Abstract
BACKGROUND
It is becoming increasingly evident that microRNAs (miRNA) are associated with the development and progression of prostate cancer (PCa).
METHODS
We examined the hypothesis that plasma miRNA levels can differentiate patients by aggressiveness in 82 PCa patients. Taqman based quantitative RT-PCR assays were performed to measure copy number of target miRNAs.
RESULTS
miR-20a was signficantly overexpressed in plasma from patients with stage 3 tumors compared to stage 2 or below (p=0.03). The expression levels for miR-20a and miR-21 were significantly increased in patients with high risk CAPRA scores (16,623 and 1,595 copies, respectively). Significantly increased miR-21 and miR-145 expression were observed for patients with intermediate or high risk D’Amico scores compared to patients with low risk scores (p=0.047 and 0.011, respectively). The relapse rates for CAPRA scores ranged from 1.9% for low risk to 9.5% for intermediate risk and to 22.2% for high risk patients (p=0.023). For D’Amico scores, the relapse rates ranged from 0.0% for low risk to 7.4% for intermediate risk and 17.6% for high risk patients (p=0.039). Expression of miR-21 and miR-221 significantly differentiated patients with intermediate risk from those with low risk CAPRA scores (AUC=0.801, p=0.002). Four miRNAs (miR-20a, miR-21, miR-145 and miR-221) could also distinguish high vs. low risk in PCa patients by D’Amico score with an AUC of 0.824.
CONCLUSIONS
These preliminary data suggest that altered plasma miRNAs may be useful predictors to distinguish PCa patients with varied aggressiveness. Further larger studies to validate this promising finding are warranted.
Keywords: miRNAs, Prostate Cancer, Aggressiveness, Prediction
INTRODUCTION
Clinically aggressive prostate cancer (PCa) has significant biological characteristics distinguishing it from indolent disease. Aggressive tumors are more likely to have increased invasive, migratory, colonization and angiogenic abilities. Accelerated cell cycle, resistance to apoptosis and androgen independent growth are also frequently observed in advanced tumors [1,2]. Using traditional clinical tools such as PSA level, TNM stage and Gleason score, the clinician is unable to withhold treatment for PCa patients who may not benefit from it because these parameters lack sufficient sensitivity and specificity to discriminate indolent from potentially lethal cancers. Approximately 14 out of 15 patients who currently receive therapy for PCa do not experience an alteration in their survival by undergoing therapy [3]. Therefore, exploration of novel biomarkers to early identify invasive PCa cases is needed.
microRNAs (miRNAs) are involved in crucial biological processes in the development of cancer, including cell differentiation, apoptosis and proliferation [4], through imperfect pairing with target messenger RNAs (mRNAs) of protein-coding genes and the transcriptional or post-transcriptional regulation of expression [5]. Each miRNA may bind to as many as 200 gene targets, and each gene may contain multiple binding sites for different miRNAs [6]. Therefore, altered miRNA expression may contribute to the development and progression of cancer, and provide additional diagnostic information for aggressive cases.
Stable miRNAs have been detected in biological fluids, including serum and plasma, and may be associated with cancer diagnosis and prognosis [7–11], but only a few studies have been conducted on PCa. Serum miR-141 was found to distinguish advanced PCa patients from healthy controls [10]. A large-scale study investigating 667 different miRNAs in PCa patients showed that miRNA-375 and miRNA-141 were associated with advanced PCa [12]. Serum miR-21 expression was elevated in metastatic hormone refractory PCa patients [13] indicating its potential role in prognosis. Deregulated miR-20a [14] and miR-221 [15] expression in tumor tissues has been associated with Gleason score and clinical recurrence of PCa. The expression of miR-100, miR-145, miR-191 and miR-let7c in tumor tissues were also correlated with the biochemical recurrence rates of PCa [16]. These encouraging results suggested that plasma miRNA may be a valuable resource to explore noninvasive biomarkers for aggressive PCa. In the current study, we selected miR-20a, miR-21, miR-141, miR-145 and miR-221 for analysis because of this prior data and their involvement in multiple cancer metastatic pathways. We tested the hypothesis that deregulation of circulating miRNAs are promising biomarkers to distinguish high risk aggressive PCa patients from localized cases.
