Abstract
Links between substance use habits, obesity, stress and the related cardiovascular outcomes can be, in part, because of loci with pleiotropic effects. To investigate this hypothesis, we performed genome-wide mapping in 119 multigenerational families from a population in the Saguenay-Lac-St-Jean region with a known founder effect using 58 000 single-nucleotide polymorphisms and 437 microsatellite markers to identify genetic components of the following factors: habitual alcohol, tobacco and coffee use; response to mental and physical stress; obesity-related traits; and heart rate (HR) and blood pressure (BP) measures. Habitual alcohol and/or tobacco users had attenuated HR responses to mental stress compared with non-users, whereas hypertensive individuals had stronger HR and systolic BP responses to mental stress and a higher obesity index than normotensives. Genetic mappings uncovered numerous shared genes among substance use, stress response, obesity and hemodynamic traits, including CAMK4, CNTN4, DLG2, FHIT, GRID2, ITPR2, NOVA1 and PRKCE, forming network of interacting proteins, sharing synaptic function and display higher and patterned expression profiles in brain-related tissues; moreover, pathway analysis of shared genes pointed to long-term potentiation. Subgroup genetic mappings uncovered additional shared synaptic genes, including CAMK4, CNTN5 and DNM3 (hypertension-specific); CNTN4, DNM3, FHIT and ITPR1 (sex-specific), having protein interactions with genes driven from general analysis. In summary, consistent with the observed phenotypic correlations, we found substantial overlap among genomic determinants of these traits in synapse, which supports the notion that the neural synapse may be a shared interface behind substance use, stress, obesity, HR, BP as well as the observed sex- and hypertension-specific genetic differences.
Keywords: obesity, stress, substance use, synapse
Introduction
Alcohol, tobacco and coffee are among the most commonly consumed psychoactive substances in the world; their concurrent use has been consistently shown across a wide variety of populations.1 Stress is also related to the initiation, intensification and relapse of substance use.2 In addition, both stress and substance use have been implicated in cardiovascular disease and cardiovascular risk factor phenotype, obesity.3, 4, 5, 6 Substance use, obesity, stress and cardiovascular disorders are complex traits that have multifactorial etiologies with substantial genetic components.1, 4, 6, 7, 8, 9 The HPA axis, the main component of body's response to stress, has been implicated in the etiology of these traits.10, 11, 12, 13, 14 For example, variations in the mu opioid receptor, a regulator of the HPA axis, influence the response to stress, food intake and substance use.3, 10, 14 However, the genetic determinants of and the relationships between these traits are still largely elusive, and further studies are required.
We hypothesized that the connections between substance use habits, obesity, stress and cardiovascular outcomes are, in part, because of shared genetic factors. To investigate this hypothesis, we performed genome-wide linkage and association scans in a cohort of 119 families from the founder population of Saguenay-Lac St-Jean, which was previously recruited to identify the genetic factors of hypertension, to identify the genetic determinants of the following: habitual alcohol, coffee and tobacco use; obesity-related anthropometric data; the cardiovascular components of the behavioral response to the mental stress of mathematical tests and changes in plasma catecholamines after an orthostatic test as biomarkers of responses to mental and physical stress; and 24-hour heart rate (HR) and blood pressure (BP).9 Furthermore, the identified genes were subjected to functional annotation to explore the underlying biological meanings.
