Abstract
Voltage-dependent ion channels open and close in response to changes in membrane electrical potential due to the motion of their voltage-sensing domains (VSDs). VSD charge displacements within the membrane electric field are observed in electrophysiology experiments as gating currents preceding ionic conduction. The elementary charge motions that give rise to the gating current cannot be observed directly, but appear as discrete current pulses that generate fluctuations in gating current measurements. Here we report direct observation of gating-charge displacements in an atomistic molecular dynamics simulation of the isolated VSD from the KvAP channel in a hydrated lipid bilayer on the timescale (10-μs) expected for elementary gating charge transitions. The results reveal that gating-charge displacements are associated with the water-catalyzed rearrangement of salt bridges between the S4 arginines and a set of conserved acidic side chains on the S1–S3 transmembrane segments in the hydrated interior of the VSD.
Voltage-sensing domains (VSDs) regulate the opening of voltage-dependent ion channels in response to changes in membrane electrical potential. They are composed of four transmembrane segments (S1–S4) and their overall architecture is conserved from Archaea to mammals (1–3). The main voltage-sensitive element in VSDs is the S4 segment, which contains four-to-eight highly conserved triplet repeats consisting of one positively charged residue (mostly arginine) followed by two hydrophobic residues. Displacements of these charges within the membrane electric field after a sudden change in the membrane potential are measured as capacitive currents, termed gating currents, in electrophysiology experiments (4). The elementary charge motions that give rise to the gating current appear as shot-like events in high-bandwidth recordings of gating current fluctuations, but cannot be observed directly due to the limitations of available recording technology (5–7). To characterize the conformational dynamics of the VSD under an applied membrane potential, we performed all-atom molecular dynamics simulations of the isolated Kv VSD from Aeropyrum pernix (KvAP) in a 1-palmitoyl-2-oleoyl-phosphatidylcholine bilayer in excess water under a constant electric field on the timescale (10-μs) expected for elementary gating charge transitions (5,6). The results suggest that shot-like events of gating current are associated with changes in the solvation environment of the S4 arginines as they move through the hydrated interior of the VSD.
We modeled the membrane potential based on a one-dimensional linearized Poisson-Boltzmann calculation performed along the transmembrane direction through the center of the VSD (8). The magnitude of the constant electric field corresponds to a potential drop of 120 mV over the linear region of the electrostatic potential profile (shown in Fig. S1 in the Supporting Material). The time evolution of the VSD total charge displacement (defined in Methods in the Supporting Material) with respect to the initial configuration is shown in Fig. 1 A.
Figure 1.
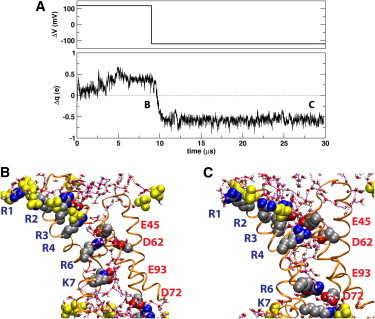
(A) (Upper panel) Time-dependence of applied membrane potential, ΔV, relative to the intracellular side of the membrane. (Lower panel) Total charge displacement within the membrane electric field with respect to the initial configuration. Switching the membrane potential from a depolarizing to a hyperpolarizing potential after 9 μs produces ∼1e of net gating charge. (B and C) Snapshots of the VSD at two membrane potentials: (B) Depolarized (ΔV = 120 mV) VSD after 8.5 μs. R4–K7 are shifted outwards with respect to the unpolarized trajectory. (C) Hyperpolarized (ΔV = –120 mV) VSD after 28.35 μs. The labels in panel A indicate the location of the snapshots within the trajectory. The VSD is shown in secondary structure representation. The conserved charged side chains (shown as filled spheres) are colored by atom type (carbon, silver; oxygen, red; nitrogen, blue). The labels (in blue) of the basic side chains in S4 and the S4-S5 linker follow the order of the triplet repeats starting at the extracellular end (corresponding to R117, R120, R123, R126, R133, and K136 in the KvAP sequence). The conserved positions for acidic side chains are labeled (in red) according to the KvAP sequence. An additional acidic side chain (E107) that forms a persistent salt-bridge with R3 is also shown. Lipid phosphate groups in the first coordination shell of the protein (filled yellow spheres). Water molecules in the first coordination shell of the protein are shown in licorice representation colored by atom type (hydrogen, white). Water-water hydrogen bonds are drawn as broken lines. For clarity, the hydrogen atoms in the protein side chain are not shown.
