Abstract
Primary cilia are nearly ubiquitous cellular appendages that provide important sensory and signaling functions. Ciliary dysfunction underlies numerous human diseases, collectively termed ciliopathies. Primary cilia have distinct functions on different cell types and these functions are defined by the signaling proteins that localize to the ciliary membrane. Neurons throughout the mammalian brain possess primary cilia upon which certain G protein-coupled receptors localize. Yet, the precise signaling proteins present on the vast majority of neuronal cilia are unknown. Here, we report that dopamine receptor 1 (D1) localizes to cilia on mouse central neurons, thereby implicating neuronal cilia in dopamine signaling. Interestingly, ciliary localization of D1 is dynamic, and the receptor rapidly translocates to and from cilia in response to environmental cues. Notably, the translocation of D1 from cilia requires proteins mutated in the ciliopathy Bardet-Biedl syndrome (BBS), and we find that one of the BBS proteins, Bbs5, specifically interacts with D1.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0603-4) contains supplementary material, which is available to authorized users.
Keywords: Ciliopathy, Neuronal cilia, Dopamine receptor 1, Bardet-Biedl syndrome
Introduction
Nonmotile primary cilia are typically solitary microtubule-based organelles present on nearly every cell type in the mammalian body. Primary cilia play critical roles in coordinating numerous cellular and developmental signaling pathways [1–3]. Neurons throughout the mammalian brain possess a primary cilium projecting from the cell body. The fact that several ciliopathies have prominent neurological phenotypes indicates that neuronal cilia provide important functions in the brain [4]. Subsets of neuronal cilia contain certain G protein-coupled receptors (GPCRs), including somatostatin receptor 3 (Sstr3) [5], serotonin receptor 6 (Htr6) [6, 7], and melanin-concentrating hormone receptor 1 (Mchr1) [8, 9]. This suggests that some neuronal cilia sense neuromodulators in the brain. Yet, the signaling pathways mediated by the vast majority of neuronal cilia in the mammalian brain are unknown.
The ciliopathy Bardet-Biedl syndrome (BBS) is characterized by obesity, retinal dystrophy, renal anomalies, hypogenitalism, polydactyly, and cognitive deficits [10]. There are at least 14 causative BBS genes [11], and seven of the BBS proteins (BBS1, 2, 4, 5, 7, 8, and 9) form a complex, called the BBSome, that constitutes a coat complex and mediates vesicular trafficking to the cilium [12, 13]. We previously reported that Sstr3 and Mchr1 fail to localize to neuronal cilia in mouse models of BBS [9], suggesting BBS proteins are required for trafficking of GPCRs to neuronal cilia. This is supported by a recent study showing that the BBSome interacts with a fragment of human SSTR3 and this interaction is required for ciliary targeting [12]. Furthermore, as Mchr1 is an important regulator of feeding behavior [14] and BBS mice develop hyperphagia-induced obesity [15, 16], we hypothesize that mislocalization of cell-type-specific ciliary signaling proteins together with disruption of signaling is the fundamental mechanism underlying the pathophysiology of the pleiotropic BBS phenotypes.
A critical step in determining the signaling pathways mediated by neuronal cilia is to identify the signaling proteins that specifically localize to cilia. Although the ciliary membrane is contiguous with the plasma membrane, ciliary localization of proteins is tightly regulated and only certain proteins localize to the ciliary membrane. We recently reported that ciliary GPCRs selectively localize to cilia when expressed in inner medullary collecting duct (IMCD) cells whereas nonciliary GPCRs do not localize to cilia [8]. Using expression of chimeric GPCRs in IMCD cells, we found that the third intracellular (i3) loops of Sstr3 and Htr6 are sufficient to mediate ciliary localization of the nonciliary receptors Sstr5 and Htr7, respectively [8]. Comparison of the i3 loops from Sstr3 and Htr6 identified a loose consensus sequence that we used to identify Mchr1 as a novel ciliary GPCR. Interestingly, the i3 loops of Sstr3 and Htr6 are sufficient but not necessary for ciliary localization [8], suggesting there are additional ciliary localization sequences within Sstr3 and Htr6.
