Abstract
The life of an mRNA from transcription to degradation offers multiple control check points that regulate gene expression. Transcription, splicing, and translation have been widely studied for many years; however, in recent years, new layers of posttranscriptional and posttranslational control have been uncovered. They involve the regulation of the metabolism of mRNA in cytoplasmic foci. They are collections of ribonucleoprotein complexes that, in most cases, remain still uncharacterized, except the processing bodies (PBs) and stress granules (SGs), which have been studied (and reviewed) in detail. A challenging prospective is to know how many different classes of foci exist, which functions they support, how are they formed, and how do they relate one to each other. Here, we present an update of the component of the different granules, a possible function, and hypothesis on their in vivo dynamics related to translational control.
1. Introduction
In recent years, several cytoplasmic foci/granules that contain proteins and RNA have been described. Two of them have been studied in more detail as they are related to mRNA silencing: stress granules (SG) and processing bodies (PB). SG are repressed mRNPs transiently induced in response to cellular stress. They range from 0,5 to 5 μm [1]. PB are discrete RNP cytoplasmic foci of 0,1-2 μm where the machinery of RNA interference, degradation and storage locates. In PB the mRNAs are forming mRNP complexes either repressing translation, in degradation complexes or stored for further use [2, 3]. SG and PB have been shown to share a growing number of proteins that are added in a day-to-day basis to the list of their components. SG, PB and other cytoplasmic foci are highly dynamic structures, although PB are quite stable over the time [4]; see also Supplementary Movie 1 available online at doi:10.1155/2012/504292. They are in a dynamic steady state with other mRNPs, such as polysomes in response to the translational state of the cell [5]. Although we do not intend to extensively review SG and PB, which have been matter of fine reviews in the last years [6–10], we will overview their functions before we address neglected issues and hypothesis.
2. Stress Granules
Translation initiation is the key regulatory step of translational control. Therefore, it is the most sensitive step to changes in the cellular environment, including stress. A key step in translation initiation inhibition is the phosphorylation of eIF2α, which results in an increase on the affinity of eIF2-GDP for eIF2B, sequestering this factor to prevent new round of translational initiation [11]. During this process, translation is inhibited and polysomes become released from the mRNA leading to the accumulation of inactive mRNPs in SG. The SG are in equilibrium with active polysomes. Protein elongation inhibitors, such as cycloheximide, prevent the assembly of SG by blocking the polysomes in an inactive state, while protein initiation inhibitors promote the formation of SG [8]. Table 1 shows the components of SG described up to now. They can be classified in three main groups as follows.
Table 1.
Components of stress granules.
| Protein | Function | Interacting proteins |
|---|---|---|
| Ago2 | Cleaves interfered RNA | RISC, FXR1 |
| APOBEC3G | Antiviral response | ? |
| Ataxin-2 | Translation | PABP-1 |
| Caprin-1 | Cell growth | G3BP |
| CPEB | mRNA repression | RCK, eIF4E, FXR1 |
| DIS1 | Unknown | eIF3h |
| eIF3 | Translation | 40S, eIF4G |
| eIF4E | Translation | CPEB, Smaug, eIF4G, 4ET |
| eIF4G | Translation | eIF4E, eIF3, PABP-1 |
| FAST | Translation | TIA-1 |
| FMRP, FXR1 | Translation | Ago2, RISC |
| FBP, KSRP | mRNA degradation | TIA-1 |
| FUS/TLS | Transcriptional control | Transcriptional machinery |
| G3BP | Ras signalling | Caprin |
| HuR | mRNA stabilization | ? |
| IP5K | Signalling | ? |
| Lin28 | Developmental control | ? |
| LINE 1 ORF1p | Transposon | ? |
| MLN51 | Splicing | Exon junction |
| PABP-1 | Translation | eIF4G, eIF3, ataxina-2 |
| RCK(p54) | mRNA degradation | GE-1, TTP |
| Plakophilin | Adhesion | G3BP, FXR1 |
| PMR1 | mRNA degradation | TIA-1 |
| Pumilio 2 | mRNA silencing | ? |
| Rap 55 | mRNA silencing | ? |
| Rpb4 | Transcription | ? |
| SRC3 | Transcription | TIA-1 |
| Staufen | mRNA silencing | ? |
| SMN | RNP assembly | Complejo SMN |
| TDP-43 | Transcription and splicing regulator | eIF4G, eIF3, eIF2, ribosomal proteins, STAU-1, Xnr |
| TIA-1(rox-8), TIAR | mRNA silencing | FAST, SRC3, PMR1, FBP |
| TRAF2 | Signalling | eIF4G |
| TTP, BRF-1 | mRNA silencing | RCK (p54) |
| YB-1 | Cold shock | ? |
| ZBP1 | Localization | ? |
(1) Core components: stalled initiation complexes (polyadenylated mRNAs and translation factors eIF4E, eIF4A, eIF4G, eIF3, eIF2, PABP, and proteins of the small ribosome subunit).