MATERIALS AND METHODS
Patients and Sample Collection
This study was approved by the Institutional Review Board of Columbia University Medical Center (CUMC). Eligible PCa patients were ascertained from men attending urology clinics at CUMC during the period of December 2008 to July 2010. The recruitment criteria were men diagnosed with primary, incident, and histologically confirmed PCa who underwent radical prostatectomy at CUMC. Cases were excluded from the current study if they had a prior history of cancer, previous prostate surgery, a recent severe weight loss, severe diseases (uncontrolled cardiovascular diseases, hepatic or renal dysfunction), on hormonal treatment or taking finasteride. A total of 82 PCa patients who provided blood samples prior to any treatment were available for the current study. Plasma samples were stored in the Herbert Irving Comprehensive Cancer Center (HICCC) Biomarkers Core. All cases had tumors with clinical stage II or III, no clinical evidence of lymph node or distant metastases, and with complete clinical and serum PSA data. Patients were routinely followed up every three months to measure serum PSA and obtain biochemical relapse data.
The aggressiveness of PCa cases were categorized by the Cancer of the Prostate Risk Assessment (CAPRA) score [17] and D’Amico’s criteria [18,19]. CAPRA score is based on the patient’s age, PSA level, tumor stage, Gleason score, and percentage of biopsy cores positive for cancer at diagnosis. Risk of aggressiveness was categorized as high, intermediate or low when CAPRA scores were 6–10, 3–5 and 0–2, respectively [17]. D’Amico score defines low risk as clinical stage T1c or T2a, a PSA level of 10ng/ml or less, and a Gleason sum of ≤ 6; intermediate risk as either clinical stage T2b, a PSA level >10ng/ml but <20ng/ml, or a Gleason sum of 7; and high risk as clinical stage T2c or greater, PSA level >20ng/ml, or a Gleason sum of ≥8 [18,19].
Epidemiological and Clinicopathological Data Collection
A standardized questionnaire was used to collect information on demographic and socioeconomic factors, self-reported personal medical history, environmental exposures (smoking, unusual work, radiation, heavy metals, and infectious exposures), lifestyle factors (alcohol consumption, dietary type, vitamin and nutritional supplements intake), physical activity, and family history of prostate and other cancers. Clinicopathological data for patients were obtained from the CAISIS, IRB approved clinical outcomes research data base in Dept. of Urology.
RNA Extraction and MiRNAs Quantification
Total RNA, including miRNAs were isolated from 250μl of plasma using the miRNeasy Mini kit (Qiagen, Frederick, MA) according to the manufacturer’s protocol. Taqman based quantitative RT-PCR assays were performed according to manufacturer’s recommended protocol (ABI, Foster City, CA) with minor modifications. Chemically synthesized RNA oligonucleotides (Invitrogen, Carlsbad, CA) corresponding to the target miRNAs were used to generate standard curves (serially diluted and expressed as copy numbers per μl). The standard curves ranged from 3 to 3×104 copies or from 30 to 3×104 copies. A standard curve for miR-145 is shown in Supplemental Fig. 1. After reverse transcription (10ng RNA per reaction), the real-time PCR was performed by adding TaqMan miRNA-specific primers and fluorescence-labeled probe. The PCR mixtures contained 1.33 μl RT products and Universal PCR Master Mix in a total volume of 20μl; reaction was carried out on an Applied BioSystems 7500 Sequence Detection System. Cycling variables were 95°C for 10 min followed by 40 cycles at 95°C for 15s and annealing/extension at 60°C for 1 min. All reactions were performed in triplicate. Data were analyzed by SDS Software (ABI, Foster City, CA) with the automatic Ct setting for assigning baseline and threshold for Ct determination. The reproducibility of duplicate assays was excellent for all four assays (Supplemental Fig. 2) with an overall R2=0.96 (p<0.01). The averages of inter- and intra- assay CVs were 1.45% and 0.97%, respectively. Supplemental Fig. 3 presents inter- and intra-assay CVs which range from 0.01% to 7.2%, indicating the high accuracy and reliability of the assay.