Methods
Phenotypes
The details about the families and the results of extensive phenotyping were previously described.9 A total of 15% of the subjects (N=135) had received antihypertensive and/or lipid-lowering medications, which were stopped before the start of phenotyping protocol.9 Phenotyping for habitual substance use was performed using standard questionnaires; those using a substance on regular basis were grouped as affected, and those who never or occasionally use a substance were considered to be unaffected. HR and BP changes during the mental stress of mathematical tests were used as markers of the response to mental stress.9 The subjects were asked to pass a 2-min arithmetic test; in parallel, the subjects' HR, diastolic blood pressure (DBP) and systolic blood pressure (SBP) were measured every 5 min before the test (three times), every 2 min during the test (two times) and every 2 min after the test (six times). The stress response was defined as the difference between the first HR and BP values during the math test and the average-resting values before the test. For the orthostatic stress test, the plasma epinephrine (EP) and norepinephrine levels were monitored in blood samples taken from the subjects during 60 min in a supine position and 10 min in a standing position; these data along the difference between standing and supine positions (delta) were used as biomarkers of response to physical stress. Phenotyping for obesity and the hemodynamic measures were previously described.9
The generalized estimating equation (implemented in R statistical package version 2.6.1), which accounts for familial correlation using a sandwich estimator of the variance under exchangeable correlation, was used to measure the relatedness of the phenotypes.15 As significant portions of variation of studied traits were attributed to sex and hypertension status,16 sex- and hypertension-specific genetic mappings were also performed. In the general and hypertension-specific analysis, age and sex were included as covariates, and in the sex-specific analysis, only age was used as a covariate. Heritability was calculated using the SOLAR software version 4.2.0.17
Genotyping
Genotype information was previously described.9, 16, 18 In summary, 469 subjects were genotyped using the GeneChip Human Mapping 50 K Array Xba240, and 537 subjects were genotyped using 437 microsatellite markers. Altogether, the 719 subjects that were genotyped using microsatellites and/or single-nucleotide polymorphisms (SNPs) were analyzed in the present study. Genotyping errors were detected and filtered (∼0.2%) using the MERLIN (version 1.1.0)19 and PLINK software (Boston, MA, USA; version 1.06). SNPs with the parameters r2⩽0.8, HWE>0.001 and MAF>0.05 were included in analysis. The WGAviewr software (Durham, NC, USA; version 1.52Z) was used to identify nearby genes for each SNP by specifying up- and downstream spans of 500 kb.20
Genetic mapping
Merging the stable markers, SNPs and potentially unstable but highly informative microsatellite markers can increase the power of genetic analysis.21 Hence, to increase the power of current study, the microsatellite and SNP genotype information were merged, and a haplotype map was created by specifying r2>0.4.22 A multipoint linkage analysis was performed on the haplotype map using MERLIN and followed by the family-based association test (FBAT) in the FBAT software version v2.0.2c.23 Unlike case–control studies, family-based association is designed to test for association in the presence of linkage. Hence, a positive finding implies both linkage and association. In addition, family-based association tests are more immune to spurious association caused by population stratification compared with case–control designs.23
To identify specific loci for each trait, loci that were within a 1 LOD-drop interval of the linkage peak and that had at least two identically annotated SNPs associated with P<10−3 were preselected. Furthermore, for each SNP, P-values were corrected by the number of SNPs within the linkage area, and P-value-adjusted SNPs that were not significant (P>0.05) were excluded. For loci that were associated with multiple traits, SNPs with association P-value<10−3 were preselected. Combined P-values were calculated using Fisher's combined probability test, and P<1.4 × 10−6 was considered to be significant after a Bonferroni correction by the number of SNPs analyzed (N=34 741).
Functional annotations
Protein interaction data were extracted from the Ingenuity Pathway Analysis database version 8.0, and DAVID Functional Annotation Tool24 version 6 was used to identify significantly enriched KEGG pathways from the list of shared genes. Genes involved in a biological process display higher expression and patterned expression profiles in the tissues in which the biological process takes place.25 Therefore; we compared the expression profiles of the shared genes across 65 normal human tissues using data from COXPRESdb database26 version c3.1. Hierarchical clustering of the expression data was performed using the GenePattern software27 version 3.1.1 based on the complete linkage method and using the Pearson's correlation coefficient as a measure of distance. The synapse databases SynDB21 release 2006 and G2Cdb28 version 07 were used to query whether a gene has synaptic function.
Results
General phenotypic characteristics
The prevalence of hypertension was significantly higher in those who had never used tobacco (P=0.02) compared with former and current smokers (Supplementary Table S1). Hypertensive subjects had a higher obesity index (P<0.05) and a stronger HR (P=0.03) and SBP (P=0.02) response to mental stress compared with normotensive subjects (Supplementary Table S2). In all, 80% of the studied traits exhibit sexual dimorphism (P<0.05). Norepinephrine levels and the hemodynamic responses to mental stress were not significantly different between males and females (Supplementary Table S2). In addition, although BP increased with age, HR changes remained insignificant (Supplementary Table S2).
Sex- and hypertension status-based differences in the heritabilities of the studied traits were observed (Supplementary Table S4). For instance, the heritabilities of average sitting HR (87 vs. 34%) and average sitting BP (0.69 vs. 33% for DBP and 48 vs. 10% for SBP) were higher in normotensive than in hypertensive subjects (Table 1). Moreover, although genetic factors appeared to be more important in the initiation (females=81%, males=56%) and persistence of tobacco use (females=97%, males=23%) in females, the heritability of alcohol use was higher in males (85%) compared with females (63% Table 1).
