Abstract
New experimental approaches are required to detect the elusive transient intermediates predicted by simulations of virus assembly or disassembly. Here, an atomic force microscope (AFM) was used to mechanically induce partial disassembly of single icosahedral T = 1 capsids and virions of the minute virus of mice. The kinetic intermediates formed were imaged by AFM. The results revealed that induced disassembly of single minute-virus-of-mice particles is frequently initiated by loss of one of the 20 equivalent capsomers (trimers of capsid protein subunits) leading to a stable, nearly complete particle that does not readily lose further capsomers. With lower frequency, a fairly stable, three-fourths-complete capsid lacking one pentamer of capsomers and a free, stable pentamer were obtained. The intermediates most frequently identified (capsids missing one capsomer, capsids missing one pentamer of capsomers, and free pentamers of capsomers) had been predicted in theoretical studies of reversible capsid assembly based on thermodynamic-kinetic models, molecular dynamics, or oligomerization energies. We conclude that mechanical manipulation and imaging of simple virus particles by AFM can be used to experimentally identify kinetic intermediates predicted by simulations of assembly or disassembly.
Introduction
Assembly of a protein shell, or capsid, is an obligate step in the life cycle of any virus; the reverse process, capsid disassembly, is required by many viruses to release their genome into the infected cell (1–7). A deep understanding of capsid assembly and disassembly may contribute to the development of antiviral drugs based on interference with either process (1–5,7–10), and to the engineering of capsids and self-assembling nanoparticles for biomedical or nanotechnological applications (11–13).
In about half of the virus families and most animal viruses, the capsid is roughly spherical and arranged with icosahedral symmetry. Unfortunately, current understanding of icosahedral capsid assembly and disassembly is limited. Important insights have been obtained through the experimental identification of populated intermediates or, rarely, transient intermediates during the scaffold-mediated assembly, maturation, or dissociation of complex icosahedral virus particles (1,2,6,14–17). However, these elaborate, multistep processes are very difficult to investigate in detail using theoretical models.
Many small icosahedral capsids do not require a protein or nucleic-acid scaffold for assembly and either are not subjected to stabilization by irreversible maturation, or can be procured in immature form. These capsids are much more amenable to thermodynamic and kinetic modeling and to detailed simulations of assembly and disassembly. The different theoretical approaches undertaken (18–33) have revealed important foundations and features of these processes. However, the predicted pathways are difficult to confirm experimentally, partly because of the transient nature of the intermediates. In vitro studies of simple viral capsids generally revealed two-state processes, with stable building blocks and complete capsids, but no substantially populated intermediate states (18,21,24,34).
We use experimental approaches to study the assembly, disassembly, and physicochemical properties of a simple model virus, the minute virus of mice (MVM). The icosahedral T = 1 capsid of MVM is one of the smallest and structurally simplest known (35) (Fig. 1), and the interactions between the 60 protein subunits are strictly equivalent. During infection by MVM, trimers of the capsid protein are formed in the cytoplasm and constitute stable building blocks (capsomers). These capsomers are then transported to the cell nucleus (36–38), where they undergo a conformational transition that makes them competent for capsid assembly (38,39).
Figure 1.
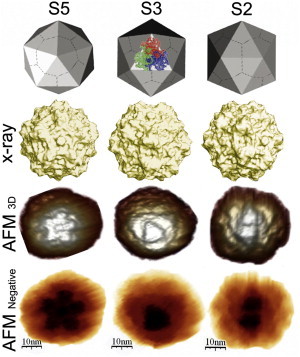
AFM imaging of MVM. (Top) Geometric model of the MVM capsid. In the central image, a ribbon diagram of a trimeric capsomer is superimposed. (Second row) Crystallographic model of the MVM capsid (35,63). (Third row) 3D images obtained by AFM of single MVM capsids. (Bottom) Negative images of the same capsids. Increased height is indicated by lighter (third row) or darker (bottom) tones. Capsids are oriented with an S5, S3, or S2 (left to right) symmetry axis on top (center of the image).
No conditions for in vitro assembly of the MVM capsid have been established yet; however, MVM capsids devoid of viral nucleic acid readily self-assemble in eukaryotic cells (40). MVM capsid assembly does not occur if a few intercapsomer interactions are removed by single mutation of a few key interfacial residues (41). However, it has been observed that once the capsid is assembled, no mutation of any other interfacial residue tested leads to any significant reduction in the high temperature (∼75°C) required for capsid dissociation (40,42), despite this capsid not being subjected to stabilization by maturation. These observations suggest that the MVM capsid, like other icosahedral virus capsids (24,43,44), may be thermodynamically unstable but kinetically stabilized (24).
Different theoretical models and simulations with simple icosahedral capsids have predicted kinetic (transient) intermediates that could also occur during MVM capsid assembly or disassembly. First, for the hepatitis B virus (HBV) capsid, hysteresis of dissociation was observed and explained within the framework of a thermodynamic-kinetic model by Zlotnick and colleagues (24). Their simulations revealed that during the early stages of dissociation, nearly complete capsids, especially those that lacked just one capsomer, accumulate transiently and create an energy barrier against further dissociation. These nearly complete capsids were slow to dissociate further, but readily reassociated with free capsomers, acting like kinetic traps. In this way, capsids that are thermodynamically unstable (because of weak interactions between capsomers (45)) can be kinetically stabilized, and their integrity preserved, even at very low concentrations (24).
