Abstract
The epidermal growth factor receptor (EGFR) is essential to multiple physiological and neoplastic processes via signaling by its tyrosine kinase domain and subsequent activation of transcription factors. EGFR overexpression and alteration, including point mutations and structural variants, contribute to oncogenesis in many tumor types. In this study, we identified an in-frame splice variant of the EGFR called mini-LEEK (mLEEK) that is more broadly expressed than the EGFR and is overexpressed in several cancers. Unlike previously characterized EGFR variants, mLEEK lacks the extracytoplasmic, transmembrane and tyrosine kinase domains. mLEEK localizes in the nucleus and functions as a transcription factor to regulate target genes involved in the cellular response to endoplasmic reticulum (ER) stress, including the master regulator of the unfolded protein response (UPR) pathways, molecular chaperone GRP78/Bip. We demonstrated that mLEEK regulates GRP78 transcription through direct interaction with a cis-regulatory element within the gene promoter. Several UPR pathways were interrogated and mLEEK expression was found to attenuate the induction of all pathways upon ER stress. Conversely, knockdown of mLEEK resulted in caspase-mediated cell death and sensitization to ER stress. These findings indicate that mLEEK levels determine cellular responses to unfavorable conditions that cause ER stress. This information, along with the overexpression of mLEEK in tumors, suggests unique strategies for therapeutic intervention. Furthermore, the identification of mLEEK expands the known mechanisms by which the EGFR gene contributes to oncogenesis and represents the first link between two previously disparate areas in cancer cell biology: EGFR signaling and the UPR.
Keywords: EGFR, GRP78, ER stress, unfolded protein response, transcription
Introduction
The epidermal growth factor receptor (EGFR) is a transmembrane glycoprotein linked to numerous physiological processes through ligand stimulation of its intrinsic kinase activity leading to the induction of signaling cascades (Yarden and Sliwkowski, 2001). Frequent overexpression or alterations of the EGFR in cancer contribute to pathogenesis through effects on cell growth, survival and motility (Yarden and Sliwkowski, 2001). The most common variant, EGFRvIII, contains a deletion of exons 2–7 in the extracellular domain and is frequently found in glioblastoma (GBM) (Wong et al., 1992). Other structural variants have been described, including EGFRvIVa and EGFRvIVb, which contain deletions of segments downstream of the tyrosine kinase (TK) domain and confer oncogenic potential in vivo (Pines et al., 2010). In addition, point mutations in the TK domain contribute to drug sensitivity in non-small-cell lung cancer (Lynch et al., 2004; Paez et al., 2004). Novel signaling mechanisms have also been identified in cancer cells where the receptor has a direct association with the sodium/glucose transporter 1 (Weihua et al., 2008), or translocates to the nucleus and associates with transcription factors (Lin et al., 2001).
Cellular homeostasis is dependent on correct folding of proteins in the endoplasmic reticulum (ER) and disruption of this process plays a role in cancer pathogenesis (Wouters and Koritzinsky, 2008). ER stress results from the accumulation of misfolded proteins and can be caused by conditions within actively growing cells or the tumor microenvironment, such as hypoxia and nutrient deprivation (Feldman et al., 2005). The unfolded protein response (UPR) is a conserved program that has evolved to meet the demands for increased protein folding (Malhotra and Kaufman, 2007). GRP78/BiP acts as a sensor within the ER and regulates the UPR through interactions with three major ER signaling proteins: IRE1, PERK and ATF6 (Malhotra and Kaufman, 2007). Pathways triggered by these proteins reduce the ER protein load through several mechanisms, including increasing ER chaperone transcription. Thus, the UPR enables cells to survive in unfavorable conditions and promote tumor growth (Wouters and Koritzinsky, 2008). GRP78 is also a molecular chaperone and participates in several phenotypes, including proliferation, chemoresistance and suppression of apoptosis through caspase inhibition (Reddy et al., 2003; Lee, 2007). Overexpression of GRP78 has been reported in numerous cancers, including lung and GBM (Moenner et al., 2007).
In this study, we identified a novel EGFR variant, mini-LEEK (mLEEK), which is overexpressed in tumors and plays a role in the UPR. In contrast to previously identified EGFR variants, mLEEK is localized in the nucleus and directly activates transcription of target genes. The ER chaperone GRP78 was identified as a transcriptional target of mLEEK. Upregulation of GRP78 as a result of mLEEK expression leads to decreased induction of the UPR upon ER stress. Furthermore, knockdown of mLEEK expression sensitizes cells to ER stress. Beyond the identification of a novel EGFR variant overexpressed in tumors, our findings represent the first link between two previously disparate areas in cancer cell biology: EGFR signaling and the UPR. These findings suggest unique strategies for therapeutic intervention.
Results
Identification of a novel EGFR variant
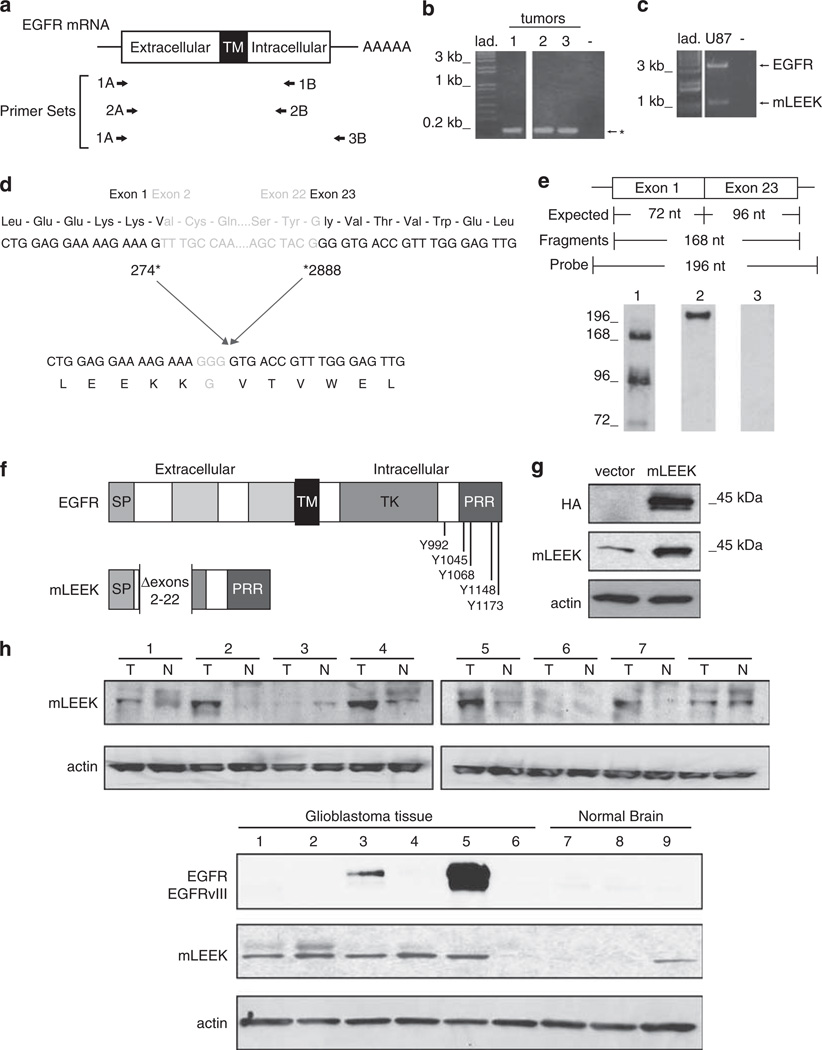
We sought to determine if EGFR mutations previously identified in the TK domain coexist with the EGFRvIII alteration (Lynch et al., 2004; Paez et al., 2004). Reverse transcription–polymerase chain reaction (RT–PCR) analysis of tumor samples using a primer set based within the first exon and immediately downstream of the TK domain consistently identified a smaller transcript than expected for wild-type EGFR or EGFRvIII (Figures 1a and b). Sequencing revealed that this product represents a junction between EGFR exons 1 and 23 (Figure 1d). RT–PCR with a primer set that amplified the entire open-reading frame and subsequent sequencing analysis confirmed that the deletion of exons 2–22 is the only alteration found in this molecule (Figure 1c). Identical products were identified in several tumor samples (8/8 breast; 8/13 ovarian; 5/7 colon). To validate the presence of an exon 1–23 junction, ribonuclease protection assays were conducted using a probe that spanned the junction (Figure 1e). Analysis in A431 cells, which demonstrate expression of the variant by RT–PCR, revealed strong hybridization of a junction spanning probe. Products corresponding to exons 1 and 23 were also detected, reflecting the high expression of wild-type EGFR in this cell line.
