Summary
T-bet is a transcription factor that drives the Th1 immune response primarily through promoting expression of the IFN-γ gene. Polymorphisms in the T-bet gene, TBX21, have been associated with immune-mediated diseases such as asthma and systemic sclerosis. We found that the TBX21 promoter polymorphism T-1993C is associated with a significant decrease in IL-4 and IFN-γ production by stimulated primary human lymphocytes from healthy participants.
Keywords: TBX21, T-bet, single nucleotide polymorphism, cytokine, Th1/Th2 cells
Introduction
The transcription factor T-bet, “T-box expressed in T cells,” is a member of the T-box family and is a master regulator of T-cell differentiation. As a potent transactivator of IFNG, T-bet stimulates production of the hallmark Th1 cytokine, IFN-γ, and drives Th1 polarization. T-bet simultaneously opposes commitment to the Th2 and Th17 lineages.1,2 T-bet function has been investigated extensively in murine knockout models. In addition to directing the CD4+ cell lineage, these models have demonstrated important roles for T-bet in dendritic cells, CD8+ T cells, B cells, natural killer (NK) cells, and NKT cells. T-bet functioning is required in these cell types for defense against intracellular organisms and for antineoplastic responses.3,4 T-bet has also been shown to play an influential role in inflammatory processes such as inflammatory bowel disease, autoimmune diseases such as rheumatoid arthritis, and immune-mediated conditions like asthma.2
In humans, several single nucleotide polymorphisms (SNPs) in TBX21 are known to affect T-bet expression levels and are associated with disease susceptibility. The TBX21 T-1993C promoter polymorphism (rs4794067) is associated with decreased T-bet expression levels.5,6TBX21 T-1993C is associated with an increased risk of developing asthma6 and aspirin-induced asthma7 and with a decreased risk of developing systemic lupus erythematosus8 and autoimmune hepatitis type 1.9 Other TBX21 SNPs have been associated with genital HSV infection (rs17244587)10 and systemic sclerosis (rs11650354).11 Additionally, T-bet status is of interest with regards to cancer development and prognosis, because Th2-dominant cytokine profiles are known to be tumor-supportive.12 Although there have been many associations with disease susceptibility in humans, to our knowledge, there have been no studies which directly measure cytokine production by primary human lymphocytes harboring TBX21 polymorphisms. In this study, we investigated the effect of the TBX21 T-1993C promoter polymorphism on production of IFN-γ and IL-4 by stimulated primary human lymphocytes from healthy donors.
Materials and Methods
Participant blood sample and questionnaire collection
Participants were recruited at the University of North Carolina at Chapel Hill (UNC-CH) Health Care Blood Donation Center in the platelet donation clinic from August 2010 to December 2010. In addition to medication and disease exposure restrictions, platelet donors are required to be adults who are in good health. Blood samples were collected from 220 donors and complete questionnaires were obtained from 210 of those people (men 90; women 120) with a mean age of 31.8 years (SD 15.5; min 17 max 73). Informed written consent for involvement in the study and for blood-product biobanking was obtained from all participants. The protocol was approved by the UNC-CH institutional review board (IRB). The blood samples were collected in two 10ml EDTA vacutainers and one pro-coagulation vacutainer. Questionnaires were completed during the platelet donation process. All blood samples were transported to the BioSpecimen Processing facility on the UNC-CH campus for processing and product storage within 3 hours of the blood draw.
TBX21 T-1993C polymorphism genotyping
DNA was isolated from coagulated whole blood and then stored by the UNC-CH BioSpecimen Processing facility on the UNC campus. The Puregene [Qiagen, Valencia, CA] modified salting-out precipitation method was used for DNA extraction in a high-throughput automated method on the Autopure DNA extraction robot. TBX21 (rs4794067) genotyping was performed by the UNC-CH Mammalian Genotyping Core using the TaqMan PCR technique and manufacturer-supplied protocols, reagents and software [Applied Biosystems, Foster City, CA; catalogue no. C_11626504_10]. Briefly, 5~10 ng of genomic DNA was amplified in 5uL reactions containing 1x Universal TaqMan Master Mix and 1x probe mix according to the manufacturer's instructions. PCR amplification was performed and allelic discrimination was conducted by comparing the post-amplification intensities of allele-specific reporter dyes to pre-amplification intensities for each sample relative to negative control reactions. End-point reading of the genotypes was performed with a 7900HT Real-Time PCR System [Applied Biosystems, Foster City, CA].