Second, Rapaport used a molecular dynamics (MD) approach to simulate reversible assembly, in the presence of solvent, of small populations and individual particles of a generic icosahedral T = 1 capsid made of just 20 triangular capsomers (30,33). This theoretical setup precisely matches actual assembly in the cell of the T = 1 capsid of MVM from 20 trimeric (triangular) capsomers. Among other findings, the results revealed that only a few transient intermediates are relatively long-lived and frequently encountered along the pathway. These included pentamers of capsomers and, as in the kinetic simulations of HBV (24), nearly complete capsids lacking only one or a few capsomers. In addition, assembled capsids did not readily dissociate, even under conditions in which they would not assemble from capsomers, again revealing hysteresis of dissociation.
Third, Reddy et al. proposed energetically favorable pathways for the reversible assembly of different virus capsids based on the calculated relative association energies in potential oligomeric intermediates (20). The calculated assembly profile for parvoviruses like MVM suggests that they assemble from multiples of the trimeric capsomer, with a significant preference for incomplete capsids missing a pentamer of trimers, which could be an assembly intermediate (6).
To sum up, different theoretical approaches predicted key kinetic intermediates that could be transiently encountered during assembly and/or disassembly of the MVM T = 1 capsid: 1), temporally stable, nearly complete capsids lacking one capsomer; 2), capsids missing one pentamer of trimers; and 3), free pentamers of trimers (Fig.2). These and other predictions stress the desirability of devising experimental approaches to detect the elusive, transient intermediates of icosahedral capsid assembly or disassembly proposed by modeling and simulation. Recently, transient intermediate states have been identified in complex viral models by kinetic trapping (46), time-resolved x-ray scattering, (47) or mass spectrometry (48,49). Here, we have used atomic force microscopy (AFM) to mechanically induce partial disassembly of MVM particles and experimentally identify theoretically predicted kinetic intermediate(s).
Figure 2.
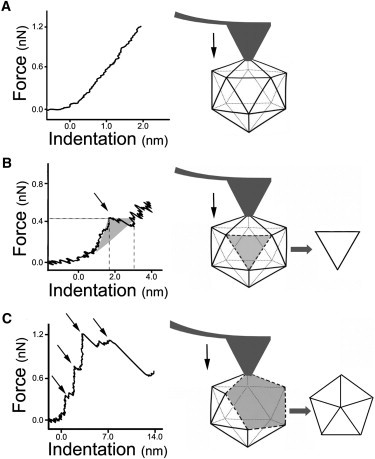
Force-versus-indentation curves corresponding to representative indentation events that produced an elastic deformation without any loss of integrity (A), or were directly associated with loss of a single capsomer (B) or a pentamer of capsomers (C). Arrows point to single or multiple drops in the force applied during the otherwise linear regime, that were associated with structural transition(s) leading to loss of a capsid subunit. In B, the shaded area was used to calculate the energy dissipated in a specific event (see text). The schemes at right show the intermediates formed in each case. The same intermediates had been predicted by theory.
In recent years, AFM has been applied to explore, in close-to-physiological conditions, the mechanical properties of virus particles ((50); reviewed by Roos et al. (51,52)), including MVM capsids and virions (53,54). Nanoindentation of individual particles using the tip of the AFM microscope generally elicited an elastic response, but deeper indentations frequently led to mechanical failure and eventual disruption of the particles (50,51). In a study with herpes simplex virus (HSV), it was possible to identify by AFM imaging sites in the capsid where capsomers were removed by chemical treatment (55–57). Further, controlled AFM indentations led to the mechanical displacement, or removal, of identifiable capsomers from HSV or bacteriophage Φ29 particles (57,58).
We reasoned that AFM could be similarly used with much simpler icosahedral particles to initiate their disassembly by indentation in physiological buffer, and to identify by high-resolution imaging any kinetic intermediate(s) that might appear. This single-particle approach applied to MVM revealed transient intermediates predicted by theory and simulation and provided strong experimental support for those models.
Materials and Methods
Recombinant plasmids
Plasmid pSVtk-VP1/2 (59) or bacmid BM-VP2 (42) were used for the production of MVM capsids containing proteins VP1 and VP2, or VP2 only, respectively. Plasmid pTRp (originally provided by P. Tattersall, Yale University, New Haven, CT, (60) and later modified (59)), was used for production of MVM virions.
Expression and purification of empty capsids and virions of MVM
For production of VP2-only capsids, H-5 cells were transfected with BM-VP2 (42), and the capsids were purified (40). For production of VP1/VP2 capsids or virions, NB324K cells were transfected with pSVtk-VP1/2 or pTRp, respectively, and the particles were purified (36,37,53). Preparations were dialyzed against phosphate-buffered saline and kept at 4°C or −70°C. Purity and integrity were assessed by electrophoresis, electron microscopy, and/or AFM.
Imaging MVM capsids and virions by AFM
AFM experiments on MVM particles in buffer were carried out as described (53,54). The AFM (Nanotec Electrónica, Madrid, Spain) was operated in jumping mode (61) in liquid. We used cantilevers (RC800PSA, Olympus, Tokyo, Japan) with a nominal spring constant of 0.1 N/m. The maximum normal force applied during AFM imaging was of the order of 100 pN (low force) to minimize particle damage and other undesirable effects. AFM images were processed using WSxM software (62). The observed structures were compared with the crystal structures of the MVMi virion (PDB ID 1Z1C) and MVMp capsid (PDB ID 1Z14) (35,63) (Fig. 1).