Statistical Analysis
miRNA expression data were assessed for the distribution of normality using Kolmogrov-Smirov test [20]. Since miRNA levels were skewed, log-transformation was used to more closely approximate normality in the final analysis. miRNAs levels were reported as the mean ± standard deviation (SD). A two-sample t test assuming equal variance was carried out to compare miRNA levels by covariates of demographic and clinic-pathological characteristics, including age at diagnosis (<60 vs. ≥60yrs), follow-up time (<19.8 vs. ≥19.8 months), family history of PCa (no vs. yes), prostate volume (<50 vs. ≥50ml), presurgery PSA level (≤6 vs. >6 ng/ml), tumor stage (T2a-T2c vs. ≥T3) and relapse status (no vs. yes). One-way ANOVA was used to compare miRNAs levels by CAPRA or D’Amico scores (high vs. intermediate vs. low risk), covariates of ethnicity (Caucasian vs. African-American vs. others) and Gleason sum score (6 vs. 7 vs. 8–9).
Logistic regression was used to construct receiver operating characteristic (ROC) curves using expression level of miRNA adjusted for age and ethnicity [21]. The maximum sensitivity and specificity and the area under the curve (AUC) were identified by numerical integration of each ROC curve. A prediction model was built by fitting all four miRNAs into the logistic regression model, and the stepwise backward model selection was performed to determine the best combinations of miRNAs to significantly discriminate PCa patients with varied risk of aggressiveness. A likelihood ratio test p value <0.05 was considered statistically significant. All statistical analyses were completed using Statistical Analysis System 9.0 (SAS Institute, Cary, NC).
RESULTS
The demographic and clinicopatholgic characteristics of the 82 PCa patients are shown in Table I. The mean age of patients was 61 years and over 56% were Caucasian. The average PSA level and mean prostate volume were 6.3ng/ml (SD=6.0) and 53.0 ml (SD=15.8), respectively. Most cases had tumors at stage 2 or below (76.8%), lymph node status of Nx or N0 (98.8%) and Gleason score 6 or 7 (76.8%). All patients were at M0 status because those with metastasis did not undergo surgery. Due to short follow-up time (mean=20.2 months), only 5 relapse cases were observed.
TABLE I.
Demographic and clinic-pathological characteristics of 82 prostate cancer patients in the current study
| Variables | No of patients (%) |
|---|---|
| Age at diagnosis | |
| Mean ± SD (yr) | 61.2 ± 6.3 |
| < median (60) | 35 (42.7) |
| ≥ median (60) | 47 (57.3) |
| Ethnicity | |
| Caucasian | 46 (56.1) |
| African-American | 10 (12.2) |
| Others | 26 (31.7) |
| Family history of prostate cancer | |
| No | 59 (72.0) |
| Yes | 20 (24.4) |
| Missing | 3 (3.6) |
| Prostate volume (ml) | |
| Mean ± SD | 53.0 (15.8) |
| < median (50) | 35 (42.7) |
| ≥ median (50) | 36 (43.9) |
| Missing | 11 (13.4) |
| PSA levels (ng/ml) | |
| Mean ± SD | 6.3 (6.0) |
| < 6 | 51 (62.2) |
| ≥ 6 | 24 (29.3) |
| Missing | 7 (8.5) |
| Tumor stage | |
| T2a | 5 (6.1) |
| T2b or T2c | 58 (70.7) |
| ≥ T3 | 19 (23.2) |
| Lymph node status | |
| Nx | 3 (3.7) |
| N0 | 78 (95.1) |
| N1 | 1 (1.2) |
| Metastasis status | |
| M0 | 82 (100.0) |
| Gleason sum score | |
| 6 | 24 (29.3) |
| 7 | 39 (47.5) |
| 8–9 | 19 (23.2) |
| Follow-up time | |
| Mean ± SD (months) | 18.3 ± 10.2 |
| < median (19.8) | 38 (46.3) |
| ≥ median (19.8) | 38 (46.3) |
| Missing | 6 (7.4) |
| Relapse status | |
| No | 71 (86.6) |
| Yes | 5 (6.1) |
| Missing | 6 (7.3) |
|
| |
Overall plasma miRNAs expression levels (absolute copy numbers/μl) were higher for miR-20a (mean=4,833, range from 194 to 128,583), miR-21 (mean=670, range from 33 to 8,253) and miR-221 (mean=770, range from 66 to 4,325) compared with miR-145 (mean=112, range from 13 to 644). Because plasma miR-141 expression was undetectable for all patients, no further analysis was conducted. When comparing miRNA copy numbers by clinicopatholgic characteristics (Table II), increased trends of miRNAs levels were almost always observed for PCa patients with prostate volumes of 50 ml or higher, PSA level 6 ng/ml or higher, tumor stage 3 or above, and Gleason score 7 or higher but were not statistically significant. miR-20a was statistically signficantly overexpressed in tumor stage 3 with a mean of 11,215 copies/μl compared to stage 2 or below (mean=2,908 copies/μl, p=0.03). However, given the number of comparisons this may be due to chance. No significant difference for miRNAs expression was observed between relapse-free and relapsed patients, probably due to the small sample size of relapsed cases.