Figure 1.
mLEEK: a novel variant of the EGFR. (a) Primer sets used in RT–PCR and nested PCRs. A, sense primer; B, antisense primer. (b) RT–PCR of coding region of human EGFR using human breast tumors (1–3) and 1A/1B primers, followed by nested PCR using 2A/2B primers. Predicted sizes for full-length EGFR and EGFRvIII are 2782 and 1981 bp, respectively. *The presence of an alternative EGFR amplification product. (c) RT–PCR using 1A/3B primers and RNA from U87MG cells as template. (d) Sequencing of the alternative product identified above reveals that mLEEK is the result of the joining of exons 1–23. Numbering refers to human EGFR cDNA sequence (Accession no. X00588). (e) RNase protection detects the exon 1–23 fusion. A 196 nt. labeled antisense probe spanning the junction was used where 72 nt. are from exon 1 and 96 nucleotides come from exon 23 with an additional 28 nt. of non-complementarity to permit discrimination between full-length probe and protected fragments. The protected fragment corresponding to the junction of exons 1 and 23 is 168 nt. RNA was from A431 cells (lane 1). Undigested probe is included as a reference (lane 2). RNase protection with yeast RNA served as a negative control (lane 3). (f) Schematic of mLEEK protein structure compared to EGFR. SP, signal peptide; TM, transmembrane domain; TK, tyrosine kinase domain; PRR, proline-rich region. The known phosphorylation sites (residues numbered) are indicated. (g) Western blot of NIH3T3 cells stably expressing empty vector or mLEEK with an HA tag, probed with anti-HA, anti-mLEEK or anti-actin. (h) Top, Western blots of colon tumor lysates (T) and paired normal tissue (N), probed with anti-mLEEK antibody. Bottom, Western blot of glioblastoma tumor tissue (1–6) and normal brain tissue lysate (7–9) probed with anti-mLEEK or anti-EGFR.
Sequence analysis shows that this variant maintains the open-reading frame of the EGFR and creates a novel glycine at the junction. We call this molecule mLEEK based on the N-terminal amino acids derived from exon 1 and because it is highly truncated relative to wild-type EGFR and EGFRvIII. mLEEK is predicted to lack the EGF ligand-binding domain, transmembrane domain and the ATP binding site of the TK domain; however, it does retain all major autophosphorylation sites and a proline-rich region in the C-terminal end (Figure 1f).
An mLEEK-specific antibody was generated by immunizing rabbits with a peptide derived from the exon 1–23 junction. Serum was affinity purified using the immunizing peptide and detected a 45 kDa protein in western blots of cell lines and primary tumors (Figures 1g and h and Supplementary Figure 1). Importantly, mLEEK antibody did not crossreact with EGFR or any known variants. The 45 kDa protein is consistent with the expected size for mLEEK based on amino-acid composition and the size of in vitro translated protein (Supplementary Figure 1). A plasmid containing mLEEK cDNA with an epitope tag at the C terminus (mLEEK-HA) also expressed a 45 kDa protein, which was detected by the mLEEK polyclonal antibody in immunoprecipitation and two-dimensional polyacrylamide gel electrophoresis–western analysis (Figure 1g and Supplementary Figure 1). The same sized protein was also detected in murine cells (Supplementary Figure 1). These data confirm that mLEEK is endogenously expressed and not the product of a pseudogene.
mLEEK is widely expressed in normal tissues and overexpressed in human tumors
Using affinity-purified mLEEK antibody, mLEEK was detected in a variety of cell types and 29/30 human tissues, including hematopoietic cells and other cell/tissue types where EGFR has not been found, indicating a much wider distribution of mLEEK (Tables 1 and 2). Interestingly, immunohistochemistry revealed increased expression in multiple human tumors, including those from ovary, lung and skin (Table 3 and Supplementary Figure 2). Western blot analysis of human brain and colon showed that mLEEK is expressed at varying degrees in normal tissue and is consistently overexpressed in tumors (Figure 1h). As ~40% of primary brain tumors (GBMs) have amplification of the EGFR gene and subsequent overexpression of the protein and about 70% of GBMs with EGFR overexpression also express mutated forms of EGFR, we probed these GBMs for EGFR expression (Libermann et al., 1984; Malden et al., 1988; Yamazaki et al., 1988; Ekstrand et al., 1991, 1992; Wong et al., 1992). Although two out of six GBMs analyzed had high EGFR expression and one of the two also expressed EGFRvIII, increased expression of EGFR did not correlate with increased mLEEK expression (Figure 1h).
Table 1.
Human cell lines screened for mLEEK expression
| Cell line | Source | mLEEK | Method |
|---|---|---|---|
| A431 | Epidermoid carcinoma | + | RT–PCR; western |
| HCT116 | Colorectal carcinoma | + | Western |
| HEK293 | Kidney | + | Western |
| HT1080 | Fibrosarcoma | + | Western |
| MDA MB 468 | Breast adenocarcinoma | + | RT–PCR; western |
| U87MG | glioblastoma | + | RT–PCR; western |
Abbreviations: mLEEK, mini-LEEK; RT–PCR, reverse transcription–polymerase chain reaction.
Cell lines were cultured from frozen stocks. RNA was extracted and analyzed by RT–PCR as described in Figure 1. Lysates were prepared for analysis by western blotting with anti-mLEEK antibody. Positive expression of mLEEK is indicated with ‘+’.
Table 2.
Expression of mLEEK in human tissues
| Tissue type (total no.) | mLEEK positive | Method |
|---|---|---|
| Adrenal | 4/4 | IHC, western |
| Bladder (urinary) | 2/2 | IHC |
| Brain | 3/6 | IHC, western |
| Breast | 4/6 | RT–PCR, IHC |
| Colon | 6/8 | Western |
| Colon | 5/6 | IHC |
| Endometrium | 2/3 | IHC |
| Esophagus | 2/2 | IHC |
| Gall bladder | 1/1 | IHC |
| Kidney | 1/1 | Western |
| Liver | 4/6 | IHC |
| Lung | 8/8 | IHC |
| Lymph node | 2/2 | IHC |
| Myometrium | 1/1 | IHC |
| Ovary | 4/4 | IHC |
| Pancreas | 1/1 | IHC |
| Placenta | 2/2 | IHC |
| Prostate | 5/5 | IHC |
| Rectum | 1/1 | IHC |
| Salivary gland | 1/1 | IHC |
| Skeletal muscle | 1/2 | IHC |
| Skin | 2/2 | IHC |
| Small bowel | 1/1 | IHC |
| Smooth muscle | 1/1 | IHC |
| Soft tissue | 1/1 | IHC |
| Spleen | 3/3 | IHC |
| Stomach | 5/5 | IHC |
| Testis | 2/2 | IHC |
| Thyroid | 1/3 | IHC, western |
| Tonsil | 1/1 | IHC |
| Uterine cervix | 0/1 | IHC |
Abbreviations: IHC, immunohistochemistry; mLEEK, mini-LEEK; RT–PCR, reverse transcription–polymerase chain reaction.
RNA was extracted from human tissue samples and analyzed by RT–PCR as described in Figure 1. Lysates were prepared for analysis by western blotting with anti-mLEEK antibody. Paraffin-embedded tissue samples were examined by IHC with anti-mLEEK antibody.
Table 3.
mLEEK is overexpressed in cancer tissues
| n | Staining intensity | ||||
|---|---|---|---|---|---|
| 3+ | 2+ | 1+ | 0 | ||
| Skin | |||||
| Normal | 5 | — | — | 80% | 20% |
| Melanoma | 30 | 10% | 37% | 53% | 0% |
| Lung | |||||
| Normal | 11 | — | 36% | — | 64% |
| Adenocarcinoma | 25 | 36% | 36% | 20% | 8% |
| Squamous | 26 | 4% | 23% | 46% | 27% |
| Ovary | |||||
| Normal | 3 | — | — | — | 100% |
| Adenocarcinoma | 31 | 52% | 35% | 13% | — |
Abbreviation: mLEEK, mini-LEEK.