Mechanically induced disassembly of MVM capsids and virions
Individual viral particles adsorbed on a silanized glass surface were located by AFM imaging. Once a particle with a fivefold (S5), threefold (S3), or twofold (S2) symmetry axis on top was identified, the lateral piezo scan was stopped when the tip was above the center of the particle. Then, a series of high-force (∼1-nN) indentations were carried out on the selected particle. For each indentation, the z-piezo was elongated until the tip established contact with the virus particle, the elongation was continued to perform the indentation, and the corresponding force-versus-distance (Fz) curve was obtained. The indentation depth, calculated from the force-versus-indentation curve (Fig. 2), ranged from 0.5 to 9 nm.
Images were obtained before and after each series of high-force indentations. The series of indentations were generally terminated when the particle underwent a detectable, permanent structural modification, or when close to 40 indentations had been carried out without observing any permanent change. On occasion, after the particle had been structurally modified, additional indentations were carried out to induce further modifications, and the particle was imaged again. In some cases, when the particle needed to be reoriented to get a symmetry axis on top, a tangential indentation close to the side of the particle was performed. The Fz curve obtained for each indentation event was processed to obtain the force applied on the particle (Eq. 1) and the indentation depth:
| (1) |
F is the force applied (nN); kc the spring constant of the cantilever (nN/nm); (d × s) is the cantilever deflection (nm) obtained as the product of the signal of the photodiode, d (volts), and the sensitivity of the photodiode, s (nm/volt). Each cantilever was calibrated as described (64), implemented online at www.ampc.ms.unimelb.edu.au/afm/theory.html#normal.
Calculation of the energy dissipated during removal of viral-particle subunits
The energy dissipated was estimated as described (65) from force-versus-indentation curves, which showed evidence of a structural transition, reflected in the punctual loss of the linear regime during the indentation (Fig. 2). Briefly, the work was estimated from the area defined by the difference in slope in the curve before and after the point where a sudden drop in the applied force took place (Fig. 2 B). In the case of curves with multiple steps (Fig. 2 C), the total energy was estimated by the sum of the energies calculated for each step.
Results and Discussion
MVM capsids are highly resistant to mechanically induced disassembly
The MVM capsids produced in H-5 or NB324K cells were made of 60 copies of capsid protein VP2, or ∼51 copies of VP2 and 9 copies of VP1, respectively. However, except for a longer N-terminus, VP1 is identical to VP2 in both sequence and structure. The N-termini are disordered and subjected to translocation through capsid pores and proteolytic removal, and they do not participate in the architecture of the icosahedral capsid shell (35,63). As expected based on this fact, the results we obtained by indentation of either capsid type were indistinguishable and are presented here together.
Single capsids submerged in physiological buffer were imaged by AFM. The high-resolution images obtained allowed us to identify topographic features (spikes at S3, dimples at S2, and cylindric prominences at S5) on the icosahedral particles (Fig.1) (53,54). These features were used to detect structural changes and the approximate point on each particle where the force was applied in indentation experiments.
Selected individual capsids were subjected to series of high-force (∼1-nN) indentations. This force is close to that previously used to disrupt different virus particles. The whole process on a single particle took up to ∼15 min, and changes could be followed over time.
In total, >800 single capsids were analyzed. Most of them withstood a high number (over 40, and up to 100 at times) of high-force indentations and repeated imaging without any detectable, permanent structural alteration. These and previous observations (53,54) revealed that the MVM capsid is quite robust compared to other virus particles (51,52) and can withstand high mechanical forces and a high number of elastic deformations without apparent damage or obvious material fatigue.
Mechanical disassembly of MVM capsids frequently results in nearly complete capsids lacking only one trimeric capsomer
Despite the high robustness of the MVM capsid, repeated high-force indentations and/or imaging eventually led to its permanent structural alteration. Very frequently, the indented capsids remained nearly complete. Seventy-seven of these particles were analyzed in detail.
Remarkably, in 53 of those 77 cases, only one capsomer (a trimer of capsid proteins) had been mechanically removed; the capsid remained otherwise intact (Fig. 3). In images of MVM particles, each trimeric capsomer on the visible particle surface (close to the center of the image) can be recognized by a centrally located, protruding spike formed by the intertwining of long loops from the three proteins in the capsomer (Fig. 1). After the indentations, one of the capsomers was clearly missing, leaving a gap at the position it had originally occupied (compare Fig. 3). In a few cases, the removed capsomer could be observed beside the incomplete capsid (Fig. 3 D). In AFM images, width cannot be precisely measured due to tip-sample dilation effects, but height is a precise measurement; the approximate expected width and measured height (∼8 nm; Fig. 3 D) of the removed subunit, observed by AFM, corresponded with those of a trimeric capsomer in the capsid crystal structure.
Figure 3.
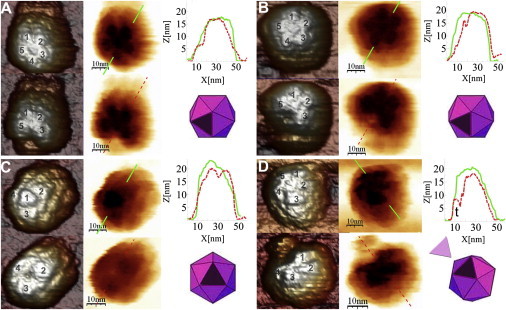
AFM images of individual MVM capsids before and after removal of a single capsomer. Each panel shows results obtained with a different particle and includes 3D (left) and negative (center) images of the same particle before (upper) and after (lower) removal of a single capsomer. In the 3D image, the capsomers around the axis closest to the top of the particle (S5 in A, B, and D, and S3 in C) are numbered. Included at right are height profiles of the capsids along the lines indicated in the negative images (upper) before (solid line) and after (dashed line) losing one capsomer, as well as a scheme of the incomplete capsid after losing one capsomer (lower). In D, the detached capsomer can be observed at 11 o'clock beside the capsid, and its equatorial height profile (t) is shown to the left of the incomplete capsid profile.