TABLE II.
miRNAs expression levels (copy number/μl) by demographic and clinic-pathological characteristics
| Variables | N | miR-20a | miR-21 | miR-145 | miR-221 |
|---|---|---|---|---|---|
| Age at diagnosis | |||||
| < median (60 yr) | 35 | 7,324 | 830 | 117 | 798 |
| ≥ median (60 yr) | 47 | 2,978 | 551 | 108 | 748 |
| p value | 0.20 | 0.35 | 0.80 | 0.78 | |
| Ethnicity | |||||
| Caucasian | 46 | 5,175 | 501 | 95 | 687 |
| African-American | 10 | 2,368 | 465 | 90 | 726 |
| Others | 26 | 5,174 | 1,049 | 153 | 945 |
| p value | 0.86 | 0.21 | 0.30 | 0.44 | |
| Family history of prostate cancer | |||||
| No | 59 | 3,784 | 630 | 118 | 824 |
| Yes | 20 | 2,023 | 445 | 65 | 538 |
| p value | 0.27 | 0.49 | 0.17 | 0.18 | |
| Prostate volume | |||||
| < median (50) | 35 | 3,735 | 636 | 123 | 685 |
| ≥ median 50 | 36 | 6,806 | 829 | 128 | 868 |
| p value | 0.43 | 0.57 | 0.90 | 0.39 | |
| PSA levels (ng/ml) | |||||
| < 6 | 51 | 3,850 | 689 | 109 | 709 |
| ≥ 6 | 24 | 7,630 | 748 | 125 | 955 |
| p value | 0.34 | 0.87 | 0.70 | 0.24 | |
| Tumor stage | |||||
| T2a-T2c | 63 | 2,908 | 529 | 103 | 812 |
| ≥ T3 | 19 | 11,215 | 1,140 | 145 | 626 |
| p value | 0.03 | 0.08 | 0.34 | 0.39 | |
| Gleason sum score | |||||
| 6 | 24 | 2,541 | 379 | 63 | 705 |
| 7 | 39 | 6,311 | 790 | 132 | 818 |
| 8–9 | 19 | 4,692 | 792 | 126 | 739 |
| p value | 0.63 | 0.45 | 0.21 | 0.86 | |
| Follow-up time (months) | |||||
| < median (19.8) | 38 | 6,141 | 599 | 106 | 817 |
| ≥ median (19.8) | 38 | 2,532 | 495 | 107 | 686 |
| p value | 0.29 | 0.67 | 0.99 | 0.50 | |
| Relapse status | |||||
| No | 71 | 4,477 | 565 | 108 | 741 |
| Yes | 5 | 2,348 | 295 | 83 | 901 |
| p value | 0.76 | 0.58 | 0.76 | 0.70 | |
|
| |||||
We compared plasma expression of the four candidate miRNAs using CAPRA and D’Amico scores to categorize PCa patients as high, intermediate or low risk of aggressiveness (Table III). Overall, expression levels of miR-20a and miR-21 were significantly increased for patients with high risk CAPRA score (16,623 and 1,595 copies, respectively) compared to patients with intermediate (4,723 and 818 copies) or low risk scores (2,836 and 450 copies). No significant associations were obtained for miR-145 and miR-221 expression and CAPRA score. The relapse rates consistently increased with CARPA scores from 1.9% for low risk to 9.5% for intermediate risk and 22.2% for high risk patients (p=0.023). A similar trend of high D’Amico score and increased relapse rate was observed from 0.0% for low risk to 7.4% for intermediate risk and 17.6% for high risk patients (p=0.039). But these results are based on only 5 relapsed cases. Significantly increased miR-21 and miR-145 expression were observed for patients with intermediate or high risk D’Amico scores compared to patients with low risk score (p=0.047 and 0.011, respectively). No significant associations were obtained for miR-20a and miR-221 expression and D’Amico scores.