(2) RNA-binding proteins associated to silencing and transcript stability: TIA-1, TIAR [12], FAST, Argonaute [13], CPEB, smaug, DExD/H-box RCK/p54 (o Dhh1), XRN1 [5].
(3) RNA-binding proteins associated to mRNA metabolism either translation of degradation such as G3BP [14] and Staufen [15].
The key concept regarding SG is that they are responsible of protecting the mRNA during cell stress, altering the composition of the mRNPs in a reversible manner. As soon as the cell recovers, the mRNPs regain their translational capacity.
3. Processing Bodies
These structures have been described many times since 1997, when Bashkirov et al. observed that the exonuclease Xrn1 is located in small granular structures in the cytoplasm of mammalian cells and call them “Xrn1 foci” [17]. Later on, the decapping enzyme Dcp2 was also described to occur in cytoplasmic foci [18]. Contemporary, Eystathioy et al. have described that a protein associated to neuropathy named GW182 occurs in cytoplasmic speckles called GW bodies [19, 20]. Other RNA-related protein, the eIF4E-transporter, was also localized in discrete cytoplasmic foci [16, 21, 22]. Short after, a seminal work of Sheth and Parker established the functional bases of the now called PB that resulted in the same structures described many times before [23]. They demonstrated that PB contains enzymes involved in the degradation of the mRNA. Later one further studies showed that they are also related to miRNA metabolism and can store mRNAs to bring them back to polysomes (reviewed in [3, 24]. They include, different than SG, neither ribosomal proteins nor translation factors, except eIF4E. They do not present either the exosome components [25]. eIF4G and PABP were found in yeast PB, although at low level and in stress conditions resulting on glucose deprivation [26]. Proteins and mRNA can reversely go in and out of PB [25]. The relationship of PB and polysomes is demonstrated by the blocking of PB formation by cycloheximide. A summary of the components in different organisms is shown in Table 2. The occurrence of such large and diverse set of proteins (and the list continuously grows up) suggests that PBs are involved in a plethora of posttranslational processes regulating gene expression, such as mRNA degradation and silencing. mRNA degradation starts with the shortening of the poly-A tail—the deadenylation. In eukaryotes, there are several complexes involved in the process: PARN2-PARN3 initiates the process, which continues with the action of the CAF1-CCR4-NOT complex. Later on, mRNA degradation continues by nucleolytic cleavage on both ends. 3′ → 5′ degradation is catalyzed by the exosome and the SKI complex, while 5′ → 3′ degradation requires previous decapping by DCP2 and the coactivator DCP1 and the action of the exonuclease XRN1. All these enzyme localize in PB. There are several evidences indicating that mRNA degradation occurs in PB.
Table 2.