Table 1. Examples of hypertension and sex status differences in heritabilities (H2r) of studied traitsa.
|
Males |
Females |
Normotensives |
Hypertensives |
All |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | N | H2r | N | H2r | N | H2r | N | H2r | N | H2r |
| HRb | 134 | 0.67 | 131 | 0.76 | 141 | 0.87 | 124 | 0.34 | 265 | 0.58 |
| DBPb | 134 | 0.53 | 133 | 0.25 | 141 | 0.69 | 126 | 0.33 | 267 | 0.41 |
| SBPb | 134 | 0.54 | 133 | 0.66 | 141 | 0.48 | 126 | 0.10 | 267 | 0.42 |
| Current vs. non-tobacco users | 377 | 0.39 | 320 | 0.62 | 323 | 0.26 | 374 | 0.45 | 697 | 0.39 |
| Former vs. never tobacco users | 291 | 0.56 | 224 | 0.81 | 216 | 0.71 | 299 | 0.43 | 515 | 0.61 |
| Current vs. former tobacco users | 187 | 0.23 | 220 | 0.97 | 191 | 0.18 | 216 | 0.62 | 407 | 0.46 |
| Alcohol use | 377 | 0.85 | 321 | 0.63 | 326 | 0.51 | 372 | 0.55 | 698 | 0.59 |
Abbreviations: DBP, diastolic blood pressure; H2r, heritability; HR, heart rate; SBP, systolic blood pressure.
Bold and underlined numbers indicate the presence of heritability differences between subgroups (e.g., between males and females).
The heritability estimates of all studied traits are presented in Supplementary Table S4.
Average of sitting.
Phenotypic relatedness
Alcohol and tobacco users had an attenuated HR response to mental stress compared with non-users (both P=0.04;. Supplementary Table S3). The analysis of both global and regional measures of obesity indicated that current tobacco users are slender compared with former users and those who had never used tobacco (Supplementary Table S3). In addition, tobacco users had higher EP levels compared with former users and those who had never used tobacco. Meanwhile, the EP levels were not significantly different between former tobacco users and those that had never used tobacco, suggesting that the use of tobacco increases the level of EP in the body (Supplementary Table S3). Coffee use appears to increase the mean ambulatory HR (P=0.01); tobacco users had a lower resting BP compared with former users or subjects with no history of tobacco use (all P<0.05). In addition, the differences in the BP values between former tobacco users and those who had never used tobacco were insignificant (Supplementary Table S3).
Genetic mappings and functional annotation
General genetic mapping
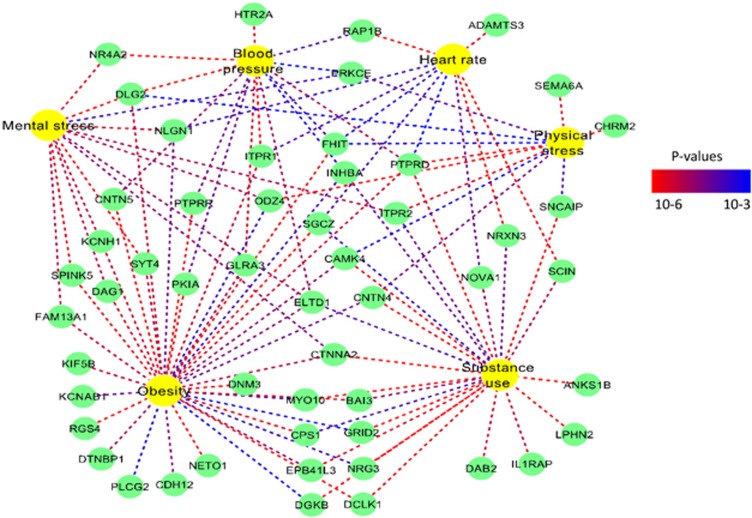
The functional annotation of the specific genes derived from the joint linkage and association mapping (Supplementary Table S5) indicated that a number of these genes share functions and interactions. Probing the protein interactions revealed that GRID2, HTR2A, LPHN2, LRP1B, MTPN and NETO1 share interactions with the synaptic protein DLG4 (Figure 3). Furthermore, a search of the synapse databases indicated that the CHRM2, DAB2, GRID2, HTR2A, ITPR2, LPHN2 and NETO1 genes have synaptic function (Figure 1 and Supplementary Table S10).
Figure 1.
Overview of the identified genes that have synaptic function. Trait nodes are yellow, and gene nodes are green. The edges are colored in a heat scale according to the strength of the association (P-values). A full color version of this figure is available at the Hypertension Research journal online.
