes we identified have been implicated in other studies examining the stimulant properties of drugs of abuse, and may be promising candidates for follow-up studies. In the case of Meth15 we replicated our previous finding regarding the gene Csnk1e, which has been shown to influence the locomotor stimulant response to methamphetamine (Palmer et al. 2005; Bryant et al. 2009a). Others have found associations between Csnk1e and methamphetamine dependence as well as heroin addiction (Veenstra-VanderWeele et al. 2006; Levran et al. 2008). In addition, expression of the gene for the galanin 3 receptor (Galr3) also mapped to the Meth15 QTL. Transgenic mice overexpressing galanin were reported to have attenuated amphetamine-induced locomotor activity, as compared to controls (Kuteeva et al. 2005). We also found that the expression of Epha3 and Epha5 mapped to the Meth16 QTL. Sieber et al. (2004) investigated the functional role of Epha signaling by overexpressing a broad-range Epha receptor antagonist in the central nervous system of transgenic mice. Transgenic mice displayed a 40–50% reduction of dopaminergic neurons in the striatum, as well as insensitivity to the locomotor activating effects of amphetamine. While co-mapping of a QTL and an eQTL does not constitute proof that the latter causes the former, it does suggests a clear and testable hypothesis -- the candidate gene can be directly manipulated using a variety of molecular or pharmacological approaches (e.g. Bryant et al. 2009a). In addition, gene expression differences in other brain regions, across multiple developmental time-points, and in a variety of cell types may further aid in the identification of genes underlying these QTLs. Finally, we identified between 12 and 64 genes with “consequential” SNPs within each of our constrained QTL intervals (Supplemental Table 2). A subset of these resulted in premature stop codons, which are especially likely to alter the function of the gene. Follow-up studies will determine if any of these SNPs result in non-conservative amino acid changes, or if they occur in evolutionarily conserved amino acids; as these SNPs are most likely to cause phenotypic differences. Taken together, these bioinformatic approaches allowed us to narrow both the size of the QTLs and to identify a smaller subset of genes that we believe are likely to cause these QTLs.
In conclusion, we have mapped a large number of QTLs associated with novel locomotor activity and methamphetamine sensitivity in an AIL. Some of the QTLs correspond to regions identified by other researchers, and in the majority of cases we have narrowed the confidence intervals quite significantly as compared to previous studies. While it is clear that the integrated analysis of the F2 and F8 AIL offers vast improvement over only using F2 mice, it is still insufficient for obtaining single gene resolution. However, the combination of high resolution mapping with sequence and expression data offers a powerful approach and permitted identification of several candidate genes that may underlie differences in these phenotypes. In summary, AILs allow GWAS to be performed in a situation where all alleles are common, and where uniform environmental conditions can be maintained, which limits the interactions between genes and environment. These advantages allowed us to map QTL with a modest sample size and identify small regions that warrant further molecular evaluation.
Supplementary Material
Narrowing of QTL intervals in integrated analyses. a) Integrated, F2 and F8 Act1 QTL results 0–5 minutes. b) Integrated, F2 and F8 day 1 QTL results for Act4 QTL during 5–10 minutes. c) Integrated, F2 and F8 ActX QTL results during 0–5 minutes. d) Integrated, F2 and F8 Meth9 QTL results 10–15 minutes. e) Integrated, F2 and F8 Meth15 QTL results 0–5 minutes. f) Integrated, F2 and F8 Meth9 QTL results 10–15 minutes.
eQTLs for Act1, Act4, ActX, Meth9, Meth15, and Meth16. Table includes gene symbol and name, as well as Mb location for each gene, mean expression, max LOD, and eQTL chromosome and Mb location.
Genes with non-synonymous coding SNPs forAct1, Act4, ActX, Meth9, Meth15, andMeth16. Table includes gene symbol and the number of non-synonymous SNPs. SNPs resulting in a frameshift mutation, stop-gain, stop-loss and SNPs located in essential splice sites are indicated with an asterisk.
Acknowledgments
Grants: This work was supported by NIH grants MH079103, DA07255, DA024845 and DA021336.