Components of processing bodies.
| Protein | Function | Organisms |
|---|---|---|
| XRN1, Sc Kem1 | 5′ → 3′ exonuclease | Human, mice, Sc |
| GW182, Ce AIN-1 | miARN function | Human, Dm, Ce |
| DCP2, Ce DCAP2 | Decapping | Human, Dm, Ce, Sc |
| DCP1, Ce DCAP1 | Decapping | Human, Dm, Ce, Sc |
| Hedls, Ge-1 | Decapping coactivator | Human, Dm |
| Dm CG5208, Pat1 | Decapping coactivator | Dm, Sc |
| EDC3 (Lsm16) | Decapping coactivator | Human, Dm, Sc |
| Lsm1-7 | Decapping coactivator | Human, Sc |
| RAP55 | Putative decapping coactivator | Human |
| RCK/p54, Dm Me31B, Ce CGH-1, Sc Dhh1 | Decapping coactivator, translational control | Human, Dm, Ce, Sc |
| eIF4E | Translation initiation | Human, rat, mouse, Dm, Sc |
| eIF4E-T | Translational repression | Human |
| SMG7 | Nonsense mediated decay | Human |
| SNG5 | Nonsense mediated decay | Human |
| UPF1, Sc Nam7 | Nonsense mediated decay | Human, Sc |
| UPF2 | Nonsense mediated decay | Human |
| UPF3 | Nonsense mediated decay | Human |
| Argonaute | siRNA/miRNA pathways | Human, Dm, Ce |
| CCR4-CAF1-NOT complex | Deadenylation | Human, Dm, Sc |
| CPEB | Translational control | Human |
| FAST | S/T phosphoprotein activator of Fas | Human |
| TTP | ARE-mediated mRNA degradation | Human |
| Staufen | mRNA localization | Human, mice, Dm |
| Rbp1 | Mitochondrial RNA degradation | Sc |
| Rbp4 | RNA pol II subunit | Sc |
| Sbp1 | Suppressor of deccaping | Sc |
| Germin 5 | Part of small nuclear RNPs | Human |
| Dcs2 | Stress-induced regulator | Sc |
| APOBEC3G, APOBEC3F | Antiviral activity | Human |
(i) The assembly of PB depends on mRNA, as RNase treatment of the cells induces the disappearing of PB [27, 28].
(ii) Inhibition or removal of the deadenylase Ccr4 reduces the number and size of PB, while the removal of the downstream-acting enzymes Xrn1 and Dcp1 does not affect the stability of PB [21].
(iii) mRNA degradation intermediates are present in PB [23].
Therefore, one can conclude that mRNA degradation occurs in PB and depends on the existence of degradation enzymes and mRNA degradation intermediates [21, 23, 25, 29]. Many of the PB components are not restricted to the foci and also are present in the soluble cytoplasm and nuclei, suggesting that the different processes might start before the mRNAs entry into PB. PBs are also related to mRNA quality control mechanisms, such as nonsense-mediated decay (NMD). The detection of premature termination in the cells by spotting an mRNA with an abnormal stop codon is mediated by a surveillance complex composed by UPF1, UPF2 y UPF3, additional proteins, namely, SMG1, and SMG5-7 [30–32]. As soon as the surveillance complex is assembled, the degradation enzymes (Dcp1, Xrn1) are recruited to the mRNA in PB. Although the degrading enzymes are located in PB, the mechanism of recruitment is unknown. In silencing, there are two types of small mRNAs that regulate posttranscriptional gene expression: siRNAs and miRNAs. Despite the different mechanism of silencing, in both cases participate the protein Argonaute (Ago) and the RISC (RNA-induced silencing complex). In the case of siRNAs, Ago produces an endonucleolytic cleavage of the mRNA to promote degradation by the 3′ → 5′ and 5′ → 3′ decay machinery in PB. In the case of the miRNAs, they recruit Ago to direct the repressed mRNA to degradation mediated by the PB proteins GW182, CCR4-CAF1-NOT1, DCP2, DCP1, and XRN1.