Immunohistochemistry was performed with anti-mLEEK antibody on paraffin-embedded normal or tumor specimens. Staining intensity was evaluated by a surgical pathologist. Representative images are presented in Supplementary Figure 2.
mLEEK is localized to the nucleus
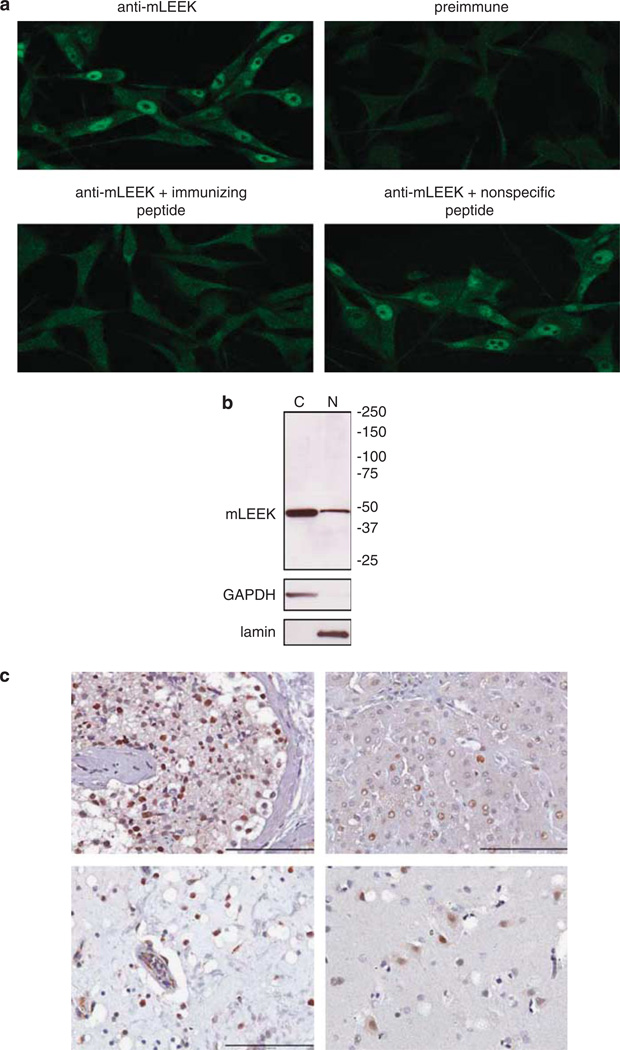
To gain insights into mLEEK’s function, we determined its subcellular localization. Immunofluorescence analysis of U87MG human glioma cells showed a distinct nuclear localization of endogenous mLEEK (Figure 2a). Peptide competition experiments and the lack of nuclear staining with preimmune sera confirmed the specificity of the staining (Figure 2a). Similar patterns were observed in HT1080 cells, where the specificity of staining with the mLEEK antibody was further confirmed by small interfering RNA (siRNA)-mediated knockdown of mLEEK (Supplementary Figure 3). Western blotting of subcellular fractions from U87MG cells verified that mLEEK was found in the nuclear fraction (Figure 2b). Immunohistochemistry also detected mLEEK in the nucleus of normal human tissue specimens (Figure 2c). To determine whether levels of mLEEK in the nucleus are regulated by an active transport mechanism, cells were treated with leptomycin B, an inhibitor of CRM1, a component of the nuclear export machinery. Leptomycin B treatment did not lead to the accumulation of mLEEK in the nucleus, suggesting that nuclear mLEEK levels are not regulated by an active nuclear transport mechanism (Supplementary Figure 3; Gorlich and Kutay, 1999; Allen et al., 2000; Macara, 2001; Stewart et al., 2001).
Figure 2.
mLEEK is localized to the nucleus. (a) Localization of mLEEK in U87MG cells was visualized by indirect immunofluorescence with mLEEK antibody. Peptide competition experiments with immunizing peptide corresponding to the exon 1–23 junction or nonspecific peptide were included as controls. As an additional control, cells were stained with preimmune sera. (b) U87MG cells were fractionated into cytoplasmic (C) and nuclear (N) fractions and lysates from equivalent amounts of cellular starting material were analyzed by western blotting with mLEEK antibody. Purity of the fractions was determined by blotting against lamin and GAPDH to distinguish the nuclear and cytoplasmic fractions, respectively. (c) Immunohistochemistry was performed on paraffin-embedded human tissue with mLEEK antibody. Testes (top left); liver (top right); urinary bladder (bottom left); and cerebrum (bottom right). Scale bar is 100 µm.
mLEEK upregulates expression of ER chaperone proteins
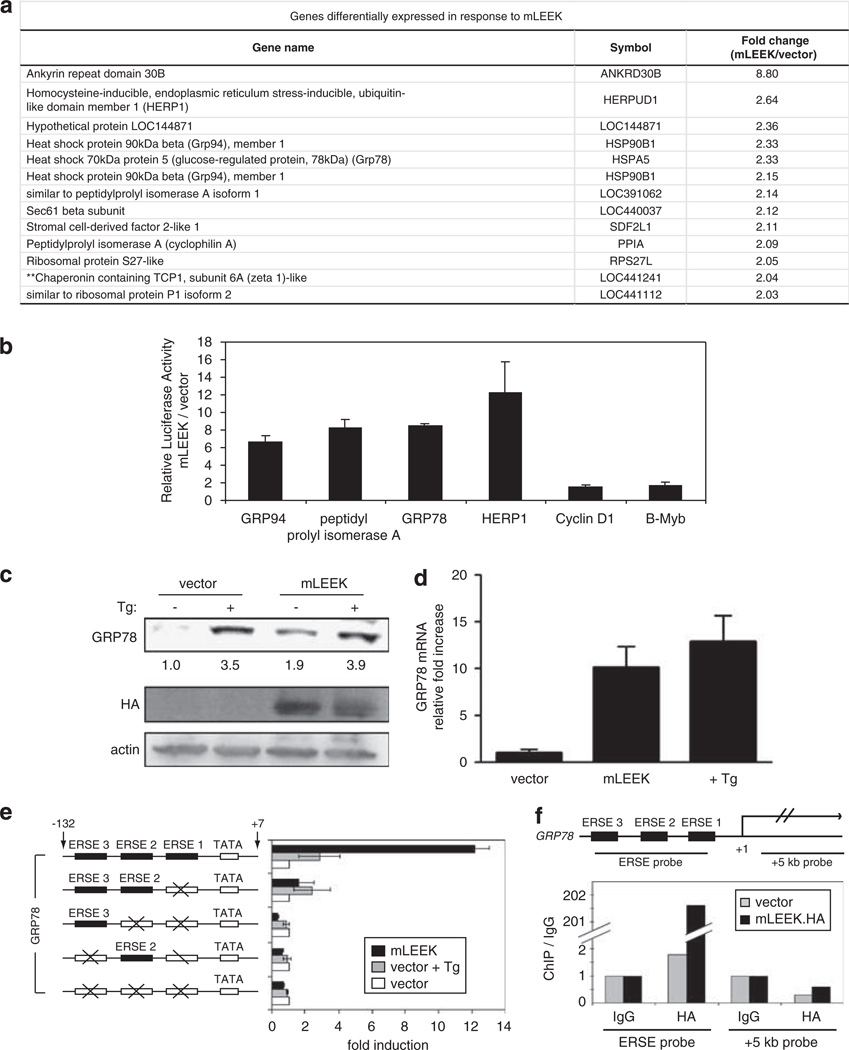
Nuclear localization suggests that mLEEK might be involved in some aspect of nuclear function. Moreover, EGFR contains a potent transactivation domain within the C terminus, which is conserved in mLEEK (Lin et al., 2001). We explored whether mLEEK could activate transcription by using microarray analysis to identify genes with differential expression upon mLEEK expression in HEK293 cells. Gene ontology analysis of the most upregulated genes in four separate experiments consistently revealed a subset of genes involved in protein folding in the ER, including HSPA5 (aka: GRP78, BiP), HSP90B1 (aka: GRP94), HERPUD1 (aka: HERP), SDF2L1 and PPIA (aka: CYPA) (Figure 3a). Previous microarray analyses of genes upregulated in response to overexpression of EGFR or EGFRvIII have not shown induction of any components of the ER protein folding machinery (Pedersen et al., 2001b, 2005).
Figure 3.
mLEEK upregulates the expression of GRP78 by interacting with the transcriptional machinery. (a) A subset of mRNAs identified in microarray experiments that were consistently >2 fold upregulated by overexpression of mLEEK relative to empty vector. (b) Induction of promoter activity by mLEEK was assayed using a panel of luciferase reporter constructs, which were co-transfected into HT1080 cells along with pcDNA/mLEEK-HA or pcDNA/HA. Averages from three independent experiments are presented with s.d. values (bars). (c) Endogenous GRP78 protein levels were assayed by western blotting of lysates from HT1080 cells transiently transfected with pcDNA/mLEEK-HA or pcDNA/HA. As a positive control, cells were treated with Tg for 6 h. (d) HT1080 cells were transiently transfected with pcDNA/mLEEK-HA or vector control. As a positive control, cells were treated with Tg for 6 h. Normalized GRP78 mRNA levels were measured by quantitative RT–PCR and shown relative to levels in vector control-transfected cells. Error bars represent s.d. from triplicate experiments. (e) Left, schematic diagram of GRP78-luciferase reporter constructs. Numbers indicate nucleotide position from transcription start site. Boxes represent ERSE motifs in the promoter. ERSEs with mutated sequences are indicated by crosses. Mutations have been described previously (Yoshida et al., 1998). Right, Relative luciferase activity upon transfection into cells. As a reference for induction of transcription, subsets of cells were treated with Tg for 6 h before determination of luciferase activity. Averages from three experiments are presented with s.d. values (bars). (f) Top, schematic of the GRP78 gene, showing regions amplified by PCR in ChIP assays corresponding to the ERSE region and a region approximately + 5 kb from the transcriptional start site. Bottom, crosslinked chromatin from U87 cells stably expressing mLEEK-HA or an expression vector containing the HA tag alone (vector) was subjected to ChIP using anti-HA or immunoglobulin G and analyzed by quantitative PCR using the primers sets described above. Data are represented as the IP fold enrichment of the assay site relative to sample specific background (immunoglobulin G).