Occasionally (for 10 out of 77 single capsids), two capsomers (in most cases adjacent to each other) were removed. On at least one occasion, we observed a capsid that lost one capsomer first and a second, adjacent capsomer after further indentations (Fig. 4), indicating that capsid disassembly may initially proceed through the successive removal of single capsomers.
Figure 4.
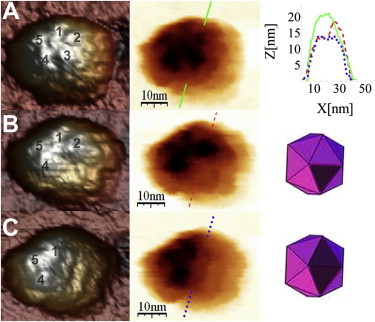
AFM images of the same MVM capsid before indentation (A) and after removal of one capsomer (B) and an additional capsomer (C). At right are a height profile of the capsid. intact (solid line) and after losing one (dashed line) or two (dotted line) capsomers (A) and a scheme of the incomplete capsid after losing one or two capsomers (B and C, respectively).
In MVM and other parvovirus capsids, interprotein interactions are weaker at the intertrimer interfaces than at interdimer or interpentamer interfaces (35); accordingly, trimers of capsid proteins constitute the stable capsomers from which the MVM capsid is assembled (36–38). The indentations may have provided enough energy to overcome the activation energy barrier of trimer dissociation, but not enough to disrupt the stronger interactions at the protein-protein interfaces within each trimer, explaining why dimers or pentamers of protein subunits were never observed.
The incomplete particles that had lost one capsomer (or even two) were fairly stable over time, and some withstood subsequent indentations without collapsing or losing further capsomers. These results would not be expected if disassembly of the MVM capsid occurred as a cooperative process with no kinetic traps. If such were the case, removal of one or two capsomers should facilitate removal of further capsomers, invariably leading in a short time to the total dissociation of the capsid into stable building blocks; no incomplete capsid intermediates would be observed.
Remarkably, the quite stable capsids we observed that lacked just one (or two) capsomer(s) after mechanically induced dissociation of single particles were exactly those predicted using two theoretical approaches: 1), the thermodynamic-kinetic model of capsid assembly and disassembly by Zlotnick (24), who predicted that nearly complete capsids lacking only one or a few capsomers would accumulate transiently and create an energy barrier against further dissociation by readily reassociating with free capsomers; and 2), the MD simulations by Rapaport on a T = 1 capsid reversibly assembled from 20 capsomers (as in the MVM capsid), which revealed nearly complete capsids as the longest-lived intermediates (30,33).
In the AFM-induced disassembly experiments, the capsomers were difficult to remove from the capsid, and the nearly complete capsids lacking one capsomer were quite stable, as would be expected if a high activation energy barrier impaired their further disassembly as predicted. Because of the single-molecule experimental setup, just one capsomer from one particle was released; also, the removed capsomer could be adsorbed on the solid surface (as in Fig. 3 D). Thus, the probability of recapture of the missing capsomer by the incomplete capsid was negligible, allowing the repeated imaging of this intrinsically stable but normally transient intermediate.
Pentamers of trimeric capsomers are sometimes removed during the mechanical disassembly of the MVM capsid
For 14 of the 77 single capsids analyzed in detail, indentations led to removal of subunits that were larger than individual capsomers (Fig. 5). Removal of these larger subunits occurred at about one-third the frequency of removal of individual capsomers, and the released subunits were most often observed beside the incomplete capsid (compare images in Fig. 5). The larger subunits removed almost invariably corresponded to pentamers of trimeric capsomers, as shown by their size, shape, fivefold symmetry, and height (Fig. 5).
Figure 5.
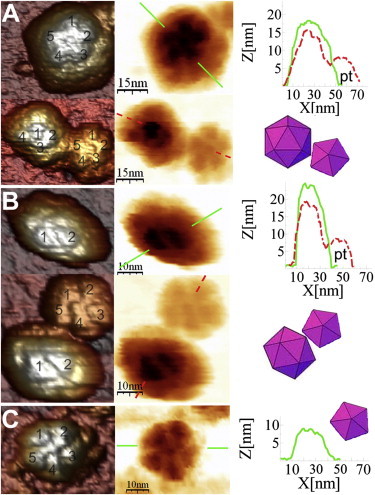
AFM images of individual MVM capsids before and after removal of a pentamer of capsomers. (A and B) Images of two different MVM particles taken before (upper) and after (lower) losing a pentamer of capsomers. Before the indentations, the capsid was oriented with an S5 axis (A) or S2 axis (B) on top. During the indentations, the capsid in A rotated, resulting in an S3 axis being positioned on top. In both A and B, the detached pentamer of capsomers can be observed beside the incomplete capsid. Also shown (right) are height profiles of the intact capsid (solid line), and of the incomplete capsid and the detached pentamer (pt) (dashed line), as well as a scheme of the incomplete capsid and the removed pentamer. (C) Closeup image of the detached pentamer of capsomers, with equatorial height profile and schematic illustration.
Even after this dramatic event that involved removal of as many as one-fourth of the capsid protein subunits, the substantially incomplete capsid remained stable over time and could be imaged. However, it was clearly less stable than the capsid lacking just one capsomer, and it usually collapsed after two further successive images had been taken.