TABLE III.
miRNAs expression levels (copy number/μl) by CAPRA (Cancer of the Prostate Risk Assessment) and D’Amico scores
| CAPRA Score | PCa cases N |
Relapse N (%) |
miR-20a | miR-21 | miR-145 | miR-221 |
|---|---|---|---|---|---|---|
| Low risk | 52 | 1 (1.9) | 2,836 | 450 | 101 | 772 |
| Intermediate risk | 21 | 2 (9.5) | 4,723 | 818 | 119 | 683 |
| High risk | 9 | 2 (22.2) | 16,623 | 1,595 | 179 | 956 |
|
| ||||||
| p | 0.023 | 0.038 | 0.046 | 0.48 | 0.72 | |
|
| ||||||
| D’Amico Score | ||||||
|
| ||||||
| Low risk | 38 | 0 (0.0) | 2,323 | 375 | 75 | 670 |
|
| ||||||
| Intermediate risk | 27 | 2 (7.4) | 10,217 | 1,177 | 186 | 972 |
|
| ||||||
| High risk | 17 | 3 (17.6) | 1,892 | 525 | 89 | 686 |
|
| ||||||
| p | 0.039 | 0.08 | 0.047 | 0.011 | 0.31 | |
|
| ||||||
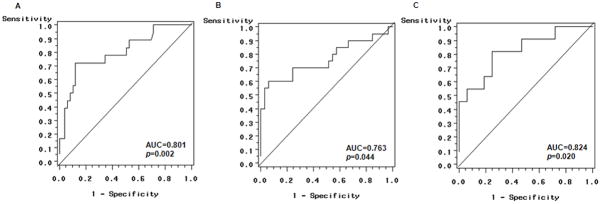
The predictive accuracies of miRNAs’ copy number for PCa aggressiveness were evaluated by logistic regression to separately construct ROC curves for CARPA and D’Amico scores. The sensitivity, specificity and AUC for significant models are presented in Fig. 1. Plasma expression of miR-21 and miR-221 significantly differentiated patients with intermediate risk from those with low risk CAPRA scores (Fig. 1A). The AUC was 0.801 with a sensitivity of 38.1% and a specificity of 94.2% (p=0.002). However, no significant miRNA was observed to predict PCa aggressiveness by comparing patients with high vs. low risk CAPRA scores. We also found that the combination of miR-20a, miR-21 and miR-145 was a significant predictor for intermediate vs. low risk D’Amico scores (Fig. 1B). The AUC is 0.763 with a sensitivity of 44.4% and a specificity of 86.8% (p=0.044). The combination of four miRNAs (miR-20a, miR-21, miR-145 and miR-221) significantly distinguished patients with high vs. low risk D’Amico scores (Fig. 1C). The AUC is 0.824 with a sensitivity of 29.4% and a specificity of 97.4% (p=0.02). Overall, the observed specificities for both CAPRA and D’Amico scores were much better than the sensitivities, indicating the specific role of miRNAs in prediction of PCa aggressiveness (Fig. 1). No signficant difference was obtained for comparing high and intermediate risk for either CARPA or D’Amico scores (data not shown).
Fig. 1.
Receiver-operator characteristic (ROC) curves plot of sensitive vs. 1 – specificity for plasma miRNAs expression (copy number/μl) and risk scores adjusted by age and ethnicity. A: ROC curve of miR-21 and miR-221 for intermediate vs. low risk of CAPRA score. An area under the curve (AUC) is 0.801 for the probability cut-point of 0.50 with a sensitivity of 38.1% (8/21) and a specificity of 94.2% (49/52). B: ROC curve of miR-20a, miR-21 and miR-145 for intermediate vs. low risk of D’Amico score. The AUC is 0.763 for the probability cut-point of 0.50 with a sensitivity of 44.4% (12/27) and a specificity of 86.8% (33/38). C: ROC curve of miR-20a, miR-21, miR-145 and miR-221 for high vs. low risk D’Amico score. The AUC is 0.824 for the probability cut-point of 0.50 with a sensitivity of 29.4% (5/17) and a specificity of 97.4% (37/38).