To confirm that candidate genes are direct transcriptional targets of mLEEK, we performed reporter gene assays (Figure 3b). GRP94 reporter activity was activated ~6.6-fold upon mLEEK expression. Remarkably, the GRP78 reporter construct was activated ~8.5-fold. The PPIA reporter showed a significant fold induction (8-fold) as did the HERP1 reporter (12-fold). As a negative control, reporters were included for genes not upregulated in our microarray analysis: cyclin D1 and B-Myb. These were not differentially regulated in response to mLEEK.
GRP78 was further examined because it has been extensively studied for its role as a master regulator of the UPR and gene ontology analysis implicates a role for mLEEK in this process (Ni et al., 2001; Fu and Lee, 2006). Endogenous GRP78 protein levels increased upon mLEEK expression (Figure 3c). As a positive control, cells were stimulated with thapsigargin to induce ER stress (Thastrup et al., 1990). It has been shown that GRP78 protein levels change modestly in response to ER stress (~2-fold) while transcript levels are more potently induced, likely due to the stability and tight regulation of the GRP78 protein (Lee, 2005). Therefore, we also measured GRP78 transcript levels upon mLEEK expression and observed a 10-fold increase (Figure 3d). Collectively, these data indicate that mLEEK has a direct effect on the expression of GRP78.
mLEEK interacts with the GRP78/GRP94 transcriptional machinery
GRP78 and GRP94 are coordinately regulated at the transcriptional level in response to ER stress (Chang et al., 1989; Liu and Lee, 1991). The ERSE, a 19-nucleotide cis-acting element in the promoters of these genes, is important for binding stress-responsive transcription factors (Yoshida et al., 1998, 2000; Parker et al., 2001). Since mLEEK mediates transcriptional activation of GRP78, we hypothesized that it is acting via the ERSE. mLEEK revealed a potent effect on a GRP78 promoter reporter when all three ERSE elements were intact (Figure 3e). Mutation of ERSE1 greatly diminished the effect, and combined elimination of all ERSE elements completely abolished the mLEEK’s ability to activate transcription. Interestingly, mLEEK’s pattern of relative dependence upon each ERSE motif parallels the dependence of thapsigargin and tunicamycin, two other known inducers of ER stress. However, the effect of mLEEK on GRP78 transcription was more potent than treatment with tunicamycin at the standard concentration used to induce ER stress (Yoshida et al., 1998).
To determine if the dependence of mLEEK upon the ERSE is via a direct interaction, chromatin immunoprecipitation (ChIP) experiments were conducted. mLEEK specifically associated with ERSE-containing portions of the GRP78 promoter, but not with regions of the GRP78 gene downstream of the transcriptional start site (Figure 3f). These data show that mLEEK physically interacts with the ERSE motif in the GRP78 promoter in vivo.
Overexpression of mLEEK does not induce the UPR nonspecifically and minimizes UPR induction upon ER stress
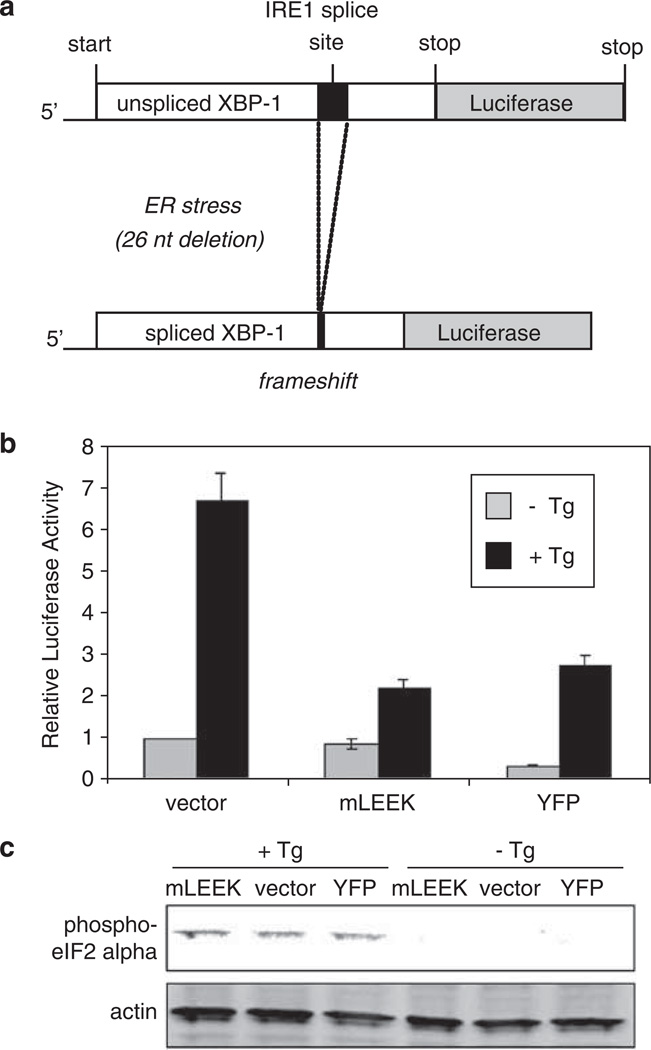
We also addressed the possibility that exogenous expression of mLEEK indirectly created ER stress. Cytomegalovirus promoter-driven expression of ER-localized proteins does not activate GRP78 or GRP94 transcription (Yoshida et al., 1998). Expression of YFP as a nonspecific control did not result in transcriptional induction (Supplementary Figure 4). To confirm that exogenous mLEEK expression does not generate ER stress nonspecifically, we utilized a construct in which the XBP1 gene is fused with luciferase (Feldman et al., 2005). Upon ER stress, IRE1 becomes activated and splices the transcript of XBP1 to produce the active form of this transcription factor, which in turn upregulates GRP94 and GRP78 (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2003). Luciferase activity is ordinarily undetectable in this system, but under ER stress, splicing of XBP1 mRNA removes a stop codon and results in a XBP1-luciferase chimera (Figure 4a). Using cells expressing this reporter, stimulation with thapsigargin led to a 7-fold increase in reporter activity, but overexpression of mLEEK did not induce luciferase expression even at high concentrations (Figure 4b). Interestingly, expression of mLEEK decreased the production of luciferase in response to thapsigargin treatment, suggesting that mLEEK could attenuate the effects of this ER stress-inducing agent (Figure 4b). We also assessed whether the PERK-activated arm of the UPR is aberrantly activated in response to mLEEK expression. Upon ER stress, PERK phosphorylates eIF2α (Harding et al., 1999). Accordingly, stimulation of cells with thapsigargin resulted in eIF2α phosphorylation. However, overexpression of mLEEK did not result in eIF2α phosphorylation (Figure 4c). These data show that exogenous expression of mLEEK does not trigger nonspecific activation of the UPR.
Figure 4.
Overexpression of mLEEK does not globally induce the UPR. (a) Schematic of the reporter system used to monitor XBP1 splicing. (b) HT1080 cells stably expressing the XBP1-luciferase reporter were transiently transfected with either pcDNA/HA, pcDNA/mLEEK-HA or pcDNA/YFP-HA as indicated. Cells were either treated with Tg or vehicle control. Luciferase activity in vehicle control transfected with vector was set at 1. Averages from three independent experiments are presented with s.d. values (bars). (c) HT1080 cells were transfected with the same plasmids used above and phosphorylation of eIF2α was monitored by western blotting of lysates. Cells were treated with Tg or vehicle control for 6 h.