PBMC isolation and cryopreservation
Peripheral blood mononuclear cells (PBMCs) were isolated from anti-coagulated whole blood by centrifugal separation using Histopaque [Sigma, St. Louis, Mo]. The cells were washed and frozen in 20% fetal bovine serum (FBS) and 10% DMSO freezing solution. The cells were stored overnight in -80°C and then transferred to liquid nitrogen for long term storage.
Assessment of intracellular cytokine production
The cryopreserved PBMCs were defrosted by rapid re-warming in a 37°C water bath and immediately washed with RPMI supplemented with FBS. The intracellular cytokine assay was performed twice. The cells for batch 1 were allowed to recover for 10 days, and for batch 2 they recovered for 17 days. The samples were monocyte-depleted by exclusion of adherent cells. Cell viability was confirmed with Trypan Blue staining.
Intracellular (IC) staining was performed according to a modified BD Pharmingen alternative IC staining protocol. Briefly, the monocyte-depleted PBMCs were assessed for viability, counted, and activated with 1ug/ml phorbol 12-myristate 13-acetate (PMA) [Sigma, St. Louis, Mo] plus 0.5ul/mL Ionomycin [Sigma, St. Louis, Mo] for 4 hours in 5% CO2 at 37°C in the presence of 1ul/ml GolgiPlug protein transport inhibitor (Brefeldin A, 1.0 ug/ml) [BD, Cat. No. 554715]. The cells were then washed and stained for cell-surface antigens using fluorochrome-conjugated antibodies against CD3 and CD16+CD56, to identify T cells and natural killer cells respectively, or the appropriate Ig negative control. Only the second batch was stained for CD3. After a 15 minute incubation at room temperature (RT) in the dark, the cells were washed then fixed and permeabilized by addition of Fixation/Permeabilization solution [BD, Cat. No.554722]. The cells were then incubated in this solution for 20 minutes at RT in the dark.
After washing, the cell pellets were resuspended in Perm/Wash buffer [BD, Cat. No. 554723] which contained the intracellular fluorochrome-conjugated anti-cytokine antibodies at optimal concentrations and incubated for 30 minutes at RT in the dark. The antibody cocktail consisted of anti-IFN-γ PerCP-Cy5.5/anti-IL6 V450/anti-IL-4 APC. Isotype matched antibodies to the cytokines were utilized to control for non-specific binding and a set of CD19 controls on separate cells was used for gating. The cells were then fixed in RPMI/2% paraformaldehyde (PFA) and analyzed immediately using a Dako Cyan flow cytometer and Summit software. Forward and side scatter properties were utilized to determine the lymphocyte population.
Statistical analysis
Statistical analysis was performed using SAS software version 9.2 (SAS Institute, Inc., Cary, NC). The nonparametric method of the Wilcoxon rank-sum test was used to assess the differences in the quantity of cytokine production between the TBX21 -1993CC and -1993TT genotypes. Analyses were conducted for the two batch datasets separately, as well as for the combined batch dataset. The linear mixed effect model for repeated measurements, using the PROC MIXED procedure, was utilized to handle standard data analysis for the combined datasets. The parameter of interest was estimated using the unstructured covariance matrix. The statistical testing of the allele effect was conducted using the CONTRAST statement in the MIXED procedure, and the test statistic and the p-value were reported. The measure of the intracellular staining was quantified by mean fluorescence intensity as determined by flow cytometry. A two-sided p-value of less than 0.05 was considered significant.
Results
TBX21 genotype analysis
SNP genotyping identified 12 participants homozygous for TBX21 -1993C (rs4794067) (5.5%), 74 participants who were -1993TC heterozygotes (34.1%), and 131 participants who were homozygous for the wild-type -1993T allele (60.4%). The genotype frequencies of rs4794067 in our group were very similar to those reported in the human dpSNP for the European group HapMap-CEU: CC 0.044, CT 0.389, TT 0.566 from 226 sample counts (http://www.ncbi.nlm.nih.gov/SNP).