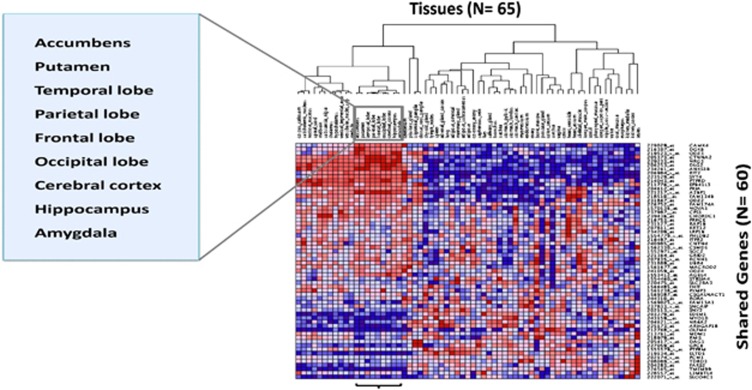
The family-based association scans identified genes commonly associated (P<0.05) with substance use, obesity, stress responses and hemodynamic traits after correction for multiple testing (Supplementary Table S6). The GRID2, ITPR2, LRP1B and PCM1 genes, identified under the linkage peaks, were also associated with multiple traits (Supplementary Table S5 and S6). Functional annotation indicated that 44% of the identified shared genes have synaptic function (Figure 1 and Supplementary Table S10), and the results of the KEGG pathway analysis of these genes significantly indicated the long-term potentiation (LTP) pathway (P=0.03). Furthermore, the gene expression analysis of the shared genes indicated that these genes are highly expressed and exhibit patterned expression in a cluster of brain-related tissues, including brain lobes, cerebral cortex, amygdala, hippocampus and putamen (Table 2 and Figure 2).
Table 2. Gene expression profilesa of shared genes driven from general, sex-specific and hypertension-specific genetic mapping.
|
General |
Sex specific |
Hypertension specific |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | N | Mean | s.e. | Tissue | N | Mean | s.e. | Tissue | N | Mean | s.e. |
| Temporal lobe | 60 | 0.33 | 0.1 | Cerebellum | 27 | 0.31 | 0.2 | Cerebellum | 16 | 0.62 | 0.3 |
| Occipital lobe | 60 | 0.32 | 0.1 | Accumbens | 27 | 0.31 | 0.1 | Temporal lobe | 16 | 0.56 | 0.3 |
| Accumbens | 60 | 0.32 | 0.1 | Occipital lobe | 27 | 0.30 | 0.2 | Amygdala | 16 | 0.52 | 0.2 |
| Frontal lobe | 60 | 0.32 | 0.1 | Parietal lobe | 27 | 0.30 | 0.2 | Accumbens | 16 | 0.52 | 0.3 |
| Parietal lobe | 60 | 0.32 | 0.1 | Frontal lobe | 27 | 0.29 | 0.1 | Parietal lobe | 16 | 0.52 | 0.3 |
| Amygdala | 60 | 0.29 | 0.1 | Putamen | 27 | 0.29 | 0.1 | Cerebral cortex | 16 | 0.50 | 0.3 |
| Cerebral cortex | 60 | 0.29 | 0.1 | Temporal lobe | 27 | 0.27 | 0.1 | Putamen | 16 | 0.50 | 0.3 |
| Dorsal root ganglia | 60 | 0.28 | 0.1 | Amygdala | 27 | 0.27 | 0.1 | Frontal lobe | 16 | 0.50 | 0.2 |
| Putamen | 60 | 0.27 | 0.1 | Cerebral cortex | 27 | 0.27 | 0.1 | Occipital lobe | 16 | 0.49 | 0.3 |
| Hippocampus | 60 | 0.26 | 0.1 | Hippocampus | 27 | 0.25 | 0.1 | Hippocampus | 16 | 0.49 | 0.2 |
Gene expression values (normalized log2 ratios) were extracted from COXPRESdb database.25 For each group of genes 65 human tissues were tested and the first 10 tissues with the highest mean expression are presented here. The entire data set is available in Supplementary Table S9.
Figure 2.
Gene expression heat map across 65 normal human tissues of the shared genes derived from the general genetic mapping. The shared genes tend to have higher expression and patterned expression profiles in the cluster of brain-related tissues, including the nucleus accumbens, putamen, brain lobes, cerebral cortex, hippocampus and amygdala. The expression values are presented in shades of red (high expression) to blue (low expression). The gene expression values were extracted from the COXPRESdb database.26 A full color version of this figure is available at the Hypertension Research journal online.