4. The Cycle of an mRNA in the Cell: SG, PB, Polysomes, and the Unknown Intermediates
From the previous analysis, one can establish many unsolved aspects on cytoplasmic foci function. One of them is the dynamic of the mRNP remodeling. The current model suggests an active movement of mRNPs from and to polysomes and from and to SG and PB [33]. However, how does it happen and the factors involved are not known. Translationally active mRNAs can interact, in response to errors in translational initiation or to specific recruitment of regulatory proteins, with translational repressors such as Dhh1, Pat 1, Lsm1-7, eIF4E-T. Those factors would promote the replacement of the translational machinery from the mRNA, promote the cap removal and determine degradation [33] or the accumulation of silenced mRNA in PB. Within PB, mRNPs could undergo further remodeling and define a path to follow, including their return to polysomes. In addition, PBs have been shown to interact and exchange components or their own nature with SG (reviewed in [6, 7]) in a process that may result in mRNPs intermediates of unknown nature. Evidence for the diversity of cytoplasmic foci and their components results from immunocytochemistry and colocalization studies. A common factor present in most cytoplasmic mRNPs is the cap-binding protein eIF4E. eIF4E occurs in active polysomes as a translation initiation factor, in SG as part of the stalled initiation complex, and in PB as the only translation factor present there in multicellular eukaryotes. We observed in Drosophila S2 cells that eIF4E colocalizes with different pairs of markers, either for PB (GW182, Lsm1, Me31B—an ortholog of the helicase rck/p54) or SG (TIA-1) and that the colocalization does not occur in all foci in the same way (PVF, CL, and RRP, unpublished data and Figure 1). In some cases, the foci contain one, the other, or both components. In the foci that show colocalization of both factors, the relative amount of each component may vary from foci to foci, as judged by confocal microscopy quantification of the colocalized factors (PVF, CL, RRP, unpublished observation). This implies that there are a diversity of granules. An appealing hypothesis is that eIF4E is a common link among different mRNPs, playing different roles depending on their interactors. One plausible function could be that the accumulation of mRNPs in eIF4E-containing foci is a way to regulate the rate of translation in different physiological states (cell cycle phases, developmental stages, circadian rhythms). Moreover, it has been reported that, in mammalian cells, eIF4E interacts in PB with at least two factors, rck/p54 and eIF4E-T [21]. These are simultaneous interactions within the PB and imply that both proteins could contact different domains of the same eIF4E molecule or that they would represent different populations of mRNPs or different functions within the same PB. In either cases, the complexity of the interactions in vivo is more diverse than it has been expected. A model for the remodeling of active mRNPs to silence and degradation based on Andrei et al. [21] is depicted in Figure 2. This might require several intermediate states that can be the maturation steps of a mRNA in the way of a PB or within a PB. This would correlate with the large diversity of components and interactions within a cytoplasmic foci and the diversity of the foci within a cell. The understanding of the dynamics of mRNP is far from clear and unpredictable paths remain to be discovered. They will need further research and more sophisticated methods for in vivo studies.
Figure 1.