Most remarkably, the fairly stable, three-fourths-complete capsids and stable free pentamers of capsomers we observed after mechanical disassembly had been also predicted by different theoretical approaches. 1), Rapaport's MD simulations of a T = 1 capsid (30,33) revealed rapid initial capsid growth that largely led to pentamers of capsomers. Pentamers of capsomers (formed by either accretion of capsomers or disassembly during the simulated process) lived much longer than did smaller clusters (including dimers and trimers). Further, loss of precisely five capsomers from a growing particle occurred more frequently than loss of a lower or higher number of capsomers. This prediction does not imply that once the capsid is fully formed, losing one pentamer should be more frequent than losing one protomer. In the fully assembled capsid all interfaces are satisfied, whereas in the assembly intermediates they are not. Thus, losing one pentamer could be more difficult when starting from a fully closed capsid, as observed in our experiments. 2), By estimation of association energies in possible oligomeric intermediates of reversible assembly, Reddy and Johnson (6) predicted that parvoviruses (like MVM) assemble from multiples of the trimeric capsomer. They found a significant preference for precisely the structure containing 45 subunits—three-fourths of a capsid that is missing a pentamer of trimers (15 subunits), suggesting that this could be an intermediate during parvovirus assembly.
Icosahedral capsid assembly involves a nucleation step; subsequent assembly essentially proceeds by the successive addition of capsomers to the nucleus. An interesting possibility that remains to be tested is that a pentamer of trimeric capsomers could act as a nucleus during assembly of the MVM capsid. Capsid reassembly may not occur in the AFM experiments because of extreme dilution and adsorption to the solid surface of the single removed pentamer.
Mechanically induced collapse of the MVM capsid
After repeated indentations and imaging, many single particles experienced a severe loss of integrity, and several fragments could be observed where the indented capsid had been. In several cases at least, this catastrophic failure could be traced to harsh mechanical treatment involving higher force, deeper indentation, particle-tip adhesion or uncontrolled cantilever movement. A few of those cases were analyzed in detail (Fig. S1 and Fig. S2 in the Supporting Material). In some instances, the gradual disruption of the capsid could be followed during a series of indentations (Fig. S2). One or two complete pentamers of capsomers, in addition to some smaller fragments or individual capsomers, could be identified also in these cases (Fig. S1 and Fig. S2).
In one of the disassembly experiments, the capsid collapsed and was fully flattened on the solid surface (Fig. 6). Fifteen or sixteen capsomers were clearly counted (Fig. 6 B). The flattened structure looked rather like a continuous sheet of triangular faces used to build an icosahedral model (Fig. 6 D), with the capsomers located in relative positions similar to those of the faces in the model (compare Fig. 6, B and D). Thus, many capsomers appear to be still connected in one or, at most, a few disconnected sets of capsomers. This particle exemplifies that even fracturing many intercapsomer interfaces in the MVM capsid leads not to a complete dissociation of the capsid into 20 individual capsomers but to fewer sets of several capsomers, among which pentamers are conspicuous.
Figure 6.
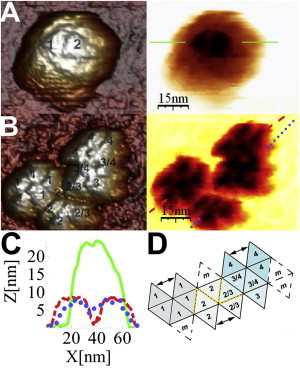
AFM images of a single collapsed MVM capsid. (A) Before indentation, the intact capsid is oriented with an S2 axis on top; in the 3D image, the capsomers around this axis are numbered. (B) After indentation, the capsid is completely flattened. Counting of capsomers reveals that only 4 or 5 of the 20 capsomers are missing; in the 3D image, the capsomers around each of four S5 axes observed are numbered 1–4. (C) Height profiles of the capsid before (solid line) and after (dashed and dotted lines) collapse. (D) A 2D-extended icosahedral model with the faces of the icosahedron labeled according to the code used for the equivalent capsomers in B. Black arrows connect intercapsomer interfaces that are not formed in the model but appear to be still formed in the collapsed capsid. The four missing capsomers (m) in the collapsed capsid are labeled. Some capsomers or pentamers of capsomers in B may be not attached (D) but just lying next to each other. Possibly disconnected interfaces are indicated by a yellow dashed line in the model.
Estimation of the energy applied for the removal of single capsomers or pentamers of capsomers
We observed a trend toward the release of a single capsomer after successive high-force indentations (30 to 100) with a depth of ∼2 nm. In contrast, removal of a pentamer of capsomers generally involved few (<15) but deeper (∼5-nm) indentations. In addition, when removal of a single capsomer could be associated to a particular indentation, the corresponding curve showed evidence of a single structural transition, reflected in a single drop in the applied force during the otherwise linear regime (Fig. 2 B). In contrast, removal of pentamers of trimers was almost invariably associated to several transitions, reflected in two or more drops in the applied force during the linear regime (Fig. 2 C).
Because images were taken just before and after indentation, a single indentation could be clearly identified as the immediate cause of the loss of a single capsomer in two cases and a pentamer of capsomers in six cases. From the curves, the average energy associated with loss of a single capsomer or a pentamer of capsomers was estimated using a procedure previously applied to other biological assemblies (58,65). The average values obtained were 3.6 × 10−19 J for removal of a capsomer from a single MVM capsid (217 kJ/mol), and 6.1 × 10−19 J for removal of a pentamer of capsomers (367 kJ/mol). Similar values were obtained by averaging 18 or 17 curves with ruptures in their linear regime due to loss of a capsomer or a pentamer of capsomers, respectively (not shown).