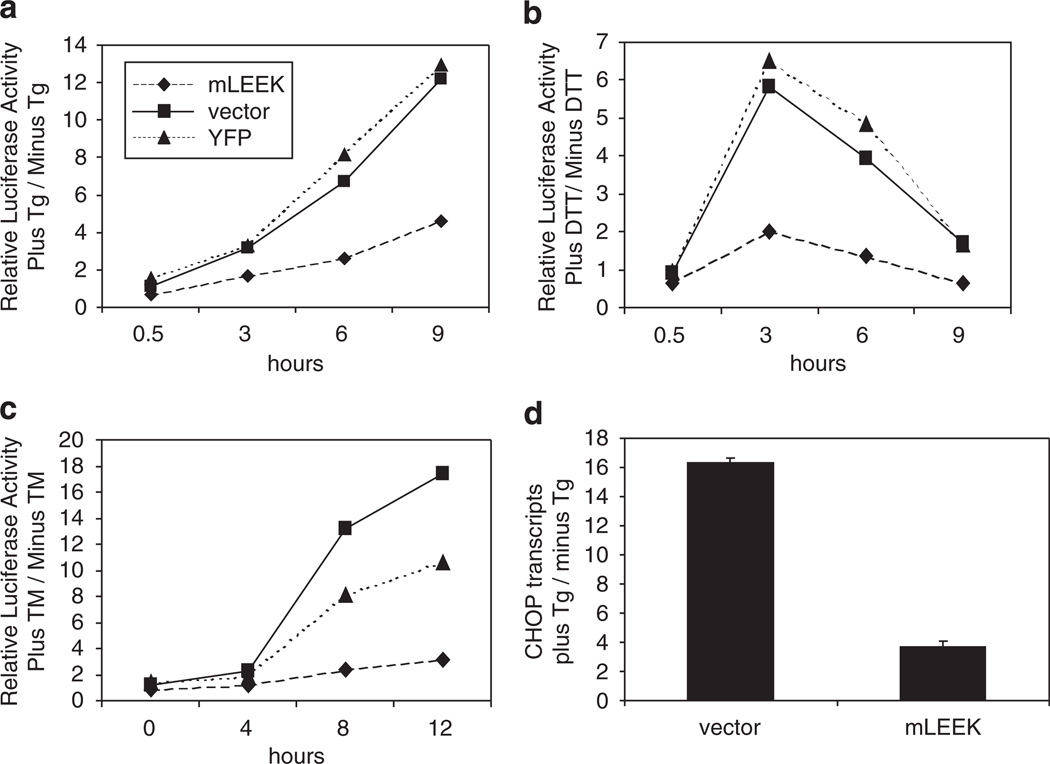
To expand upon the observation that mLEEK expression diminishes induction of the IRE1-mediated arm of the UPR, time-course experiments were performed utilizing XBP1-luciferase reporter cells. In response to thapsigargin-mediated ER stress, cells expressing mLEEK show lower induction of luciferase activity than control cells (Figure 5a). mLEEK has a similar protective effect when ER stress is induced by dithiothreitol or tunicamycin (Figures 5b and c). These results suggest that mLEEK interferes with XBP1 activation upon ER stress.
Figure 5.
Expression of mLEEK diminishes induction of the UPR. (a) HT1080/XBP1-luciferase cells were transiently transfected with pcDNA/HA, pcDNA/mLEEK-HA or pcDNA/YFP-HA. Cells were treated with Tg or vehicle control for the time points indicated before determination of luciferase activity. Averages from triplicate experiments were calculated and data from treated cells are normalized to untreated cells. (b) Same as (a), except that cells were treated with dithiothreitol or vehicle control. (c) Same as (a), except cells were treated with tunicamycin (Tm) or vehicle control. (d) HT1080 cells were transiently transfected with pcDNA/mLEEK-HA or pcDNA/HA. Cells were treated with Tg or vehicle control for 7 h before extraction of RNA. CHOP mRNA levels were measured by qRT–PCR and are reported as the ratio of plus Tg/− Tg for each data point.
To determine if mLEEK’s ability to minimize UPR induction upon ER stress extends to other arms of the UPR, we analyzed induction of CHOP transcription. CHOP transcription is under the control of the ATF4 transcription factor, which is selectively translated during ER stress as a result of PERK activation. Induction of CHOP transcription upon ER stress was reduced in cells exogenously expressing mLEEK (Figure 5d). This is consistent with a recent finding that GRP78 suppresses CHOP induction (Pyrko et al., 2007). These data show that expression of mLEEK minimizes UPR induction upon ER stress. This is likely because direct upregulation of molecular chaperones circumvents the need for UPR induction upon ER stress.
mLEEK knockdown results in caspase-mediated cell death and sensitization to ER stress
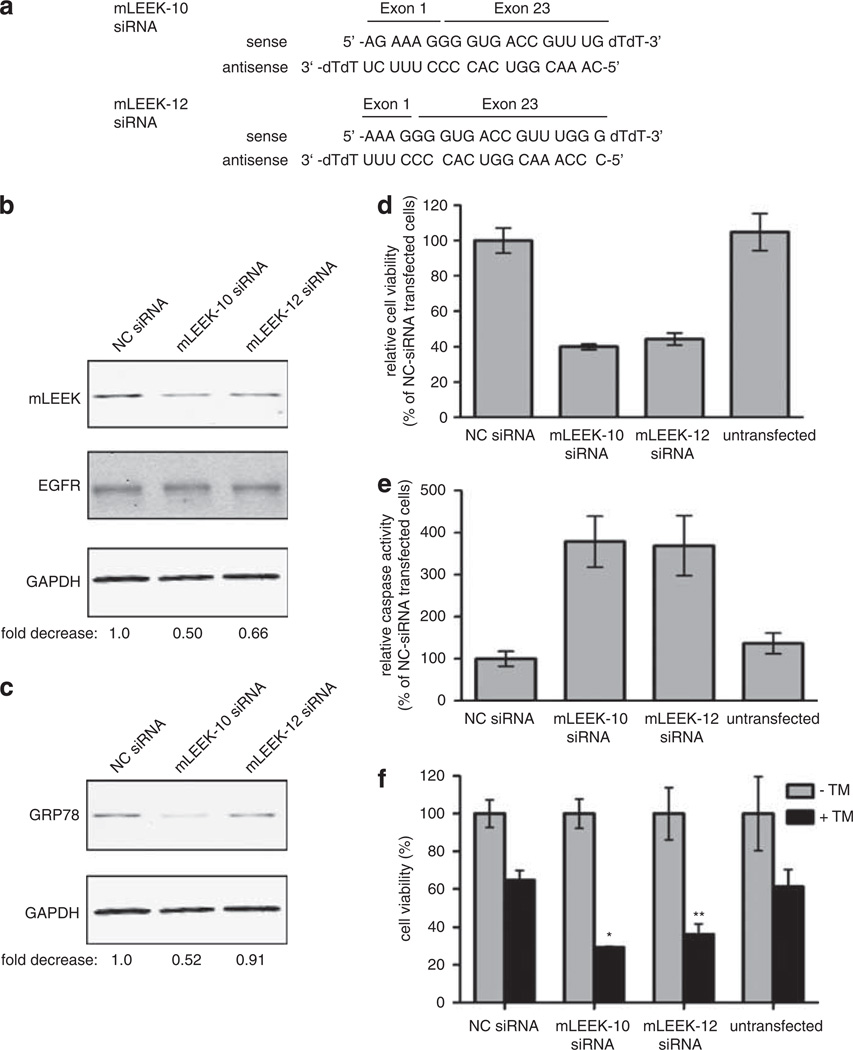
To study the physiological effects of mLEEK loss-of-function, we identified siRNAs that reduce mLEEK expression. To specifically target mLEEK without affecting EGFR levels, we designed siRNAs targeting the exon 1–23 junction (Supplementary Figure 5). After testing 13 siRNAs, two were found to specifically reduce mLEEK expression without altering EGFR levels (Figure 6a). mLEEK siRNA-10 consistently reduces mLEEK expression ~40%, while mLEEK siRNA-12 reduces it ~30% (Figure 6b). We studied the effect of mLEEK knockdown in HeLa cells, which express moderate amounts of endogenous mLEEK. Knockdown of mLEEK by siRNA resulted in reduced GRP78 expression, corroborating the findings associated with mLEEK overexpression (Figure 6c). Thus, GRP78 expression is dependent on mLEEK levels, as knockdown of mLEEK reduces GRP78 levels and exogenous introduction of mLEEK increases GRP78 levels.
Figure 6.
mLEEK is essential for cell viability and mLEEK knockdown leads to caspase-mediated cell death and sensitization to ER stress. (a) Sequence of mLEEK-10 and mLEEK-12 siRNA. (b) Western blot of HeLa cells transiently transfected with mLEEK-10, mLEEK-12 or non-targeting control (NC) siRNA. Cells were harvested at 36 h and lysates blotted with indicated antibodies. Westerns were performed using fluorescently labeled secondary antibodies and detected with the Odyssey infrared imager. The signals from mLEEK and GAPDH were quantified and the ratio of mLEEK/GAPDH was determined. These ratios were normalized relative to the NC siRNA control (designated 1.0) to obtain the fold decrease. (c) Membranes from blots shown above were reprobed with anti-GRP78 antibody and quantified as above. (d) Decrease in HeLa cell viability as a result of mLEEK siRNA transfection. Cell viability was determined 72 h post-transfection using the CellTiter-Blue assay. Averages from three experiments are presented with s.d. values (bars). (e) Induction of apoptosis in cells transfected with mLEEK siRNA. Caspase-3/7 activity was measured 3 days after transfection using the Caspase-Glo 3/7 Assay. Values represent the average percentage of caspase 3/7 activity relative to that of NC siRNA-transfected cells. Results are averages with s.d. of four independent experiments. (f) HeLa cells were transfected with mLEEK-10, mLEEK-12 or non-targeting controls siRNA (NC). The cells were treated with tunicamycin or vehicle control for 24 h. Cell viability was determined as above. *P<0.001; **P<0.005, significantly different from the viability in Tm-treated non-targeting siRNA-transfected cells.