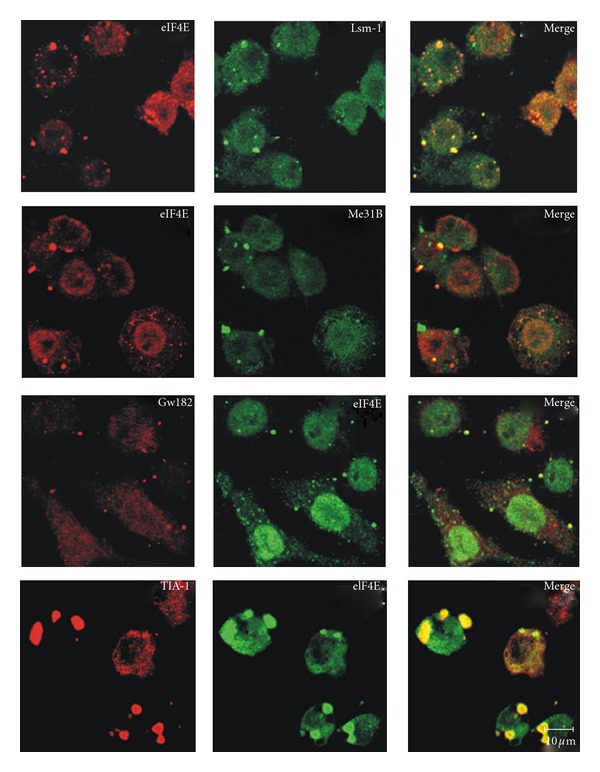
Colocalization of eIF4E with components of PBs and SGs shows a diversity of cytoplasmic foci quality. The experiment shows that, in every case, the granules contain both components or either one or the other in different quantities. This would represent intermediates or different forms of eIF4E-containing foci. Drosophila melanogaster S2 cells were transfected with proteins fusion CFP-Lsm-1 or CFP-Me31B. GW182, eIF4E, and Rox8 (the TIA-1 ortholog in Drosophila) were revealed using antibodies against GW182, anti-V5, and anti-TIA-1, respectively. In the bottom panel (row 4), the cells were prestressed with arsenite for 30 minutes.
Figure 2.
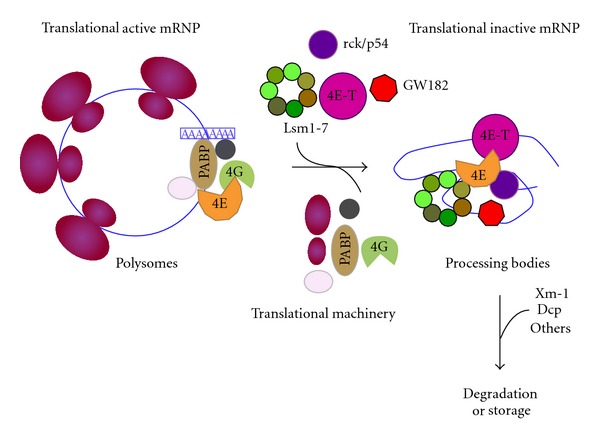
Relationship among active polysomes and PBs. The recruitment of active polysomes to PB implies the removal from the mRNA of the translation factors by translational repressors. Some of them have been demonstrated to interact with eIF4E in vivo (rck/p54 and eIF4E-T, [16]). They further interact and/or recruit the enhancers of decapping Lsm1-7 or the miRNA-related protein GW182 to form the PB. Later on, they assemble the decapping and degradation enzymes and/or the proteins required for silencing and storage into PB. All the intermediate steps of this process can represent different populations of granules coexisting in the cell and visible with different morphology that might reflect a variety of components and/or diverse stoichiometry.
Supplementary Material
Colocalization of eIF4E with components of PBs and SGs shows a diversity of cytoplasmic foci quality. The experiment shows that, in every case, the granules contain both components or either one or the other in different quantities. This would represent intermediates or different forms of eIF4E-containing foci. Drosophila melanogaster S2 cells were transfected with proteins fusion CFP-Lsm-1 or CFP-Me31B. GW182, eIF4E, and Rox8 (the TIA-1 ortholog in Drosophila) were revealed using antibodies against GW182, anti-V5, and anti-TIA-1, respectively. In the bottom panel (row 4), the cells were prestressed with arsenite for 30 minutes.
Relationship among active polysomes and PBs. The recruitment of active polysomes to PB implies the removal from the mRNA of the translation factors by translational repressors. Some of them have been demonstrated to interact with eIF4E in vivo (rck/p54 and eIF4E-T, [20]). They further interact and/or recruit the enhancers of decapping Lsm1-7 or the miRNA-related protein GW182 to form the PB. Later on, they assemble the decapping and degradation enzymes and/or the proteins required for silencing and storage into PB. All the intermediate steps of this process can represent different populations of granules coexisting in the cell and visible with different morphology that might reflect a variety of components and/or diverse stoichiometry.