Subgroup genetic mappings
The hypertension-specific genetic analysis revealed that SNP rs4687150 (HWE P=0.3, MAF=0.3), which is inside the IL1RAP gene, is associated with coffee use with a negative association signal (Z=−3.5, P=0.0004) in hypertensive subjects and a positive association signal (Z=+3.8 P=0.0002) in normotensive subjects. The sex-specific genetic analysis revealed that SNP rs4888197 (HWE P=1, MAF=0.2), which is inside the PLCG2 gene, was associated with thigh skinfold and that rs847936 (HWE P=0.7, MAF=0.8), which is near the SCIN gene, was associated with tobacco use with negative association signals (both P⩽0.0009, −Z) in females and positive association signals (both P⩽0.0009, +Z) in males. We found that all three genes, IL1RAP, PLCG2 and SCIN, share synaptic function (Figure 1 and Supplementary Table S10). In addition, SNP, rs847936 was not only positively associated with never smoking in males, but was also positively associated with both wake HR (P=0.00095) and sleep HR (P=0.000055) in this group (Supplementary Table S8).
Subgroup specific genes that were identified through joint linkage and association mapping, namely DNM3, ADAMTS3, CDH12, DTNBP1, KCNAB1, KIF5B, RGS4 and SEMA6A, had synaptic function (Supplementary Table S7, Figure 1 and Supplementary Table S10). The CAMK4, CNTN4, CSMD1, FHIT, OLFM4, PTPRD and RORA genes, which were identified through the general genetic mapping, also appeared in the subgroup genetic mapping results. The genes BAI3, CNTN4, DCLK1, FHIT, INHBA, ITPR1, NLGN1, NRXN3, PTPRR and SCIN, which were identified in the sex-specific analysis, and CAMK4, CNTN5, GLRA3, PTPRD and SPINK5, which were identified in the hypertension-specific analysis, had synaptic function (Supplementary Table S8 and S10). Furthermore, the pathway analysis using the commonly associated genes derived from subgroup genetic mappings also significantly indicated the LTP (P=0.02) and GnRH-signaling pathways, which are two interconnected pathways (P=0.04).
The gene expression profiles of the shared genes derived from the general and subgroup mappings were compared across 65 normal human tissues. The results indicated that the shared genes that were identified through the sex-specific, hypertension-specific or general genetic mappings display higher expression values in brain-related tissues (Table 2).
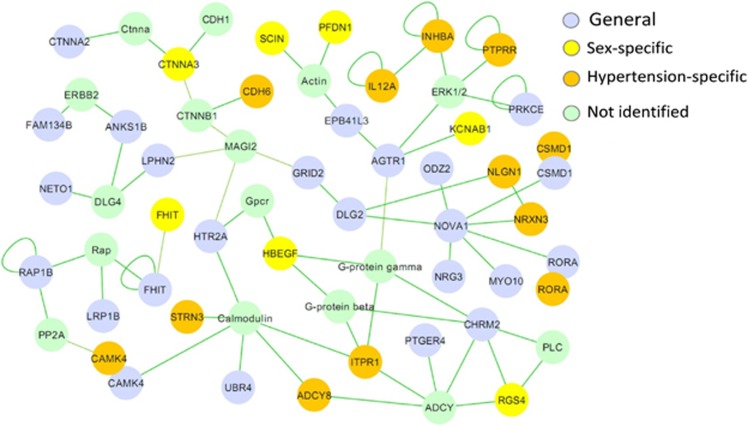
Protein interaction analysis
Candidate genes derived from the general (n=76), sex-specific (n=31) and hypertension-specific genetic mappings (n=38) were pooled, and a connectivity diagram was built based on protein interactions. Figure 3 shows an overview of these interactions. Although these genes were derived from different analyses, we found numerous interactions among their encoded proteins. We also checked for the associations of SNPs within the missing genes of the constructed network (Figure 3). In this manner, we identified the following SNPs: rs6938572 (P=0.0006) and rs2327017 (P=0.0003) upstream of the BMP6 gene, which were associated with average sitting DBP; rs2370413 (P=0.0003) and rs2887780 (P=0.0004) inside the CACNA1C gene, which were associated with subscapular skinfold measurements in males and the SBP response to mental stress, respectively; rs10492133 (P=0.0009) inside GRIN2B, which was associated with tobacco use; rs10491321 (P=0.00035) upstream of and rs7705319 (P=0.0006) inside the PPP2CA gene, which were associated with tobacco use; rs953944 (P=0.0007) and rs1029819 (P=0.0008), which were associated with tobacco use, and rs10485912 (P=0.00075), which was associated with overall SBP, inside the MAGI2 gene; rs93059 inside the NFKB1 gene, which was associated with the average sitting HR (P=0.00098) and the DBP response to mental stress (P=0.0006); and SNPs inside the ITPR1 gene, which were associated with (0.0004⩽P⩽0.0007) the BP and SBP response to mental stress.