T-bet -1993CC homozygote status is associated with decreased IFN-γ and IL-4 production
The homozygous -1993TT and -1993CC cells were analyzed twice in separate batches. Batch 1 had 10 controls with the -1993TT genotype and 8 cases with the -1993CC genotype, and batch 2 had 7 controls and 5 cases. Four samples were excluded from analysis due to poor recovery as indicated by having a < 50% viability count by Trypan Blue staining. An additional 2 samples were not analyzed in batch 2, due to a limited number of total cryopreserved cells.
Analysis of batch 1 revealed that both IL-4 and IFN-γ production by stimulated lymphocytes was significantly decreased in cells with the TBX21 -1993CC compared to the -1993TT genotype (P = 0.004 and P = 0.03 respectively) (Figure 1). In batch 2, the levels of IL-4 and IFN-γ cytokine production were also significantly lower in the -1993CC compared to -1993TT group (P = 0.01 and P = 0.02 respectively) (Figure 2).
Figure 1.
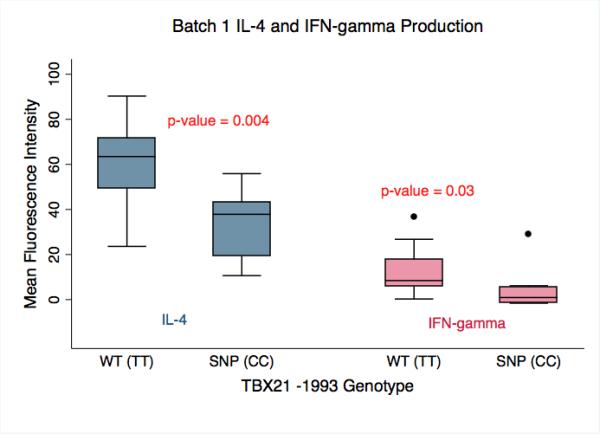
Batch 1 analysis of IL-4 and IFN-γ production by primary human lymphocytes stimulated with PMA/Ionomycin. Cells harboring the SNP genotype (SNP(CC), n = 8) produced significantly less IL-4 and IFN-γ than cells with the wild-type genotype (WT(TT), n = 10) (p-value = 0.004 and 0.03 respectively). The amount of IL-4 and IFN-γ produced is presented as the Mean Fluorescence Intensity as determined by flow cytometry.
Figure 2.
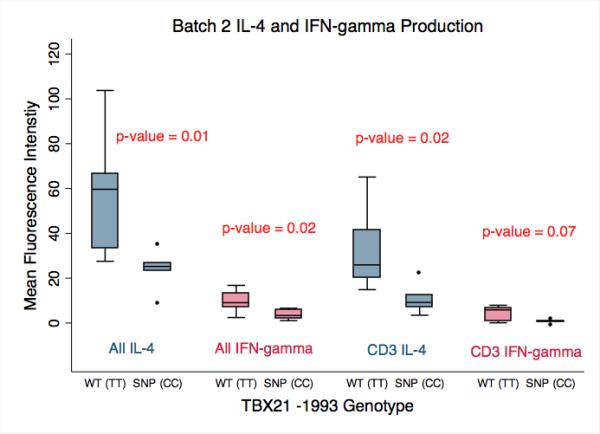
Batch 2 analysis of IL-4 and IFN-γ production by primary human lymphocytes stimulated with PMA/Ionomycin performed 1 week after Batch 1. When evaluating the entire lymphocyte population, the cells harboring the SNP genotype (SNP(CC), n = 5) produced significantly less IL-4 and IFN-γ than cells with the wild-type genotype (WT(TT), n = 7) (p = 0.01 and 0.02 respectively). CD3+ T-lymphocytes were stained in Batch 2 to allow for separate analysis of the CD3+ population. CD3+ lymphocytes with the SNP(CC) genotype produced significantly less IL-4 than cells with the WT(TT) genotype (p-value = 0.02), and produced less IFN-γ, which trended towards significance (p-value = 0.07). The amount of IL-4 and IFN-γ produced is presented as the Mean Fluorescence Intensity as determined by flow cytometry.