Under a depolarizing membrane potential (i.e., positive on the intracellular side), the total gating charge displacement increases steadily after ∼2 μs and reaches a plateau after ∼5 μs (Fig. 1). The underlying conformational change involves a substantial movement of the S3b-S4 helix-turn-helix (paddle) motif relative to S1-S2 (see Fig. S2) without any significant perturbation of the lipid bilayer (see Fig. S3). The resulting configuration of the paddle motif appears to be consistent with existing KvAP biotin-avidin accessibility data (9,10) (see Fig. S4). A comparison with a 5-μs unpolarized control simulation reveals an outward shift of all the basic side chains in S4 and the S4-S5 linker that were not already embedded in the lipid bilayer headgroup region in the initial configuration (8,11) (see Fig. 1 B and Fig. S5). At the end of this conformational change, the first four arginines in S4 (R1–R4) are exposed to the lipid bilayer headgroup region. The innermost arginine in S4 (R6) moves from the center of the VSD to the bottom of the extracellular crevice and forms a (mostly water-mediated) hydrogen-bond interaction with E45, a conserved acidic residue in the S1 segment. A lysine in the center of the S4-S5 linker (K7) is engaged in salt-bridge with E93, a conserved acidic residue in the S3a segment (see Fig. 1 B).
Switching the sign of the applied electric field to hyperpolarize the membrane after 9 μs induced a net gating charge displacement (corresponding to a shot of gating current) of ∼1e that occurred over 1.6 μs (Fig. 1). The concerted motion of R4, R6, and K7 (Fig. 1 C) contributes 75% of the total gating charge (see Fig. S6). Specifically, R6 traversed the interior of the VSD from the bottom of the extracellular crevice to the center of the intracellular crevice (Fig. 2), whereas R4 moved from the lipid headgroup region into the extracellular crevice, and K7 moved from the intracellular crevice into the lipid headgroup region on the intracellular side (see Fig. S7). Under the hyperpolarizing potential, R4 is engaged in a salt-bridge with E45 and a water-mediated hydrogen bond with D62, while R6 forms a persistent salt-bridge interaction with E93 and D72 (Figs. 1 C and Fig. 2, and see Fig. S7). R1–R3, which remain within the extracellular headgroup region throughout the 30-μs simulation trajectory (Fig. 1), only contribute ∼5% to the gating charge (see Fig. S6). Thus, the gating-charge displacement that we observed is associated with the water-catalyzed rearrangement of salt bridges between the S4 basic side chains and the set of conserved acidic side chains on the S1–S3 transmembrane segments in the hydrated interior of the VSD. The full activation of the VSD appears to be required for the generation of the gating charge event, because a trajectory initiated under a hyperpolarized potential without preequilibration under a depolarized potential failed to produce the incursion of R6 into the intracellular crevice within 14 μs (see Fig. S8).
Figure 2.
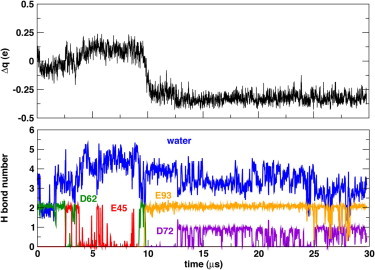
Contribution of R6 to the total charge displacement (top panel) follows closely the changes in solvation environment as the side chain moves from the center of the VSD into the extracellular cavity (under a depolarizing potential) and from the extracellular cavity to the intracellular cavity (under a hyperpolarizing potential).
Salt-bridges between conserved charged side chains have been identified as a key structural feature in VSD folding and function (12–16), and their role in the gating mechanism has been suggested in previously reported molecular dynamics simulations run for shorter times (17,18). Our results indicate that specific contributions by S4 basic side chains to the gating charge are primarily dependent on their interactions with the local electric field, which is shaped by the overall solvation of the VSD (8).
The experimental evidence regarding contributions to the gating charge by specific side chains in eukaryotic Kv channels suggests that R1–R4 carry out most of the gating charge during activation (19–21). The main determinant of the voltage gating function is the conformation of S4 in the membrane, as clearly illustrated by the functional analysis on several Shaker deletion mutants reported by Xu et al. (22), who found that only a minimum of two S4 triplet repeats (out of five in wild-type Shaker) were necessary to maintain the voltage-gating function, irrespective of their specific location in the protein sequence. In contrast to the KvAP VSD, the Kv1.2 paddle-chimera structure (2) shows the S4 segment spanning the whole transmembrane region, whereas R6 is located at end of the intracellular crevice as part of a sharp kink that leaves the S4-S5 link parallel to the membrane surface and R4 is located near the constriction separating the VSD crevices in an equivalent location to R6 in KvAP (see Fig. S9 and Long et al. (2)). This configuration strongly suggests that R6 would not cross the membrane electric field during gating in Kv1 channels, a notion consistent with the current view on VSD movements in relation to channel opening in Kv1 channels (23,24).