References
- Bennett B, Carosone-Link P, Zahniser NR, Johnson TE. Confirmation and fine mapping of ethanol sensitivity quantitative trait loci and candidate gene testing in the LXS recombinant inbred mice. J Pharmacol Exp Ther. 2006;319:299–307. doi: 10.1124/jpet.106.103572. [DOI] [PubMed] [Google Scholar]
- Bryant CD, Graham ME, Distler MG, Munoz MB, Li D, Vezina P, Sokoloff G, Palmer AA. A role for casein kinase 1 epsilon in the locomotor stimulant response to methamphetamine. Psychopharm. 2009a;203:703–711. doi: 10.1007/s00213-008-1417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CD, Chang HP, Zhang J, Wiltshire T, Tarantino LM, Palmer AA. A major QTL on chromosome 11 influences psychostimulant and opiod sensitivity in mice. Genes Brain Behav. 2009b;8:795–805. doi: 10.1111/j.1601-183X.2009.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical implications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 2008;180:2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185:1033–1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wan J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Jarvik LF, Liston EH, Jenden DJ. Behavioral responses to amphetamine in identical twins. Acta Genet Med Gemellol Roma. 1983;32:139–49. doi: 10.1017/s0001566000006425. [DOI] [PubMed] [Google Scholar]
- DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. doi: 10.1016/j.tig.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci USA. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C, Shen EH, Simpson VJ, Johnson TE. Mapping quantitative trait loci mediating sensitivity to etomidate. Mamm Genome. 2003;14:367–375. doi: 10.1007/s00335-002-2235-y. [DOI] [PubMed] [Google Scholar]
- Downing C, Carosone-Link P, Bennett B, Johnson T. QTL mapping for low-dose ethanol activation in the LXS recombinant inbred strains. Alcohol Clin Exp Res. 2006;30:1111–1120. doi: 10.1111/j.1530-0277.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- Fergusson DM, Horwood LJ, Lynskey MT, Madden PA. Early reactions to cannabis predict later dependence. Arch Gen Psychiatry. 2003;60:1033–1039. doi: 10.1001/archpsyc.60.10.1033. [DOI] [PubMed] [Google Scholar]
- Flint J. Mapping quantitative traits and strategies to find quantitative trait genes. Methods. 2011;53:163–174. doi: 10.1016/j.ymeth.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Belknap JK, O’Toole LA, Helms ML, Wenger CD, Crabbe JC. Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci. 1997;17:745–754. doi: 10.1523/JNEUROSCI.17-02-00745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haertzen CA, Kocher TR, Miyasato K. Reinforcements from the first drug experience can predict later drug habits and/or addiction: results with coffee, cigarettes, alcohol, barbiturates, minor and major tranquilizers, stimulants, marijuana, hallucinogens, heroin, opiates and cocaine. Drug Alcohol Depend. 1983;11:147–165. doi: 10.1016/0376-8716(83)90076-5. [DOI] [PubMed] [Google Scholar]
- Hart AB, de Wit H, Palmer AA. Genetic factors modulating the response to stimulant drugs in humans. In: Reif A, Cryan J, editors. Current Topics in Behavioral Neurosciences: Behavioral Neurogenetics. Spring; Heidelberg, Germany: in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, Phillips T, Crabbe J. On the integration of alcohol-related quantitative trait loci and gene expression analyses. 1. Alcohol Clin Exp Res. 2004;28:1437–48. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Hofstetter JR, Hitzemann RJ, Belknap JK, Walter NAR, McWeeney SK, Mayeda AR. Characterization of the quantitative trait locus for haloperidol-induced catalepsy on distal mouse chromosome 1. Genes Brain Behav. 2008;7:214–223. doi: 10.1111/j.1601-183X.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Sensitivity to psychostimulants in mice bred for high and low stimulation to methamphetamine. Genes Brain Behav. 2005;4:110–125. doi: 10.1111/j.1601-183X.2004.00101.x. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, de Wit H, McNamara PJ, Cao D. Rewarding, stimulant, and sedative alcohol responses and relationship to future binge drinking. Arch Gen Psychiatry. 2011;68:389–399. doi: 10.1001/archgenpsychiatry.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa N, Mori E, Oka K, Nomura H, Yatsushige N, Maruyama Y. Effects of SCH23390 injection into the dorsal striatum and nucleus accumbens on methamphetamine-induced gnawing and hyperlocomotion in rats. J Nihon Univ Sch Dent. 1989;31:451–457. doi: 10.2334/josnusd1959.31.451. [DOI] [PubMed] [Google Scholar]