Combining measurements from the two batches also demonstrated significant differences in cytokine production by each genotype while accounting for batch effect. Both IL-4 and IFN-γ production were significantly decreased in the TBX21 -1993CC group compared to -1993TT group (P = 0.001 and P = 0.02).
In batch 2, CD3+ T-lymphocytes were specifically labeled to allow for separate analysis. The level of IL-4 production by CD3+ T-lymphocytes was significantly decreased in the -1993CC group (P = 0.02), and the decrease in IFN-γ trended towards significance (P = 0.07) (Figure 2).
Discussion
In this study, we found that the TBX21 -1993CC versus -1993TT genotype was associated with attenuation of both IFN-γ and IL-4 release by stimulated human lymphocytes, including CD3+ T-lymphocytes, from healthy participants. To our knowledge, this is the largest study performed with primary human lymphocytes and the only study to directly measure the quantity of cytokine produced by these cells.
Our results confirm recent findings which demonstrate lower production of IFN-γ by human T-cell lines transfected with the T-1993C promoter polymorphism.5,6 The results are also consistent with murine knock-out models which demonstrate the animals’ inability to produce IFN-γ. The significant decrease in IL-4 production was an unexpected finding. We anticipated IL-4 levels would be elevated, because there should be less T-bet mediated repression of the Th2 lineage. Although no studies have specifically investigated the expression of Th2 genes in human cell lines transfected with T-bet polymorphisms, other evidence led us to expect an increase in IL-4 production. In T-bet deficient mice, elevated production of Th2 cytokines such as IL-4 and IL-5 as well as Th17 cytokines like IL-17A and IL-23 has been well-documented.2 In two human studies, one with systemic sclerosis patients and the other with gastric cancer patients, Th2-dominant cytokine profiles were found in patients with T-bet polymorphisms.11,12 Th2-dominant profiles are distinguished by elevated levels of cytokines such as IL-4, IL-5 and IL-13. Lastly, the only study of primary human lymphocytes also found an increased percentage of IL-4 producing CD4+ cells with a T-bet polymorphism. The team had 3 samples with the -1993CC genotype, 7 with the -1993TC genotype, and 7 with the -1993TT genotype. They stimulated the cells with PHA and ionomycin along with co-stimulatory monoclonal antibodies anti-CD28/anti-CD49d.5
Although it is unclear why the Th2 pathway was attenuated in our study, there are reports of T-bet -/- mice that also failed to demonstrate the anticipated shift toward the Th2 response. Mice challenged with Schistosoma mansoni responded with lower expression of Th2 genes, including IL4, and higher expression of Th17 genes. The authors concluded that the Th2 response is not necessarily a default pathway in T-bet deficiency.13 Exposure to Myobacterium tuberculosis and Salmonella typhinurium also failed to generate an increase in Th2 cytokines, with an overproduction of IL-10 instead.14,15 These studies suggest that the absence of T-bet alone is not enough to generate Th2 cells and other components of the inflammatory environment are required for this response. The same concept may apply to humans. Host inflammatory conditions in addition to T-bet deficiency may have been responsible for the Th2 dominance observed in human gastric cancer and systemic sclerosis patients.
In summary, the current study found that the T-1993C TBX21 SNP is associated with decreased production of IFN-γ and IL-4 by stimulated human lymphocytes. The decrease in IFN-γ was expected based on murine models and T-bet expression assays, and it is reasonable to hypothesize that the inability to adequately produce IFN-γ plays a direct role in the association with disease susceptibility. The finding that the polymorphism also decreases IL-4 production was unexpected based on the role that T-bet plays as a repressor of the Th2 lineage. The results, however, agree with some murine models which suggest that more than an absence of T-bet is required to generate Th2 profiles. Further studies on human cells or cell lines are needed to clarify the effects of TBX21 SNPs on Th2 cytokines like IL-4.
Acknowledgements
This research was supported in part by The University Cancer Research Fund, The North Carolina Translational and Clinical Sciences Institute (2KR261104) and grants from the National Cancer Institute (R01 CA112243 and CA112243-05S1) and from the National Institute of Environmental Health Sciences (P30ES10126 and ES014635). The authors would also like to acknowledge support from the Doris Duke Charitable Foundation, the UNC Mammalian Genotyping Core for SNP genotyping, the UNC BioSpecimen core for our DNA extractions and blood processing (https://genome.unc.edu/bsp), the UNC Biostatistics Department, and the UNC Health Care Blood Donation Center for participant recruitment.