A dramatic reduction in cell viability was the initial physiological observation seen with mLEEK knockdown (Figure 6d and Supplementary Figure 6). This is consistent with the increased cell death seen with GRP78 knockdown (Pyrko et al., 2007; Suzuki et al., 2007). We reasoned that mLEEK knockdown may involve caspase activation, as apoptosis resulting from GRP78 knockdown is caspase-mediated (Reddy et al., 2003; Suzuki et al., 2007). Consistently, we found that mLEEK knockdown resulted in ~3-fold induction of caspase-3/7 activity (Figure 6e). This finding, alongside data demonstrating an association of GRP78 with procaspase-7 that blocks caspase-7 activation, suggests a mechanism for mLEEK’s prosurvival function (Reddy et al., 2003). These data show that mLEEK knockdown increases cell death in a caspase-dependent manner.
Since mLEEK overexpression prevents induction of the UPR via upregulation of GRP78, we predicted that mLEEK knockdown and concomitant GRP78 reduction would sensitize cells to ER stress. HeLa cells transfected with siRNA were exposed to tunicamycin after siRNA transfection and cell viability was assayed. As expected, tunicamycin treatment resulted in a global reduction in cell viability of ~40% (Figure 6f). Interestingly, cells pretreated with mLEEK siRNA were further sensitized to ER stress and experienced increased cell death upon tunicamycin treatment (Figure 6f). A similar sensitivity to ER stress upon knockdown was also observed in HT1080 cells (Supplementary Figure 6). These data, in combination with mLEEK overexpression data, show that mLEEK is essential for the cell’s ability to adapt to ER stress and that tight regulation of mLEEK levels is crucial for modulation of the cellular stress response.
Discussion
The identification of mLEEK expands the repertoire by which the EGFR gene participates in signal transduction processes in cancer. The EGFR gene is amplified in a high percentage of glial tumors, and rearrangement of the gene often occurs along with amplification (Wong et al., 1987). The most common structural variant of the EGFR, EGFRvIII, contributes to oncogenesis through constitutive activation and altered signal transduction (Moscatello et al., 1996, 1998). The EGFRvIV variants utilize constitutive dimerization and increased basal activation as an oncogenic mechanism (Pines et al., 2010). Although structural alterations are often implicated as the source of these variants, the detection of EGFRvIII in tumors devoid of gene amplification or rearrangement suggests that alternative splicing also contributes to EGFRvIII formation (Moscatello et al., 1996; Pedersen et al., 2001a). We have identified mLEEK, a new EGFR variant that is produced in the absence of genomic rearrangement and amplification, implying that it also arises from alternative splicing. mLEEK is devoid of the canonical domains required for oncogenic transformation and directly participates in a transcription complex to upregulate gene programs controlling the UPR, thus revealing an unexpected role for EGFR in regulating gene expression in tumors. It would be of great interest to define the conditions and mechanisms under which the EGFR locus is capable of producing multiple transcripts with distinct signaling effects. A possible explanation is that the EGFR gene has a very large (> 100 kb) first intron and bioinformatic analyses as well as experimental evidence have shown that large introns promote alternative splicing (Haley et al., 1987; Roy et al., 2008; Kandul and Noor, 2009). The presence of a large first intron in the EGFR gene may explain the existence of multiple exon 1 splice variants, including mLEEK and EGFRvIII.
We have presented evidence showing that our EGFR variant mLEEK localizes to the nucleus. There is precedence for nuclear EGFR function and the discovery of mLEEK sheds light on some earlier observations. A large correlative study of nuclear EGFR expression in breast cancer demonstrated that the presence of EGFR in the nucleus may depend upon the antibody used for detection. Only antibodies against the C terminus were able to detect nuclear localization of EGFR in breast tumors, whereas antibodies against the N terminus failed to detect nuclear protein. Interestingly, the authors speculated that a truncated form of nuclear EGFR may exist in primary tumors and the discovery of mLEEK supports this hypothesis (Lo et al., 2005). Notably, nuclear EGFR has been detected with antibodies against the N terminus in head and neck squamous cell carcinoma, suggesting that full-length EGFR is found in the nucleus of other types of tumor tissues (Psyrri et al., 2005). An additional observation that warrants revisiting is that synthetic constructs have shown that the strongest transactivation domain is present in the C terminus (Lin et al., 2001). The discovery of mLEEK, a naturally occurring variant, provides an explanation for these two findings and suggests that the predominant nuclear form of EGFR is mLEEK.
Complex active transport mechanisms and excision from the lipid bilayer have been proposed for the translocation of full-length receptor to the nucleus, but mLEEK is small enough to passively diffuse into the nucleus and is not predicted to be membrane-bound (Lo et al., 2006; Liao and Carpenter, 2007). Small molecules may pass between the cytoplasmic and nuclear compartments by passive diffusion through the nuclear pore complex, but macromolecules greater than ~50 kDa require active transport mechanisms (Gorlich and Kutay, 1999; Allen et al., 2000; Macara, 2001; Stewart et al., 2001). Active transport of proteins through the nuclear pore complex usually requires interaction with transport receptors importins α/β and CRM1 and nuclear localization sequences present within proteins targeted for nuclear transport mediate complex formation with importins α/β (Gorlich and Kutay, 1999). Bioinformatic analysis does not predict a nuclear localization sequence within mLEEK. Moreover, treatment of cells with leptomycin B, which blocks nuclear export by targeting CRM1, did not alter levels of mLEEK in the nucleus (Supplementary Figure 3; Gorlich and Kutay, 1999). These data, in consideration with the 45 kDa size of mLEEK, suggest that mLEEK enters the nucleus through a passive diffusion mechanism. Intriguingly, conservation of the EGFR signal peptide along with deletion of the transmembrane domain suggests that mLEEK is secreted from cells. Preliminary data from our lab support this hypothesis and suggest a novel method of paracrine communication between cells (Supplementary Figure 7). This raises the question of whether nuclear localization arises from an intracellular event or nuclear translocation of mLEEK that has entered the cell by endocytosis. A potential mechanism involves the Sec61 translocon (Tsai et al., 2002). EGFR trafficking from the ER to the nucleus has been shown to be mediated by an association of EGFR with Sec61β, a component of the translocon (Liao and Carpenter, 2007; Wang et al., 2010). Since mLEEK is likely synthesized in the ER because it maintains a signal peptide, the Sec61 translocon could contribute to an intracellular mechanism of nuclear localization. Alternatively, nuclear mLEEK may arise from a mechanism involving endocytosis. These possibilities are not mutually exclusive and will require future experimentation to resolve.
The prevalence of mLEEK RNA and protein in cell lines and human tissues suggests that mLEEK represents an important part of EGFR biology, which has been overlooked previously. It is likely that mLEEK was not detected before this study because original experimental approaches to study the sequence of EGFR transcripts did not permit detection of a deletion encompassing such a large portion of the reading frame. The initial experiments that identified genomic variants of the EGFR or aberrant mRNAs utilized probes directed against the 5′ end of the gene (Wong et al., 1992). A set of experiments aimed at studying alterations at the 3′ end of the gene used methods directed specifically at this region (Eley et al., 1998). In our study, we optimized assays to specifically detect mLEEK transcript and protein. The cell’s dependence upon mLEEK expression for viability along with the prevalence of mLEEK expression suggests that mLEEK plays an essential role in cell physiology.
We have shown that mLEEK is overexpressed in human tumors and that overexpression of mLEEK causes induction of GRP78. Induction of GRP78 is known to promote cell survival upon ER stress and has been observed in many different tumor types (Lin et al., 2007; Moenner et al., 2007). Conversely, reducing mLEEK expression causes an increase in caspase-dependent apoptosis and sensitizes cells to ER stress via reduction in GRP78. Our data suggest an important implication for mLEEK in linking the UPR and cancer. Fluctuations in glucose and oxygen supplies to solid tumors create conditions of ER stress in tumor cells (Koumenis et al., 2007). GRP78 plays a prosurvival role, while CHOP is a crucial factor in the induction of the proapoptotic arm of the UPR (Oyadomari and Mori, 2004). We have shown that mLEEK induces the transcription of GRP78 and prevents the induction of CHOP in ER stress conditions. These data place mLEEK at the crux of the balance between expression of proapoptotic and prosurvival genes, which ultimately establish cellular fate during ER stress (Lin et al., 2007).
In addition, the identification of mLEEK has revealed a new means for controlling the expression of molecular chaperones important for the UPR and the protection of cells from ER stress. The current understanding is that ER-resident sensors, such as IRE1, respond to stress by initiating signaling pathways that increases transcription of chaperones. However, the upregulation of chaperones by mLEEK is extrinsic to any changes within the ER. An ER-independent mechanism by which cells can increase chaperone production has not been identified previously. Many investigators have suggested that manipulating the UPR would be of therapeutic benefit for a wide variety of diseases (Lawson et al., 1998; Boyce et al., 2005; Kim et al., 2005; Kudo et al., 2008; Mu et al., 2008). Thus far, research has centered on chemical approaches to controlling chaperones. In the case of cancer, it has been shown that inhibition of chaperones might be used to prolong survival and increase chemosensitivity (Jamora et al., 1996; Fu and Lee, 2006; Pyrko et al., 2007). The discovery of mLEEK suggests new avenues for developing drugs that act upon the UPR. Interference with mLEEK expression and function is a promising strategy to block production of chaperones and thus tumor growth and survival.