Our observation of a single charge displacement event over a 30-μs sampling interval is compatible with electrophysiological measurements on eukaryotic channels. A fast early component of the gating current with a time constant of ∼12 μs has been detected for the Shaker channel (25). The main component of the gating current in Shaker and Nav1.2 has been estimated to consist of at least one discrete transition carrying 2–3e of charge (5,6). However, these discrete events can only be resolved as charge packets that may not represent a single charge displacement event but rather the superposition of many events occurring faster than the instrument response (5–7). Our simulation suggests that concerted motions of S4 arginines generate displacements of ∼1e charge on a timescale of ∼1 μs within a single VSD.
Acknowledgments
This work was supported by the National Institutes of Health grants No. GM74637 and GM86685 and National Science Foundation grant No. CHE-0750175. The computations were carried out on the Anton computer at the National Resource for Biomedical Supercomputing at the Pittsburgh Supercomputing Center, which is funded by National Institutes of Health grant No. RC2GM093307.
Supporting Material
References and Footnotes
- 1.Jiang Y.X., Lee A., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 2.Long S.B., Tao X., MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 3.Payandeh J., Scheuer T., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong C.M., Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 5.Conti F., Stühmer W. Quantal charge redistributions accompanying the structural transitions of sodium channels. Eur. Biophys. J. 1989;17:53–59. doi: 10.1007/BF00257102. [DOI] [PubMed] [Google Scholar]
- 6.Sigg D., Stefani E., Bezanilla F. Gating current noise produced by elementary transitions in Shaker potassium channels. Science. 1994;264:578–582. doi: 10.1126/science.8160016. [DOI] [PubMed] [Google Scholar]
- 7.Crouzy S.C., Sigworth F.J. Fluctuations in ion channel gating currents. Analysis of nonstationary shot noise. Biophys. J. 1993;64:68–76. doi: 10.1016/S0006-3495(93)81341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krepkiy D., Mihailescu M., Swartz K.J. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y.X., Ruta V., MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 10.Ruta V., Chen J., MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 11.Freites J.A., Tobias D.J., White S.H. A voltage-sensor water pore. Biophys. J. 2006;91:L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papazian D.M., Shao X.M., Wainstock D.H. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari-Woodruff S.K., Schulteis C.T., Papazian D.M. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Sato Y., Uozumi N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. USA. 2007;104:8263–8268. doi: 10.1073/pnas.0611007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D., Delaloye K., Cui J. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 2010;135:595–606. doi: 10.1085/jgp.201010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeCaen P.G., Yarov-Yarovoy V., Catterall W.A. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. USA. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalili-Araghi F., Jogini V., Schulten K. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 2010;98:2189–2198. doi: 10.1016/j.bpj.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delemotte L., Tarek M., Treptow W. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2011;108:6109–6114. doi: 10.1073/pnas.1102724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal S.K., MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 20.Seoh S.A., Sigg D., Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 21.Ahern C.A., Horn R. Specificity of charge-carrying residues in the voltage sensor of potassium channels. J. Gen. Physiol. 2004;123:205–216. doi: 10.1085/jgp.200308993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Ramu Y., Lu Z. A Shaker K+ channel with a miniature engineered voltage sensor. Cell. 2010;142:580–589. doi: 10.1016/j.cell.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao X., Lee A., MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacroix J.J., Bezanilla F. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. USA. 2011;108:6444–6449. doi: 10.1073/pnas.1103397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigg D., Bezanilla F., Stefani E. Fast gating in the Shaker K+ channel and the energy landscape of activation. Proc. Natl. Acad. Sci. USA. 2003;100:7611–7615. doi: 10.1073/pnas.1332409100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.-Y., Lee A., MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw D.E., Chao J.C., Bowers K.J. Anton, a special-purpose machine for molecular dynamics simulation. ACM SIGARCH Computer Arch. News. 2007;35:1. [Google Scholar]
- 28.MacKerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 29.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan Y., Klepeis J.L., Shaw D.E. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005;122:054101–054113. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