Removal of a single capsomer from the MVM capsid involves the disruption of three intercapsomer interfaces. Assuming additivity, a value of 217/3 = 72 kJ/mol for the energy dissipated during the disruption of a single intercapsomer interface could be obtained from the AFM data. Removal of a pentamer of capsomers involves the disruption of five intercapsomer interfaces, and under the above assumption, it should require 72 × 5 = 360 kJ/mol, a value almost identical to that experimentally obtained. Zlotnick and colleagues determined experimentally that the interaction energy between subunits in the HBV or cowpea chlorotic mottle virus (CCMV) capsids is on the order of −12 to −18 kJ/mol (32). These values are about fivefold lower than the dissipated energy during disruption of a capsomer-capsomer interface in MVM, which is rather consistent with the relative association energies calculated from the corresponding atomic structures, and listed in VIPER (66). The possibility must be noted also that not all of the dissipated energy associated with the nonlinear event was used to disrupt the intercapsomer interface.
The above results suggest that relatively shallow indentations similar to the capsid thickness disrupt fewer intercapsomer interfaces and eventually lead to the removal of a single capsomer. Substantially deeper indentations disrupt a higher number of intercapsomer interfaces and may lead to the removal of a pentamer of capsomers. The relative energy required for removal of a subunit appears to be directly proportional to the number of intercapsomer interfaces that must be disrupted, leading to the easier, more frequent removal of single capsomers compared to pentamers of capsomers.
Mechanical pushing is being revealed as a method of providing the energy required to trigger conformational transitions in a viral capsid. We have found that both individual trimers and pentamers of trimers can be released from the capsid irrespective of where the force is applied on the particle. Simulation and experiment using other virus capsids have already provided evidence that fracture lines do not have to occur around the point where the force was directly applied. Further, in more complex viral particles, removal of some capsomers was achieved either by indentation at different points or by chemical treatment (57). In short, the outcome was not dependent on the direction of the force or the point in the capsid where it was applied. Moreover, the kinetic intermediates we observed had been predicted without any regard to a directional application of energy. These observations suggest that although the energy is directionally applied in indentation experiments, the same conformational transitions may occur as when energy is nondirectionally applied by thermal or chemical treatment in vitro or through the action of natural factors in vivo. This point remains to be further examined.
Mechanically induced disassembly of infectious MVM virions
In the MVM virion, segments of the ssDNA bound to the capsid inner wall (35) act like molecular buttresses that are responsible for the higher mechanical stiffness of the virion at regions closer to the DNA-binding sites (53,54). To assess whether the presence of the DNA could affect the disassembly of the virion compared to that of the empty capsid, over 200 individual virions were analyzed for mechanical disassembly. As with capsids, single virions generally withstood a high number of indentations, but repeated high-force indentations and/or imaging eventually led to their partial disassembly.
Nine of these single virions were analyzed in detail (Fig. S3). In four of those nine cases, only one capsomer was removed (Fig. S3 A). In one case, a virion that had already lost one capsomer lost a second one after further indentations. In another case, a pentamer of capsomers was lost. In all those cases, the nearly complete virion remained stable over time. In the three remaining cases, substantial particle fragmentation occurred, but a pentamer of capsomers could still be observed among the fragments (Fig. S3 B). The number of individual virions analyzed in detail was not enough to justify a statistical analysis. However, the above observations reveal that regarding the outcome of their mechanically induced disassembly, and the intermediates observed, no qualitative differences between virions and empty capsids are apparent. The resilience of the MVM virion against dissociation argues for the exit of the viral DNA without substantial capsid disassembly, either through one of the capsid pores or by removal of one capsomer (57).
Biological implications
Disassembly
The results of this study have provided the first direct observation, to our knowledge, of predicted, transient disassembly intermediates that act like kinetic traps and mediate the biological survival of some virions during the extracellular phase by kinetic stabilization, and not by maturation (24,33). In addition, the results indicate that predictions from studies of empty capsids are also applicable to the biologically relevant disassembly of a nucleic-acid-filled, infectious virion.
Assembly
Theory and simulation, supported by experiment, have shown that assembly of even the simplest virus capsids is an ordered process subjected to regulation. However, the required transient intermediates predicted by theory have been rarely observed. By inducing the reverse, a disassembly process, under special conditions that do not allow reassembly of the removed capsid subunits, this study allowed the direct observation of transient intermediates that had been predicted to participate in the assembly of simple T = 1 capsids, such as pentamers of capsomers, and capsids lacking one pentamer, or one capsomer. The results support further the biologically required hierarchical assembly of a simple virus capsid.
Conclusions
This study demonstrates the possibility of using nanoindentation and AFM imaging of single viral particles to identify transient intermediates of reversible assembly/disassembly. The intermediates most frequently observed after mechanically inducing the partial disassembly of single MVM capsids and virions are nearly complete particles lacking one capsomer, followed by three-fourths-complete particles missing one pentamer of capsomers, and free pentamers of capsomers. These experimentally observed intermediates correspond nicely with kinetic intermediates of the reversible assembly and disassembly of simple icosahedral capsids that had been theoretically predicted based on thermodynamic-kinetic models, MD simulations, or calculation of association energies. The results provide further experimental support for those models. Experimental identification of kinetic intermediates by the approach followed here may also help in the development of refined models and simulations of assembly and disassembly of viral capsids.