DISCUSSION
There is an urgent need to identify novel plasma miRNAs as non-invasive markers for prediction of aggressive PCa, but most previous stuides have shown inconsistent results. No reproducible miRNA markers related to PCa prognosis have been identified to date. A recent study evaluated miRNA profiles in 36 untreated PCa patients with different risks of progression according to CAPRA scores. Although miR-19a and miR-451 were significantly different between high-risk cases and healthy controls, no significant difference was observed for patients with low- and high-risk CAPRA scores [22]. In the current study, we provide the first evidence to suggest that plasma miRNAs may be valuable predictors to differentiate PCa patients with varied risks of aggressiveness. miR-20a and miR-21 were significantly upregulated in patients with intermediate and high CAPRA scores compared to those with low score. The expression levels of miR-21 and miR-145 were significantly higher in patients with intermediate and high D’Amico scores than those with low scores (Table III). The combination of miR-20a, miR-21, miR-145 and miR-221 expression significantly distinguished intermediate or high risk patients from those with low risk scores for aggressiveness (Fig. 1). The AUCs were ranged from 0.76 to 0.82.
Expression of miR-20a and miR-21 has been associated with clinicopathological variables of PCa. miR-20a was significantly higher in patients with a Gleason score of 7–10 compared with 0–6 (p=0.0082) [14]. Higher levels of serum miR-21 were detected in both androgen-dependent PCa (ADPC) and hormone-refractory PCa (HRPC) patients with PSA levels >4ng/ml [13]. A consistent finding was observed for miR-21 expression in patients diagnosed with metastatic PCa [23]. Our data using CAPRA and D’Amico scores to categorize PCa aggressiveness further confirmed the role of miR-20a and miR-21 in prediction of PCa prognosis. Two mechanisms may explain the observed correlations between miR-20a and miR-21 expression and PCa aggressiveness. E2F1, a transcriptional activator for DNA replication, is a target of miR-20a [24,25]. When miR-20a is activated, it may inhibit apoptosis in PCa cell lines [24,25]. Over-expressed miR-21 may induce tumor angiogenesis through targeting PTEN, and activate AKT and ERK1/2 signaling pathways, and thereby enhance HIF-1α and VEGF expression. HIF-1α is a key downstream target of miR-21 in regulating tumor angiogenesis [26]. A wider spectrum of genes involved in suppressing cell migration and invasion may also be altered by miR-21 over-expression in different cancer cells [27].
As a known tumor suppressive miRNA, miR-145 was previously found to be down-regulated in many solid tumors compared with adjacent non-tumor tissue, including PCa [28–30]. miR145 might function through direct repression of the c-MYC [31] oncogene or mucin-1 [32] and inhibition of multiple tumor survival effectors in proliferation, migration and invasion pathways [33,34]. However, few studies had explored the role of miR-145 deregulation in prediction of PCa prognosis. By examining 273 detectable miRNAs, Wang et al. identified a significant association between miR-145 down regulation and PCa grade using lymphoblastoid cell lines derived from prostate cancer patients with tumors of different aggressiveness [33], while another study measuring tumor tissues found no significant association for PCa clinicopathological features, such as tumor stage and Gleason score [28]. There are no data to date suggesting miR-145 up-regulation is associated with aggressive prostate cancer. Only one previous study of epithelial ovarian cancer found that miR-145 was significantly upregulated in tumor tissues of relapsers compared with non-relapsers [35]. Although miR-145 levels in prostate cancer tissues were generally low compared with that in non-tumor tissues, the variations of miR-145 expression were large with a 10–30 fold range in tumor tissues [29,36]. This phenomenon suggests that expression of miR-145 within prostate tumor tissues may display a heterogeneous pattern that can be used as a biomarker to differentiate PCa patients with tumors of diverse aggressiveness.
Up-regulation of miR-221 has been reported in several human cancers, including PCa [37–39]. Significantly higher levels of miR-221 were also found in tumors of metastatic PCa patients compared with localized cases [23]. However, conflicting results were found in other studies. A significant down-regulation of miR-221 was associated with aggressive PCa [15] or its biochemical recurrence [40]. miR-221 expression exhibited the most dramatic difference between androgen-dependent and androgen-independent PCa cell lines, but with inconsistent patterns [39,41]. One study found significantly increased miR-221 expression in castration-resistant PCa cells [41], but another found up-regulation in androgen-dependent patients [39]. We did not find an independent role for miR-221 on prediction of PCa aggressiveness by either CAPRA or D’Amico score. But the combination pattern of miR-20a, miR-21, miR-145 and miR-221 may significantly distinguish high vs. low risks of PCa patients by D’Amico score with an AUC of 0.824 (Fig. 1). These contradictory results suggested that different mechanisms or pathways may contribute to PCa progression due to miR-221 acting on multiple targets involved in fundamental cellular processes. For example, miR-221 expression is known to negatively regulate the cell cycle inhibitor p27Kip1 [37] and up-regulate DVL2 involved in migration [39]. miR-221 might directly inhibit ARHI, a tumor suppressor gene and stimulate cell proliferation and invasion [42]; its expression was also associated with androgen-independent growth [38].