Figure 3.
Overview of the protein interactions of genes derived from the general, sex-specific and hypertension-specific genetic mappings. The encoded proteins of the genes derived from the general, sex-specific or hypertension-specific genetic mappings share numerous protein interactions. The genes identified through two analyses (e.g., CAMK4) are displayed twice with different colors. A full color version of this figure is available at the Hypertension Research journal online.
Discussion
In this study, we investigated the connections between substance use, obesity and responses to mental and physical stress and hemodynamic traits in families from the founder population of the SLS region. The founder effect provides several benefits for mapping the genomic determinants of complex traits, including limited heterogeneity, the low likelihood of population admixture and stratification, the large size of the LD blocks and good genealogical records.9, 29
Our findings suggest that smoking may lower the BP through several mechanisms. First, obesity increases the risk of hypertension;7 therefore, the observed inverse correlation between obesity and tobacco use in our study could partly explain the lower BP in tobacco users. Second, stress is related to hypertension, which is consistent with the finding that hypertensive subjects had stronger HR and SBP responses to mental stress than normotensive subjects in our study. It has been already reported that a high-magnitude cardiovascular response to environmental stressors is a predictor of hypertension.4, 12 Therefore, our finding that habitual tobacco users displayed significantly attenuated HR responses to mental stress compared with non-users suggest that the damping effect of tobacco use on response to environmental stress may also account for the lowering effect of tobacco use on BP.
In fact, the relief from stress and negative emotion and enhancing positive moods are among the main reasons for substance use.30, 31 Physiological studies have also shown that both substance use and stress influence synaptic plasticity and that substance use can change the sensitivity of synaptic plasticity to stressors. For example, concurrent chronic nicotine treatment prevents the stress-induced impairment of the LTP pathway.32, 33 Synaptic plasticity is also implicated in the regulation of energy homeostasis as an important pathway through which peripheral metabolic hormones influence brain functions.34 Moreover, synaptic sensitization is involved in both stress- and salt-related hypertension as well as obesity-related hypertension. The repeated stimulation of the defense pathway sensitizes the synapse to respond to ever milder stressors such that hypertension eventually becomes permanent.7
In agreement with the above findings, our genetic analyses indicated that the neural synapse is a shared interface behind the studied traits. Genetic mapping uncovered numerous common genes among substance use, obesity, the response to mental and physical stress and HR and BP that had synaptic function. Furthermore, we found that the identified genes form a network of interacting proteins. The pathway analysis identified the LTP pathway, which is important in synaptic plasticity. In addition, gene expression findings revealed that these genes tend to be highly expressed and have patterned expression profiles in brain-related tissues.
Sex-specific and hypertension-specific genetic mappings identified IL1RAP, PLCG2 and SCIN as genes that were significantly negatively associated with a trait in one group and significantly positively associated with the same trait in the opposite group. All three genes had synaptic function, and in agreement with the identification of the SCIN gene as associated with tobacco use, it has been reported that nicotinic receptor stimulation induces the redistribution of the SCIN protein.35 The results of the pathway analysis and the comparison of the gene expression profiles of the shared genes derived from the sex-specific and hypertension-specific genetic mappings were similar to those from the general analysis. Moreover, we identified protein interactions and similar genes among the gene sets derived from the general and sex- and hypertension-specific genetic mappings, suggesting that sex-specific and hypertension-specific genetic differences are based on similar biological process to those identified by the general analysis. Therefore, by increasing genetic homogeneity, subgroup genetic mapping can uncover additional components of functional modules that otherwise remain obscure.