Acknowledgments
We acknowledge Prof. J. Gómez-Herrero for support and advice and Prof. J. M. Almendral for providing biological clones and expertise, Prof. A. Zlotnick for first calling our attention to hysteresis of disassembly, Dr. C. Carrasco and M. Hernando-Pérez for technical advice, and A. Rodríguez-Huete and M. A. Fuertes for technical assistance.
Work in M.G.M.'s laboratory is funded by grants from the Spanish Ministerio de Ciencia e Innovación (BIO2009-10092) and Comunidad de Madrid (S-2009/MAT/1467), and by an institutional grant from Fundación Ramón Areces. Work in P.J.deP.'s laboratory is funded by National Science Foundation grants PIB2010US-00233 and FIS2011-29493. M.G.M. is an associate member of the Institute for Biocomputation and Physics of Complex Systems, Zaragoza, Spain.
Supporting Material
References
- 1.Chiu W., Burnett R.M., Garcea R.L., editors. Structural Biology of Viruses. Oxford University Press; New York, NY: 1997. [Google Scholar]
- 2.Agbandje-McKenna M., McKenna R., editors. Structural Virology. RSC Publishing; Cambridge, UK: 2011. [Google Scholar]
- 3.Prevelige P.E., Jr. Inhibiting virus-capsid assembly by altering the polymerisation pathway. Trends Biotechnol. 1998;16:61–65. doi: 10.1016/s0167-7799(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 4.Smyth M.S., Martin J.H. Picornavirus uncoating. Mol. Pathol. 2002;55:214–219. doi: 10.1136/mp.55.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zlotnick A., Stray S.J. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol. 2003;21:536–542. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Reddy V.S., Johnson J.E. Structure-derived insights into virus assembly. Adv. Virus Res. 2005;64:45–68. doi: 10.1016/S0065-3527(05)64003-1. [DOI] [PubMed] [Google Scholar]
- 7.Zlotnick A., Mukhopadhyay S. Virus assembly, allostery and antivirals. Trends Microbiol. 2011;19:14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinlay M.A., Dutko F.J., Rossmann M.G. Rational design of antipicornavirus agents. In: Brinton M.A., Heinz F.X., editors. New Aspects of Positive-Strand RNA Viruses. American Society for Microbiology; Washington, D.C.: 1990. pp. 366–372. [Google Scholar]
- 9.Neira J.L. The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly. FEBS J. 2009;276:6110–6117. doi: 10.1111/j.1742-4658.2009.07314.x. [DOI] [PubMed] [Google Scholar]
- 10.Prevelige P.E., Jr. New approaches for antiviral targeting of HIV assembly. J. Mol. Biol. 2011;410:634–640. doi: 10.1016/j.jmb.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas T., Young M. Viruses: making friends with old foes. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 12.Mastrobattista E., van der Aa M.A., Crommelin D.J. Artificial viruses: a nanotechnological approach to gene delivery. Nat. Rev. Drug Discov. 2006;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]
- 13.Mateu M.G. Virus engineering: functionalization and stabilization. Protein Eng. Des. Sel. 2011;24:53–63. doi: 10.1093/protein/gzq069. [DOI] [PubMed] [Google Scholar]
- 14.Fane B.A., Prevelige P.E., Jr. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 15.Johnson J.E. Virus particle dynamics. Adv. Protein Chem. 2003;64:197–218. doi: 10.1016/s0065-3233(03)01005-2. [DOI] [PubMed] [Google Scholar]
- 16.Steven A.C., Heymann J.B., Conway J.F. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J.E. Virus particle maturation: insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz R., Shor P.W., Berger B. Local rules simulation of the kinetics of virus capsid self-assembly. Biophys. J. 1998;75:2626–2636. doi: 10.1016/S0006-3495(98)77708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy V.S., Giesing H.A., Johnson J.E. Energetics of quasiequivalence: computational analysis of protein-protein interactions in icosahedral viruses. Biophys. J. 1998;74:546–558. doi: 10.1016/S0006-3495(98)77813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlotnick A., Johnson J.M., Endres D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 22.Rapaport D.C., Johnson J.E., Skolnick J. Supramolecular self-assembly: molecular dynamics modeling of polyhedral shell formation. Comput. Phys. Commun. 1999;121:231–235. [Google Scholar]
- 23.Bruinsma R.F., Gelbart W.M., Zandi R. Viral self-assembly as a thermodynamic process. Phys. Rev. Lett. 2003;90:248101. doi: 10.1103/PhysRevLett.90.248101. [DOI] [PubMed] [Google Scholar]
- 24.Singh S., Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 2003;278:18249–18255. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 25.Zlotnick A. Theoretical aspects of virus capsid assembly. J. Mol. Recognit. 2005;18:479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- 26.Wales D.J. The energy landscape as a unifying theme in molecular science. Philos. Transact. A Math. Phys. Eng. Sci. 2005;363:357–375. doi: 10.1098/rsta.2004.1497. discussion 375–377. [DOI] [PubMed] [Google Scholar]
- 27.Hagan M.F., Chandler D. Dynamic pathways for viral capsid assembly. Biophys. J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zandi R., van der Schoot P., Reiss H. Classical nucleation theory of virus capsids. Biophys. J. 2006;90:1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen H.D., Reddy V.S., Brooks C.L., 3rd Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7:338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 30.Rapaport D.C. Role of reversibility in viral capsid growth: a paradigm for self-assembly. Phys. Rev. Lett. 2008;101:186101. doi: 10.1103/PhysRevLett.101.186101. [DOI] [PubMed] [Google Scholar]
- 31.Chen C., Kao C.C., Dragnea B. Self-assembly of brome mosaic virus capsids: insights from shorter time-scale experiments. J. Phys. Chem. A. 2008;112:9405–9412. doi: 10.1021/jp802498z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katen S., Zlotnick A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapaport D.C. Studies of reversible capsid shell growth. J. Phys. Condens. Matter. 2010;22:104115. doi: 10.1088/0953-8984/22/10/104115. [DOI] [PubMed] [Google Scholar]