By ROC analyses, a recent study found miR-145 was able to correctly classify 71% of PCa tumor tissues from non-tumor tissues [28]. Another independent study obtained an AUC of 0.74 for the ability of miR-145 expression to discriminate PCa tumor and non-tumor tissues [40]. miR-221 alone also displayed a similar AUC of 0.75 for its ability to significantly differentiate PCa tumor and non-tumor tissues [40]. A combination of deregulated miRNAs showed improved ability to correctly discriminate tumor and non-tumor tissues. Wach et al. observed an AUC of 0.78 for the combination of miR-143, miR-145 and miR-375 for correctly categorizing PCa tissue from non-tumor tissue [28]. A pattern of six miRNAs (miR-96, miR-149, miR-181b, miR-182, miR-205 and miR-375) had a significant AUC of 0.88 to discriminate normal and tumor tissue [40]. Our study, although having a different aim, indicates that miR-21 and miR-221 over-expression can significantly differentiate PCa patients with intermediate risk from those with low risk CAPRA scores (AUC=0.801, p=0.002). The combination of four miRNAs (miR-20a, miR-21, miR-145 and miR-221) significantly distinguished PCa patients with high risk of aggressiveness by D’Amico score with an AUC of 0.824. The observed predictive specificity in the current study was consistently much higher than the sensitivity, suggesting that the plasma miRNAs may be a specific biomarker in prediction PCa aggressiveness, but the sensitivity needs further improvement.
One strength of the current study is the inclusion of PCa patients at different risk of aggressive disease as determined by CAPRA or D’Amico score. Both risk scores were able to accurately predict PCa metastases and mortality in large retrospective cohort studies [17–19], although CAPRA score may be better than D’Amico in predicting prognostic outcome of PCa [43]. Another strength is the quantitative RT-PCR approach. This method measures absolute copy numbers for each miRNA using a standard curve eliminating potential bias due to the use of different miRNAs as endogenous controls to normalize data and minimizes the variations of input RNAs on quantitative data. Several limitations need to be recognized when interpreting the observed results. The main weakness is the cross sectional study design that lacks of prospectively collected samples. Due to the relative short follow-up time (mean=20.2 months), disease outcome data have not been obtained to directly indicate aggressiveness of PCa. However, the ongoing follow-up already showed that the relapse rates are increased for PCa patients with high-risk CAPRA or D’Amico scores (22.2%, 17.6% respectively) compared with intermedian-risk (9.5%, 7.4%) and low-risk patients (1.9%, 0.0%). The single time-point measurement of plasma miRNAs restricts our ability to determine causality related to altered miRNAs and PCa aggressiveness. Plasma samples were all derived from patients prior to or at the time of treatment when the tumor is present. It would be useful to confirm that the over-expressed miRNAs disappear after removing the target tumor tissues. Therefore, measuring post-surgery plasma samples may help to confirm the current findings. Another limitation is only five candidate miRNAs were evaluated based on previous data. Further genome-wide miRNAs screening may provide an opportunity to discover novel deregulated miRNAs that correlate with invasive PCa.
In conclusion, our results further highlight the great potential of circulating miRNAs as prognostic biomarkers to identify patients with aggressive PCa, especially to support the predictive role of circulating miR-20a, miR-21 and miR-145 in differentiating aggressive PCa patients. A large prospective study to comprehensively screen miRNAs profilings is warranted to identify novel circulating miRNAs and improve clinical prediction of PCa prognosis and enhance the quality of life of patients.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants P30 ES009089 and P30 CA013696, and TJ Marell Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors declare that no competing financial interests exist.