In summary, we have identified a novel EGFR variant that is overexpressed in tumors. mLEEK contributes to cell survival through upregulation of GRP78, which allows cells to survive in unfavorable conditions. Thus, inhibiting mLEEK expression and function is a promising strategy to control cell survival. Furthermore, the identification of mLEEK expands the known mechanisms by which the EGFR gene contributes to oncogenesis and sheds light on earlier findings related to nuclear EGFR function.
Materials and methods
RT–PCR
RNA was isolated from tumor samples and cell lines using Trizol reagent (Promega, Madison, WI, USA). First-strand synthesis and PCR amplifications were carried out using a One-Step RT–PCR system (Qiagen, Valencia, CA, USA). Primer sequences are available upon request. PCR products were excised from gels and identified by sequencing.
Ribonuclease protection assays
To generate an antisense RNA probe for mLEEK, a T7 promoter was introduced via PCR. Labeled probe was generated by in vitro transcription with biotinylated UTP (Enzo Biochem, New York, NY, USA). RNase protection experiments were conducted using the RPA III RNase Protection Assay Kit (Ambion, Austin, TX, USA). RNA was applied to a 5% TBE/urea gel (Biorad, Hercules, CA, USA) and transferred to a nylon membrane (Ambion). Biotinylated probe was detected using the Bright Star Biodetect kit (Ambion).
Antiserum preparation
Anti-mLEEK antiserum was raised by immunizing rabbits with a peptide corresponding to the mLEEK junction (LEEKKGVTVWELC), which contains an extra cysteine for conjugation to maleimide-activated keyhole-limpet hemocyanin (Pierce, Rockford, IL, USA). Injections and bleeds were performed by PickCell Laboratories (Amsterdam, The Netherlands). A crude serum was purified by affinity chromatography using the same peptide.
Cell culture and transfection
The cell lines U87MG, HEK293, HeLa S3 and A431 (from American Type Culture Collection (ATCC), Manassas, VA, USA) were cultured in Dulbecco’s modified essential medium supplemented with 10% fetal bovine serum and antibiotics. NIH3T3 cells (ATCC) were cultured in identical media, except for supplementation with calf serum. HT1080 cells (ATCC) and HT1080/XBP1-Luc (described previously; Feldman et al. (2005)) were grown in Eagle’s minimum essential medium (ATCC) supplemented with 10% fetal bovine serum and antibiotics. Where indicated, cells were incubated with thapsigargin (Tg), tunicamycin (Tm) or dithiothreitol from Sigma (St Louis, MO, USA). Cells were transfected using TransIT LT1 (Mirus, Madison, WI, USA). Stable expression of mLEEK was established in U87MG cells by retroviral infection as described previously (Xie et al., 1997). Stable expression of mLEEK in NIH3T3 was obtained using previously described methods (Moscatello et al., 1996).
Plasmids
mLEEK cDNA was engineered by blunt ligation of PCR products corresponding to exon 1 and exons 23–28, which encompass the start and stop codons for EGFR. PCR template was an expression plasmid containing the previously described full-length EGFR cDNA. Expand Long Template system (Roche, Branford, CT, USA) was used for PCR and a hemagglutinin epitope was introduced at the C terminus. mLEEK PCR product was cloned into the pMSCV plasmid (Clontech, Mountain View, CA, USA) and subcloned into the pcDNA1.1 vector (Invitrogen, Carlsbad, CA, USA). Luciferase reporter constructs containing the GRP78 promoter and mutations to the ERSE motifs were the kind gift of K Mori (Yoshida et al., 1998). A luciferase reporter containing the HERP promoter was the kind gift of K Kokame (Kokame et al., 2001). The following luciferase reporter constructs were from Switchgear Genomics (Menlo Park, CA, USA): HSP90B1, PPIA, HSPA5, CCND1 and MYBL2.
Protein analysis and immunoprecipitations
Protein extracts from cells were harvested and immunoblotted as described previously (Holgado-Madruga and Wong, 2003). The following antibodies were used: KDEL (Abcam, Cambridge, MA, USA), actin (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA, USA), mouse immunoglobulin G, rat immunoglobulin G (Sigma), hemagglutinin ascites (Covance Inc., Princeton, NJ, USA) and hemagglutinin high-affinity antibody used for immunoprecipitations (IP) (Roche). Infrared signals were quantified using Odyssey IR imaging system (Licor Biosciences, Lincoln, NE, USA). For IP, cells were lysed in lysis buffer (50 mm HEPES (pH 7.5), 0.5% NP-40, 150 mm sodium chloride with the addition of one protease inhibitor mixture tablet (Roche)/10 ml lysis buffer immediately before use) and cell lysates were cleared by centrifugation. Protein extract from each sample was incubated with protein G-Sepharose (GE Scientific, Piscataway, NJ, USA) for 1 h at 4 °C. Beads were pelleted and precleared supernatant was used for IP. Precleared cell lysates were combined with antibody pre-bound to protein G-Sepharose overnight at 4 °C with rotation. IPs were washed and released from beads as described elsewhere (Li et al., 2000).
QRT–PCR analysis
RNA was isolated from cell lines using Trizol. The reverse transcription and quantitative PCR were carried out using the Brilliant II Sybr Green QRT–PCR Master Mix Kit (Stratagene, La Jolla, CA, USA). Primers to human CHOP mRNA and cycling conditions have been described (Lin et al., 2007). All samples were normalized to Rpl19.
Luciferase assay
Using TransIT LT1 (Mirus), HT1080 cells were transiently co-transfected with cDNA expression plasmid and reporter plasmids. The luciferase activity was measured using the Steady-Glo Luciferase Assay system (Promega). For induction of UPR, cells were treated with 300 nm Tg for 6 h before harvesting, unless otherwise indicated. For experiments using the HT1080/XBP1-Luc reporter cell line, cells were transiently transfected with plasmid 24 h before induction of ER stress with 300 nm Tg, 2 µg/ml Tm or 1 mm dithiothreitol for the time points indicated.
Immunofluorescence and peptide competition
Cells were grown on glass coverslips, fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS) solution and permeabilized with 0.1% Triton X-100/PBS. Coverslips were blocked with 5% milk/PBS and incubated with mLEEK antisera in 5% milk/PBS for 1 h, rinsed in PBS and incubated with fluorescein isothiocyanate-conjugated secondary antibody (Jackson Immunoresearch, West Grove, PA, USA). Coverslips were rinsed in PBS and mounted onto microscope slides. Images were taken using Leica SP2 AOBS confocal microscope. Images were collected using equal exposure times and processed similarly. For peptide competition experiments, mLEEK antiserum was preincubated with a ~30-fold molar excess of free peptide at 4 °C overnight. Samples were centrifuged for 15 min at 4 °C to pellet any immune complexes and the supernatant was used for immunolocalization studies.
Immunohistochemistry
Tissue microarray slides containing cores from human tissue were acquired from Imgenex (San Diego, CA, USA) and Fox Chase Cancer Center (Philadelphia, PA, USA). Slides were preblocked with Background Sniper blocking reagent (Biocare Medical, Concord, CA, USA) for 15 min at room temperature, and incubated with affinity-purified mLEEK antisera (1:10 000) in Da Vinci Green Diluent (Biocare Medical) at 4°C overnight. Slides were washed with PBS and incubated with Mach 2 Rabbit HRP polymer (Biocare Medical) at room temperature for 40 min. After washing with PBS, cores were incubated with Betazoid DAB chromogen (Biocare Medical). Slides were washed with distilled water and counterstained with hematoxylin.
Microarray
cDNA microarray analysis for gene expression was carried out at the Stanford Functional Genomics Facility. HEK293 cells were transfected with pcDNA/mLEEK-HA or pcDNA/HA and RNA was harvested using Trizol. cDNA was prepared from total RNA using either Cy3 or Cy5 labeling and hybridized to an oligonucleotide array containing 44 544 70-mer probes constructed by the facility (the Human Exonic Evidence Based Oligonucleotide array). Scanning and analysis was carried out according to Stanford Functional Genomics Facility protocols. Scanned image files were visually inspected for artifacts and normalized using the GenePix software (Axon Instruments, Foster City, CA, USA). Expression data were analyzed using data analysis tools available at the Stanford Microarray Database (Demeter et al., 2007). Comparisons were made for each mLEEK-transfected cell line using expression levels from control vector-transfected cells as the reference. The fold change values, indicating the relative change in expression levels between mLEEK and control transfected cells, were used to identify genes differentially expressed.