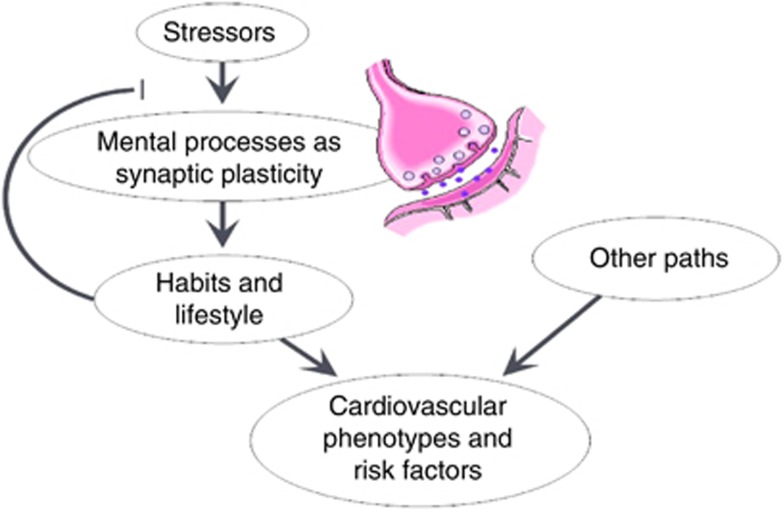
On the basis of above findings, we propose a model that (Figure 4) once an environmental stressor is perceived at synapses, it can alter the efficacy of synaptic plasticity; however, other environmental factors as substance use can modify the sensitivity of synaptic plasticity to the stressors. In the long term, the combined effects of genetic variations in the synapses and environmental factors shape an individual's lifestyles and habits. Conversely, the established habits and lifestyle influence the body and eventually modify health status outcomes, including cardiovascular disease and cardiovascular risk factor phenotypes, such as obesity. Nonetheless, because different means can lead to the same end, other factors can influence the cardiovascular system through different mechanisms.
Figure 4.
Hypothetical model of the interaction between stress, synaptic plasticity, substance use, obesity and related cardiovascular outcomes. Stressors can change the efficacy of mental processes such as synaptic plasticity. However, other environmental factors, such as substance use, modify the sensitivity of synaptic plasticity to the stressors. In the long term, the combined effects of genetic variations in the synapses and environmental factors shape an individual's lifestyles and habits. Conversely, the established habits and lifestyle influence the body and eventually modify health status outcomes, including cardiovascular disease and cardiovascular risk factors phenotypes as obesity. Nonetheless, there are other factors that modify cardiovascular outcomes through different mechanisms. A full color version of this figure is available at the Hypertension Research journal online.
The identification of shared genetic factors among the studied traits suggests that complex phenotypes may not be entirely independent of one another and has implications for the design of gene-mapping studies that jointly examine complex traits. Such studies can provide a basis for disease classification and profiling as well as creating a map of human diseases based on underlying pleiotropic genetic factors. The constructed disease map can subsequently be overlaid with drug-targeted networks to explore potential therapeutic avenues.25, 36, 37, 38, 39 The substantial overlap between the genomic determinants of substance use, stress, obesity and hemodynamic traits in genes related to synapses provides evidence to support additional studies on the involvement of genetic variations in the synapses in the etiology of complex traits in which stress and lifestyle are contributing factors.
Acknowledgments
This work was supported by funds from the National Institutes of Health Specialized Center of Research HL-54998 and the Canadian Institutes of Health Research (Cardio gene Consortium, MT-11463 and MT-14654) and by grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada to the GENESIS ICE Team.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Hypertension Research website (http://www.nature.com/hr)
Supplementary Material
References
- Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- Piazza PV, Le Moal M. The role of stress in drug self-administration. Trends Pharmacol Sci. 1998;19:67–74. doi: 10.1016/s0165-6147(97)01115-2. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wise RA. How can drug addiction help us understand obesity. Nat Neurosci. 2005;8:555–560. doi: 10.1038/nn1452. [DOI] [PubMed] [Google Scholar]
- Hamet P, Tremblay J. Genetic determinants of the stress response in cardiovascular disease. Metabolism. 2002;51:15–24. doi: 10.1053/meta.2002.33186. [DOI] [PubMed] [Google Scholar]
- Adam TC, Epel ES. Stress, eating and the reward system. Physiol Behav. 2007;91:449–458. doi: 10.1016/j.physbeh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Korner PI. Essential hypertension and its causes: neural and non-neural mechanisms. New York: Oxford University Press; 2007. [Google Scholar]
- Prescott CA, Madden PAF, Stallings MC. Challenges in genetic studies of the etiology of substance use and substance use disorders: Introduction to the special issue. Behav Genet. 2006;36:473–482. doi: 10.1007/s10519-006-9072-9. [DOI] [PubMed] [Google Scholar]
- Musani SK, Erickson S, Allison DB. Obesity-still highly heritable after all these years. Am J Clin Nutr. 2008;87:275–276. doi: 10.1093/ajcn/87.2.275. [DOI] [PubMed] [Google Scholar]