- 34.Prevelige P.E., Jr., Thomas D., King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agbandje-McKenna M., Llamas-Saiz A.L., Rossmann M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 36.Lombardo E., Ramírez J.C., Almendral J.M. A β-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 2000;74:3804–3814. doi: 10.1128/jvi.74.8.3804-3814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombardo E., Ramírez J.C., Almendral J.M. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 2002;76:7049–7059. doi: 10.1128/JVI.76.14.7049-7059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riolobos L., Reguera J., Almendral J.M. Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J. Mol. Biol. 2006;357:1026–1038. doi: 10.1016/j.jmb.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Pérez R., Castellanos M., Mateu M.G. Molecular determinants of self-association and rearrangement of a trimeric intermediate during the assembly of a parvovirus capsid. J. Mol. Biol. 2011;413:32–40. doi: 10.1016/j.jmb.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 40.Hernando E., Llamas-Saiz A.L., Almendral J.M. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology. 2000;267:299–309. doi: 10.1006/viro.1999.0123. [DOI] [PubMed] [Google Scholar]
- 41.Reguera J., Carreira A., Mateu M.G. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carreira A., Menéndez M., Mateu M.G. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Biol. Chem. 2004;279:6517–6525. doi: 10.1074/jbc.M307662200. [DOI] [PubMed] [Google Scholar]
- 43.Silva J.L., Weber G. Pressure-induced dissociation of brome mosaic virus. J. Mol. Biol. 1988;199:149–159. doi: 10.1016/0022-2836(88)90385-3. [DOI] [PubMed] [Google Scholar]
- 44.Da Poian A.T., Oliveira A.C., Weber G. Reversible pressure dissociation of R17 bacteriophage. The physical individuality of virus particles. J. Mol. Biol. 1993;231:999–1008. doi: 10.1006/jmbi.1993.1347. [DOI] [PubMed] [Google Scholar]
- 45.Zlotnick A. Are weak protein-protein interactions the general rule in capsid assembly? Virology. 2003;315:269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 46.Katen S.P., Chirapu S.R., Zlotnick A. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem. Biol. 2010;5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuma R., Tsuruta H., Prevelige P.E. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J. Mol. Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knapman T.W., Morton V.L., Ashcroft A.E. Determining the topology of virus assembly intermediates using ion mobility spectrometry-mass spectrometry. Rapid Commun. Mass Spectrom. 2010;24:3033–3042. doi: 10.1002/rcm.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uetrecht C., Barbu I.M., Heck A.J. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat. Chem. 2011;3:126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 50.Ivanovska I.L., de Pablo P.J., Wuite G.J. Bacteriophage capsids: tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roos W.H., Wuite G.J.L. Nanoindentation studies reveal material properties of viruses. Adv. Mater. 2009;21:1187–1192. [Google Scholar]
- 52.Roos W.H., Bruinsma R., Wuite G.J.L. Physical virology. Nat. Phys. 2010;6:733–743. [Google Scholar]
- 53.Carrasco C., Carreira A., de Pablo P.J. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. USA. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrasco C., Castellanos M., Mateu M.G. Manipulation of the mechanical properties of a virus by protein engineering. Proc. Natl. Acad. Sci. USA. 2008;105:4150–4155. doi: 10.1073/pnas.0708017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newcomb W.W., Brown J.C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plomp M., Rice M.K., Malkin A.J. Rapid visualization at high resolution of pathogens by atomic force microscopy: structural studies of herpes simplex virus-1. Am. J. Pathol. 2002;160:1959–1966. doi: 10.1016/S0002-9440(10)61145-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roos W.H., Radtke K., Wuite G.J. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. USA. 2009;106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanovska I.L., Miranda R., Schmidt C.F. Discrete fracture patterns of virus shells reveal mechanical building blocks. Proc. Natl. Acad. Sci. USA. 2011;108:12611–12616. doi: 10.1073/pnas.1105586108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramírez J.C., Santarén J.F., Almendral J.M. Transcriptional inhibition of the parvovirus minute virus of mice by constitutive expression of an antisense RNA targeted against the NS-1 transactivator protein. Virology. 1995;206:57–68. doi: 10.1016/s0042-6822(95)80019-0. [DOI] [PubMed] [Google Scholar]
- 60.Gardiner E.M., Tattersall P. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J. Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Herrero F.J., de Pablo P., Baró A. Scanning force microscopy jumping and tapping modes in liquids. Appl. Phys. Lett. 2002;81:2620–2622. [Google Scholar]
- 62.Horcas I., Fernández R., Baro A.M. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007;78:013705–013708. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 63.Kontou M., Govindasamy L., Agbandje-McKenna M. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sader J.E., Chon J.W.M., Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 1999;70:3967–3969. [Google Scholar]
- 65.Schaap I.A.T., Carrasco C., Schmidt C.F. Elastic response, buckling, and instability of microtubules under radial indentation. Biophys. J. 2006;91:1521–1531. doi: 10.1529/biophysj.105.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carrillo-Tripp M., Shepherd C.M., Reddy V.S. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37(Database issue):D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