Chromatin immunoprecipitation
U87/mLEEK-HA or U87/vector cells were used for ChIP assays carried out using the ChIP-IT assay kit (Active Motif, Carlsbad, CA, USA). DNA from the input and immunoprecipitated samples was subjected to PCR or quantitative PCR. Immunoprecipitated or 100-fold diluted input DNA was prepared with RT2 Real-Time SYBR green/ROX quantitative PCR Master Mix (Qiagen). GRP78 ERSE and GRP78 + 5 kb primer sets have been described (Cai et al., 2007).
Cell fractionation
Cells were lysed in a lysis buffer (50 mm HEPES (pH 7.9), 0.5% NP-40, 0.5 mm dithiothreitol, 10 µg aprotinin perml, 10µg leupeptin per ml, 100 µg phenylmethanesulfonyl fluoride per ml) and incubated for 20 min at 4 °C. Lysates were centrifuged at 800 g for 10 min at 4 °C and supernatant formed the cyto-plasmic fraction. The nuclear pellet was washed four times and resuspended in the same buffer containing 0.4 m NaCl, 5 mm EDTA and 25% glycerol. Samples were incubated for 30 min at 4 °C, extracted material was sedimented at 16 000 g for 20 min at 4 °C and resulting supernatant was termed the nuclear fraction.
siRNA transfection
Custom-designed siRNAs targeting mLEEK were obtained from Qiagen. Non-targeting control siRNAs were purchased from Dharmacon (Lafayette, CO, USA). Sequences are shown in Supplementary Figure 5. Cells were transfected using the DharmaFECT1 transfection reagent (Dharmacon).
Cell viability assay
Cell viability was assessed with the CellTiter-Blue Cell Viability Assay (Promega). Fluorescence was measured at 565/595 nm.
Caspase activity assay
Caspase-3 and -7 activities were measured in siRNA-transfected cells using the Caspase-Glo 3/7 Assay kit (Promega).
Statistical analysis
Where indicated, statistical significance was determined using the unpaired Student’s t-test.
Supplementary Material
Acknowledgements
We thank R Nitta, G Li, S Mitra, C Del Vecchio and M Holgado-Madruga for comments. We thank K Mori, K Kokame, R Prywes and G Nolan for reagents. This work was supported by the NIH under Ruth L Kirschstein National Research Service Award F31 NS056581 from the NINDS to ECP, the Ovarian SPORE at FCCC (P50 CA083638) to AKG and AJW, NIH Grants R01 CA69495, R01 CA096539 and R01 CA124832, as well as a research grant from the National Brain Tumor Foundation and Grant 15IB-0080 from the California Breast Cancer Program to AJW.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113(Part 10):1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chang SC, Erwin AE, Lee AS. Glucose-regulated protein (GRP94 and GRP78) genes share common regulatory domains and are coordinately regulated by common trans-acting factors. Mol Cell Biol. 1989;9:2153–2162. doi: 10.1128/mcb.9.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;5:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley G, Frederick L, Wang XY, Smith DI, James CD. 3′ end structure and rearrangements of EGFR in glioblastomas. Genes Chromosomes Cancer. 1998;23:248–254. doi: 10.1002/(sici)1098-2264(199811)23:3<248::aid-gcc7>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Chauhan V, Koong AC. The unfolded protein response: a novel component of the hypoxic stress response in tumors. Mol Cancer Res. 2005;3:597–605. doi: 10.1158/1541-7786.MCR-05-0221. [DOI] [PubMed] [Google Scholar]
- Fu Y, Lee AS. Glucose regulated proteins in cancer progression, drug resistance and immunotherapy. Cancer Biol Ther. 2006;5:741–744. doi: 10.4161/cbt.5.7.2970. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Haley J, Whittle N, Bennet P, Kinchington D, Ullrich A, Waterfield M. The human EGF receptor gene: structure of the 110 kb locus and identification of sequences regulating its transcription. Oncogene Res. 1987;1:375–396. [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Holgado-Madruga M, Wong AJ. Gab1 is an integrator of cell death versus cell survival signals in oxidative stress. Mol Cell Biol. 2003;23:4471–4484. doi: 10.1128/MCB.23.13.4471-4484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, Dennert G, Lee AS. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. Proc Natl Acad Sci USA. 1996;93:7690–7694. doi: 10.1073/pnas.93.15.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul NP, Noor MA. Large introns in relation to alternative splicing and gene evolution: a case study of Drosophila bruno-3. BMC Genet. 2009;10:67. doi: 10.1186/1471-2156-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Adam RM, Freeman MR. Trafficking of nuclear heparin-binding epidermal growth factor-like growth factor into an epidermal growth factor receptor-dependent autocrine loop in response to oxidative stress. Cancer Res. 2005;65:8242–8249. doi: 10.1158/0008-5472.CAN-05-0942. [DOI] [PubMed] [Google Scholar]
- Kokame K, Kato H, Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J Biol Chem. 2001;276:9199–9205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Bi M, Ye J, Feldman D, Koong AC. Hypoxia and the unfolded protein response. Methods Enzymol. 2007;435:275–293. doi: 10.1016/S0076-6879(07)35014-3. [DOI] [PubMed] [Google Scholar]
- Kudo T, Kanemoto S, Hara H, Morimoto N, Morihara T, Kimura R, et al. A molecular chaperone inducer protects neurons from ER stress. Cell Death Differ. 2008;15:364–375. doi: 10.1038/sj.cdd.4402276. [DOI] [PubMed] [Google Scholar]
- Lawson B, Brewer JW, Hendershot LM. Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. J Cell Physiol. 1998;174:170–178. doi: 10.1002/(SICI)1097-4652(199802)174:2<170::AID-JCP4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, et al. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Razon N, Bartal AD, Yarden Y, Schlessinger J, Soreq H. Expression of epidermal growth factor receptors in human brain tumors. Cancer Res. 1984;44:753–760. [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Liu ES, Lee AS. Common sets of nuclear factors binding to the conserved promoter sequence motif of two coordinately regulated ER protein genes, GRP78 and GRP94. Nucleic Acids Res. 1991;19:5425–5431. doi: 10.1093/nar/19.19.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear–cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malden LT, Novak U, Kaye AH, Burgess AW. Selective amplification of the cytoplasmic domain of the epidermal growth factor receptor gene in glioblastoma multiforme. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Montgomery RB, Sundareshan P, McDanel H, Wong MY, Wong AJ. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, III, Segatori L, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. Gamma-secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Parker R, Phan T, Baumeister P, Roy B, Cheriyath V, Roy AL, et al. Identification of TFII-I as the endoplasmic reticulum stress response element binding factor ERSF: its autoregulation by stress and interaction with ATF6. Mol Cell Biol. 2001;21:3220–3233. doi: 10.1128/MCB.21.9.3220-3233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001a;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Pedersen N, Damstrup L, Villingshoj M, Sonder SU, Rieneck K, et al. Analysis of the epidermal growth factor receptor specific transcriptome: effect of receptor expression level and an activating mutation. J Cell Biochem. 2005;96:412–427. doi: 10.1002/jcb.20554. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Thykjaer T, Orntoft TF, Damstrup L, Poulsen HS. Profile of differentially expressed genes mediated by the type III epidermal growth factor receptor mutation expressed in a small-cell lung cancer cell line. Br J Cancer. 2001b;85:1211–1218. doi: 10.1054/bjoc.2001.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines G, Huang PH, Zwang Y, White FM, Yarden Y. EGFRvIV: a previously uncharacterized oncogenic mutant reveals a kinase autoinhibitory mechanism. Oncogene. 2010;29:5850–5860. doi: 10.1038/onc.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Roy M, Kim N, Xing Y, Lee C. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. RNA. 2008;14:2261–2273. doi: 10.1261/rna.1024908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Baker RP, Bayliss R, Clayton L, Grant RP, Littlewood T, et al. Molecular mechanism of translocation through nuclear pore complexes during nuclear protein import. FEBS Lett. 2001;498:145–149. doi: 10.1016/s0014-5793(01)02489-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Lu J, Zahed M, Kita K, Suzuki N. Reduction of GRP78 expression with siRNA activates unfolded protein response leading to apoptosis in HeLa cells. Arch Biochem Biophys. 2007;468:1–14. doi: 10.1016/j.abb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Thastrup O, Cullen PJ, Drobak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci USA. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Wang YN, Yamaguchi H, Huo L, Du Y, Lee HJ, Lee HH, et al. The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc NatlAcad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Xie W, Su K, Wang D, Paterson AJ, Kudlow JE. MDA468 growth inhibition by EGF is associated with the induction of the cyclin-dependent kinase inhibitor p21WAF1. Anticancer Res. 1997;17:2627–2633. [PubMed] [Google Scholar]
- Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, et al. Amplification of the structurally and functionally altered epidermal growth factor receptor gene (c-erbB) in human brain tumors. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.