- Kuteeva E, Hokfelt T, Ogren SO. Behavioural characterisation of transgenic mice overexpressing galanin under the PDGF-B promoter. Neuropeptides. 2005;39:299–304. doi: 10.1016/j.npep.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, Adelson M, Kreek MJ. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng H. Systems genetics, bioinformatics and eQTL mapping. Genetica. 2010;138:915–924. doi: 10.1007/s10709-010-9480-x. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Cheng R, Lim JE, Palmer AA, Blizard DA. Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol Genomics. 2010;42A:33–38. doi: 10.1152/physiolgenomics.00100.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. Plos Genet. 2008;4:1–18. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLos Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Gerson ES, Simmons S, Ebert M, Kessler LR, Dibble ED, Jimerson SS, Brown GM, Gold P, Jimerson DC, Guroff JJ, Storch FI. Behavioral, biochemical and neuroendocrine responses to amphetamine in normal twins and ‘well-state’ bipolar patients. Psychoneuroendocrinology. 1982;7:163–76. doi: 10.1016/0306-4530(82)90009-9. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhart-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, Phillips TJ. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Parker CC, Palmer AA. Dark matter: are mice the solution to missing heritability? Front Genet. 2011 doi: 10.3389/fgen.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Cheng R, Sokoloff G, Lim JE, Skol AD, Abney M, Palmer AA. Fine-mapping alleles for body weight in LG/J x SM/J F2 and F34 advanced intercross lines. Mamm Genome. 2011;22:563–571. doi: 10.1007/s00335-011-9349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;9:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Kamens HM, Wheeler JM. Behavioral genetic contributions to the study of addiction-related amphetamine effects. Neurosci Biobehav Rev. 2008;32:707–759. doi: 10.1016/j.neubiorev.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F(2) and advanced intercross lines. Genes Brain Behav. 2010;9:759–767. doi: 10.1111/j.1601-183X.2010.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. Onset and course of alcoholism over 25 years in middle class men. Drug Alcohol Depend. 2011;113:21–28. doi: 10.1016/j.drugalcdep.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Sinasac DS, Burrage LC, Hodges CA, Supelak PJ, Palmert MR, Moreno C, Cowley AW, Jr, Jacob HJ, Nadeau JH. Analyzing complex traits with congenic strains. Mamm Genome. 2010;21:276–286. doi: 10.1007/s00335-010-9267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber BA, Kuzmin A, Canals JM, Danielsson A, Paratcha G, Arenas E, Alberch J, Ogren SO, Ibanex CF. Disruption of EphA/ephrin-a signaling in the nigrostriatal system reduces dopaminergic innervation and dissociates behavioral responses to amphetamine and cocaine. Molecular and Cellular Neuroscience. 2004;26:418–428. doi: 10.1016/j.mcn.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Sokoloff G, Parker CC, Lim JE, Palmer AA. Anxiety and fear in a cross of C57BL/6J and DBA/2J mice: mapping overlapping and independent QTL for related traits. Genes Brain Behav. 2011;10:604–614. doi: 10.1111/j.1601-183X.2011.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Vaccarino FJ, Amalric M, Koob GF. The neural substrates for the motor-activating properties of psychostimulants: a review of recent findings. Pharmacol Biochem Behav. 1986;25:233–248. doi: 10.1016/0091-3057(86)90261-3. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Qaadir A, Palmer AA, Cook EH, Jr, de Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacology. 2006;31:1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: Web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Wise RA, Bozarth MA. A psychomotor stimulant theory of addiction. Psychol Rev. 1987;94:469–492. [PubMed] [Google Scholar]
- Yalcin B, Wong K, Gam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Narrowing of QTL intervals in integrated analyses. a) Integrated, F2 and F8 Act1 QTL results 0–5 minutes. b) Integrated, F2 and F8 day 1 QTL results for Act4 QTL during 5–10 minutes. c) Integrated, F2 and F8 ActX QTL results during 0–5 minutes. d) Integrated, F2 and F8 Meth9 QTL results 10–15 minutes. e) Integrated, F2 and F8 Meth15 QTL results 0–5 minutes. f) Integrated, F2 and F8 Meth9 QTL results 10–15 minutes.
eQTLs for Act1, Act4, ActX, Meth9, Meth15, and Meth16. Table includes gene symbol and name, as well as Mb location for each gene, mean expression, max LOD, and eQTL chromosome and Mb location.
Genes with non-synonymous coding SNPs forAct1, Act4, ActX, Meth9, Meth15, andMeth16. Table includes gene symbol and the number of non-synonymous SNPs. SNPs resulting in a frameshift mutation, stop-gain, stop-loss and SNPs located in essential splice sites are indicated with an asterisk.