Acknowledgments
This work was supported by grants from CONICET (PIP 00318 to P. Ferrero) and ANPCyT (PICT-2008-1237 to RRP and P. Ferrero). P. Ferrero and R. Rivera-Pomar are investigators and CL doctoral fellow of the CONICET.
References
- 1.Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. Mammalian staufen 1 is recruited to stress granules and impairs their assembly. Journal of Cell Science. 2009;122(4):563–573. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan SP, Slack FJ. MicroRNA-mediated silencing inside P-bodies. RNA Biology. 2006;3(3):97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 3.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Molecular and Cellular Biology. 2007;27(11):3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aizer A, Shav-Tal Y. Intracellular trafficking and dynamics of P bodies. Prion. 2008;2(4):131–134. doi: 10.4161/pri.2.4.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Journal of Cell Biology. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cellular Signalling. 2011;23(2):324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Molecular Cell. 2009;36(6):932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends in Biochemical Sciences. 2008;33(3):141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nature Reviews Molecular Cell Biology. 2007;8(1):9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 10.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Molecular Cell. 2007;25(5):635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Current Opinion in Cell Biology. 2008;20(2):222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. Journal of Cell Biology. 1999;147(7):1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tourrière H, Chebli K, Zekri L, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. Journal of Cell Biology. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Thomas MG, Martinez Tosar LJ, Loschi M, et al. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Molecular Biology of the Cell. 2005;16(1):405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. Journal of Cell Biology. 2000;148(2):239–246. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. Journal of Cell Biology. 1997;136(4):761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. The European Molecular Biology Organization Journal. 2002;21(24):6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Molecular Biology of the Cell. 2002;13(4):1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eystathioy T, Jakymiw A, Chan EK, Séraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9(10):1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11(5):717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. Journal of Cell Biology. 2005;170(6):913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300(5620):805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Molecular Cell. 2007;25(5):635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310(5747):486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae . Molecular Biology of the Cell. 2007;18(7):2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde J-M, Heyer W-D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. The Journal of Cell Biology. 1997;136(4):761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA. 2002;8(12):1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 29.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. Journal of Cell Biology. 2004;165(1):31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Current Opinion in Cell Biology. 2005;17(3):316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nature Reviews Molecular Cell Biology. 2006;7(6):415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 32.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Current Opinion in Cell Biology. 2005;17(3):309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Current Opinion in Cell Biology. 2009;21(3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colocalization of eIF4E with components of PBs and SGs shows a diversity of cytoplasmic foci quality. The experiment shows that, in every case, the granules contain both components or either one or the other in different quantities. This would represent intermediates or different forms of eIF4E-containing foci. Drosophila melanogaster S2 cells were transfected with proteins fusion CFP-Lsm-1 or CFP-Me31B. GW182, eIF4E, and Rox8 (the TIA-1 ortholog in Drosophila) were revealed using antibodies against GW182, anti-V5, and anti-TIA-1, respectively. In the bottom panel (row 4), the cells were prestressed with arsenite for 30 minutes.
Relationship among active polysomes and PBs. The recruitment of active polysomes to PB implies the removal from the mRNA of the translation factors by translational repressors. Some of them have been demonstrated to interact with eIF4E in vivo (rck/p54 and eIF4E-T, [20]). They further interact and/or recruit the enhancers of decapping Lsm1-7 or the miRNA-related protein GW182 to form the PB. Later on, they assemble the decapping and degradation enzymes and/or the proteins required for silencing and storage into PB. All the intermediate steps of this process can represent different populations of granules coexisting in the cell and visible with different morphology that might reflect a variety of components and/or diverse stoichiometry.