- Hamet P, Merlo E, Seda O, Broeckel U, Tremblay J, Kaldunski M, Gaudet D, Bouchard G, Deslauriers B, Gagnon F, Antoniol G, Pausov Z, Labuda M, Jomphe M, Gossard F, Tremblay G, Kirova R, Tonellato P, Orlov SN, Pintos J, Platko J, Hudson TJ, Rioux JD, Kotchen TA, Cowley AW., Jr Quantitative founder-effect analysis of french canadian families identifies specific loci contributing to metabolic phenotypes of hypertension. Am J Hum Genet. 2005;76:815–832. doi: 10.1086/430133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen AG, Rutters F. The hypothalamic-pituitary-adrenal-axis in the regulation of energy balance. Physiol Behav. 2008;94:169–177. doi: 10.1016/j.physbeh.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Imumorin IG, Dong YB, Zhu HD, Poole JC, Harshfield GA, Treiber FA, Snieder H. A gene-environment interaction model of stress-induced hypertension. Cardiovasc. Toxicol. 2005;5:109–132. doi: 10.1385/ct:5:2:109. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction. Nat Neurosci. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, LaForge KS. Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Interv. 2007;7:74–78. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Seda O, Tremblay J, Gaudet D, Brunelle P-L, Gurau A, Merlo E, Pilote L, Orlov SN, Boulva F, Petrovich M, Kotchen TA, Cowley AW, Jr, Hamet P. Systematic, genome-wide, sex-specific linkage of cardiovascular traits in french canadians. Hypertension. 2008;51:1156–1162. doi: 10.1161/HYPERTENSIONAHA.107.105247. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausova Z, Gaudet D, Gossard F, Bernard M, Kaldunski ML, Jomphe M, Tremblay J, Hudson TJ, Bouchard G, Kotchen TA, Cowley AW, Hamet P. Genome-wide scan for linkage to obesity-associated hypertension in french canadians. Hypertension. 2005;46:1280–1285. doi: 10.1161/01.HYP.0000188049.23233.fb. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ge DL, Zhang KL, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB. Wgaviewer: Software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WX, Zhang Y, Zheng H, Zhang C, Xiong W, Olyarchuk JG, Walker M, Xu WF, Zhao M, Zhao SQ, Zhou Z, Wei LP. Syndb: A synapse protein database based on synapse ontology. Nucleic Acids Res. 2007;35:D737–D741. doi: 10.1093/nar/gkl876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: Pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane HC, Lempicki R. David: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: A database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. Genepattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Croning MDR, Marshall MC, McLaren P, Armstrong JD, Grant SGN. G2cdb: The genes to cognition database. Nucleic Acids Res. 2009;37:D846–D851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:9. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel JD, Stroud LR, Paronis CA. Smoking, stress, and negative affect: Correlation, causation, and context across stages of smoking. Psychol Bull. 2003;129:270–304. doi: 10.1037/0033-2909.129.2.270. [DOI] [PubMed] [Google Scholar]
- Larissa AP. Stress and alcohol interaction: An update of human research. Alcohol Clin Exp Res. 1991;15:438–459. doi: 10.1111/j.1530-0277.1991.tb00543.x. [DOI] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC. Synaptic plasticity and addiction. Nat Rev Neurosci. 2007;8:844–858. doi: 10.1038/nrn2234. [DOI] [PubMed] [Google Scholar]
- Aleisa AM, Alzoubi KH, Gerges NZ, Alkadhi KA. Nicotine blocks stress-induced impairment of spatial memory and long-term potentiation of the hippocampal ca1 region. Int J Neuropsychopharmacol. 2006;9:417–426. doi: 10.1017/S1461145705005912. [DOI] [PubMed] [Google Scholar]
- Horvath TL. Synaptic plasticity in energy balance regulation. Obesity. 2006;14:228S–233S. doi: 10.1038/oby.2006.314. [DOI] [PubMed] [Google Scholar]
- Vitale ML, Castillo ARD, Tchakarov L, Trifaró JM. Cortical filamentous actin disassembly and scinderin redistribution during chromaffin cell stimulation precede exocytosis, a phenomenon not exhibited by gelsolin. J Cell Biol. 1991;113:1057–1067. doi: 10.1083/jcb.113.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A-L, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Kaplow IM, Sirota M, Do CB, Butte AJ, Batzoglou S. A classifier-based approach to identify genetic similarities between diseases. Bioinform. 2009;25:I21–I29. doi: 10.1093/bioinformatics/btp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.