References
- 1.Coppola V, De Maria R, Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2010;17(1):F1–17. doi: 10.1677/ERC-09-0172. [DOI] [PubMed] [Google Scholar]
- 2.Le XF, Merchant O, Bast RC, Calin GA. The Roles of MicroRNAs in the Cancer Invasion-Metastasis Cascade. Cancer Microenviron. 2010;3(1):137–147. doi: 10.1007/s12307-010-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bill-Axelson A, Holmberg L, Ruutu M, Garmo H, Stark JR, Busch C, Nordling S, Haggman M, Andersson SO, Bratell S, Spangberg A, Palmgren J, Steineck G, Adami HO, Johansson JE. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med. 2011;364(18):1708–1717. doi: 10.1056/NEJMoa1011967. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da PI, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 8.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2009;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–331. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 14.Pesta M, Klecka J, Kulda V, Topolcan O, Hora M, Eret V, Ludvikova M, Babjuk M, Novak K, Stolz J, Holubec L. Importance of miR-20a expression in prostate cancer tissue. Anticancer Res. 2010;30(9):3579–3583. [PubMed] [Google Scholar]
- 15.Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, Strobel P, Riedmiller H, Kneitz B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127(2):394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 16.Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sanudo A, Dall’oglio MF, Camara-Lopes LH, Srougi M. MicroRNA-100 Expression is Independently Related to Biochemical Recurrence of Prostate Cancer. J Urol. 2011 doi: 10.1016/j.juro.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 17.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst. 2009;101(12):878–887. doi: 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. Jama. 1998;280(11):969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 19.D’Amico AV, Whittington R, Malkowicz SB, Cote K, Loffredo M, Schultz D, Chen MH, Tomaszewski JE, Renshaw AA, Wein A, Richie JP. Biochemical outcome after radical prostatectomy or external beam radiation therapy for patients with clinically localized prostate carcinoma in the prostate specific antigen era. Cancer. 2002;95(2):281–286. doi: 10.1002/cncr.10657. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarti IM, Laha RG, Roy J. Handbook of methods of applied statistics. John Wiley; 1967. pp. 392–394. [Google Scholar]
- 21.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 22.Moltzahn F, Olshen AB, Baehner L, Peek AS, Fong L, Stoppler HJ, Simko J, Hilton JF, Carroll PR, Blelloch R. Microfluidic based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in sera of prostate cancer patients. Cancer Res. 2011;71(2):550–560. doi: 10.1158/0008-5472.CAN-10-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32(3):583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 24.Sylvestre Y, De GV, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282(4):2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, Jiang Y, Kung HF, Lai L, Jiang BH. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS ONE. 2011;6(4):e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbuzova A, Schmitz AA, Vergeres G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362(Pt 1):1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Dyrskjot L, Eltze E, Wieland W, Keck B, Ekici AB, Grasser F, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2011 doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 29.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27(12):1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 30.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 31.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ, Thibodeau SN. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 2009;69(24):9490–9497. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, Chun KH. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011 doi: 10.1016/j.jconrel.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Marchini S, Cavalieri D, Fruscio R, Calura E, Garavaglia D, Nerini IF, Mangioni C, Cattoretti G, Clivio L, Beltrame L, Katsaros D, Scarampi L, Menato G, Perego P, Chiorino G, Buda A, Romualdi C, D’Incalci M. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 36.Zaman MS, Chen Y, Deng G, Shahryari V, Suh SO, Saini S, Majid S, Liu J, Khatri G, Tanaka Y, Dahiya R. The functional significance of microRNA-145 in prostate cancer. Br J Cancer. 2010;103(2):256–264. doi: 10.1038/sj.bjc.6605742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 38.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafre SA. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng C, Yinghao S, Li J. MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2. Med Oncol. 2011 doi: 10.1007/s12032-011-9934-8. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 41.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69(8):3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Zaman MS, Deng G, Majid S, Saini S, Liu J, Tanaka Y, Dahiya R. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila) 2011;4(1):76–86. doi: 10.1158/1940-6207.CAPR-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lughezzani G, Budaus L, Isbarn H, Sun M, Perrotte P, Haese A, Chun FK, Schlomm T, Steuber T, Heinzer H, Huland H, Montorsi F, Graefen M, Karakiewicz PI. Head-to-head comparison of the three most commonly used preoperative models for prediction of biochemical recurrence after radical prostatectomy. Eur Urol. 2010;57(4):562–568. doi: 10.1016/j.eururo.2009.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.