The funding sources had no role in study design, collection, analysis, interpretation of data, or in writing of the report.
Abbreviations
- SNP
single nucleotide polymorphism
- IFN-γ
interferon-γ
- IL-4
interleukin-4
- Th cells
T helper cells
- T-bet
T-box expressed in T cells
- NK
natural killer
- HSV
herpes simplex virus
- UNC-CH
University of North Carolina at Chapel Hill
- IRB
institutional review board
- EDTA
ethylenediaminetetraacetic acid
- DNA
deoxyribonucleic acid
- PCR
polymerase chain reaction
- PBMC
peripheral blood mononuclear cell
- FBS
fetal bovine serum
- IC
intracellular
- PMA
phorbol 12-myristate 13-acetate
- RT
room temperature
- PFA
paraformaldehyde
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We have no conflict of interest to disclose.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 2.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werneck MB, Lugo-Villarino G, Hwang ES, Cantor H, Glimcher LH. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J Immunol. 2008;180:8004–10. doi: 10.4049/jimmunol.180.12.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 5.Li JR, Li JG, Deng GH, et al. A common promoter variant of TBX21 is associated with allele specific binding to Yin-Yang 1 and reduced gene expression. Scand J Immunol. 2011;73:449–58. doi: 10.1111/j.1365-3083.2011.02520.x. [DOI] [PubMed] [Google Scholar]
- 6.Suttner K, Rosenstiel P, Depner M, et al. TBX21 gene variants increase childhood asthma risk in combination with HLX1 variants. J Allergy Clin Immunol. 2009;123:1062–8. 8, e1–8. doi: 10.1016/j.jaci.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 7.Akahoshi M, Obara K, Hirota T, et al. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet. 2005;117:16–26. doi: 10.1007/s00439-005-1285-0. [DOI] [PubMed] [Google Scholar]
- 8.You Y, Zhao W, Chen S, et al. Association of TBX21 gene haplotypes in a Chinese population with systemic lupus erythematosus. Scand J Rheumatol. 2010;39:254–8. doi: 10.3109/03009740903347983. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Zhao W, Tan W, et al. Association of TBX21 promoter polymorphisms with type 1 autoimmune hepatitis in a Chinese population. Hum Immunol. 2011;72:69–73. doi: 10.1016/j.humimm.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Svensson A, Bergin AM, Lowhagen GB, et al. A 3'-untranslated region polymorphism in the TBX21 gene encoding T-bet is a risk factor for genital herpes simplex virus type 2 infection in humans. J Gen Virol. 2008;89:2262–8. doi: 10.1099/vir.0.2008/001305-0. [DOI] [PubMed] [Google Scholar]
- 11.Gourh P, Agarwal SK, Divecha D, et al. Polymorphisms in TBX21 and STAT4 increase the risk of systemic sclerosis: evidence of possible gene-gene interaction and alterations in Th1/Th2 cytokines. Arthritis Rheum. 2009;60:3794–806. doi: 10.1002/art.24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang P, Qiu G, Wang S, et al. The mutations of Th1 cell-specific T-box transcription factor may be associated with a predominant Th2 phenotype in gastric cancers. Int J Immunogenet. 2010;37:111–5. doi: 10.1111/j.1744-313X.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 13.Rutitzky LI, Smith PM, Stadecker MJ. T-bet protects against exacerbation of schistosome egg-induced immunopathology by regulating Th17-mediated inflammation. Eur J Immunol. 2009;39:2470–81. doi: 10.1002/eji.200939325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan BM, Jobe O, Lazarevic V, et al. Increased susceptibility of mice lacking T-bet to infection with Mycobacterium tuberculosis correlates with increased IL-10 and decreased IFN-gamma production. J Immunol. 2005;175:4593–602. doi: 10.4049/jimmunol.175.7.4593. [DOI] [PubMed] [Google Scholar]
- 15.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175:4603–10. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]