Here, we use the presence of putative ciliary targeting sequences in the i3 loop and carboxy terminal tail to predict novel ciliary GPCRs. We show, for the first time, that dopamine receptor 1 (D1) localizes to mouse neuronal primary cilia. Notably, this localization is dynamic and D1 is recruited to cilia in response to an increase in cAMP levels and treatment with receptor ligand causes a rapid decrease in D1 ciliary localization. Interestingly, D1 ciliary localization is increased in neurons from mice lacking BBS proteins and does not decrease in response to ligand treatment. Furthermore, we find that a subunit of the BBSome, Bbs5, interacts with D1. Together, these results suggest that D1 dynamically localizes to neuronal cilia and that translocation of D1 out of cilia in response to agonist requires the BBSome and involves interaction with Bbs5.
Materials and methods
Plasmid construction
Coding sequences of the D1 receptor, BBSome subunits, and Gapdh were amplified from reverse-transcribed mouse whole brain RNA using the Superscript First-Strand Synthesis RT-PCR Kit (Invitrogen, Carlsbad, CA, USA) and cloned into a TA cloning vector (pSTBlue-1; Novagen, San Diego, CA, USA). Primers corresponding to the N-terminal and C-terminal regions of the receptors were designed with 5′ restriction sites for directional cloning. Primers were used to add an HA tag to the C-terminus of the Bbs5 protein. All amplifications were performed with Accuprime Pfx Taq Polymerase (Invitrogen). The final PCR products were cloned into the pEGFP-N vector (Clontech, Mountain View, CA, USA), pcDNA3.1(−) (Invitrogen), pcDNA3.1/myc-His (Invitrogen), and/or pGADT7 (Clontech). All DNA constructs were sequence verified.
Cell culture and transient transfections
IMCD-3 cells (ATCC, Manassas, VA, USA) were maintained in DMEM:F12 medium supplemented with 10% FBS, 1.2 g/l of sodium bicarbonate, and 0.5 mM sodium pyruvate (Invitrogen). Cells (n = 5 × 106) were electroporated with 10 μg DNA and plated at high density on glass coverslips. Cells were harvested 48 h after transfection for immunocytochemistry.
Mice and tissue preparation
The generation and characterization of Bbs2- and Bbs4-null mice has been previously described [15, 16]. All procedures were approved by the Institutional Animal Care and Use Committee at The Ohio State University. Wild-type (WT), Bbs2-null (Bbs2 −/−), and Bbs4-null (Bbs4 −/−) littermates were generated by intercrossing heterozygous animals. Brains were isolated and processed as previously described [9]. For immunofluorescence procedures, cryoprotected brains were sectioned on a freezing microtome at a thickness of 40 μm.
Neuronal cell culturing and treatments
Primary amygdala neurons from postnatal day 0–1 mouse pups were cultured as previously described [17]. Experiments were performed on neurons 7 days post-plating. Neurons were treated with SKF81297 (10 μM) alone for 15 min, pretreated with SCH23390 (10 μM) for 15 min prior to agonist addition, or forskolin (10 μM) for 15 min (all from Sigma-Aldrich, St. Louis, MO, USA). All control conditions received an equivalent volume of vehicle.
Immunofluorescence
Day 7 neurons were fixed and processed for immunofluorescence as previously described [9]. Primary antibodies included anti-adenylyl cyclase III (C-20; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-dopamine receptor 1 (SG2-D1a; Santa Cruz Biotechnology). Secondary antibodies included Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa Fluor 546-conjugated goat anti-rabbit IgG (Invitrogen). Nucleic acids were stained with DRAQ5 (Axxora, San Diego, CA, USA). All samples were imaged on a Zeiss LSM 510 laser scanning confocal microscope at the Hunt-Curtis Imaging Facility in the Department of Neuroscience at The Ohio State University. Multiple consecutive focal planes (Z-stack), spaced at 0.5–1 μm intervals, were captured. For all collected images, the brightness and contrast of each channel was adjusted using the Zeiss LSM Image Browser program.
Data analysis
Quantitative analysis of ciliated neurons in each experiment and under every condition was performed on at least three independent coverslips generated from three to four different animals. For each coverslip, the 40× microscope objective was centered on the coverslip, and three different fields were imaged. The number of D1-positive ciliated neurons and the number of ACIII-positive ciliated neurons in each image were counted by individuals blinded to the experimental conditions. The results were expressed as the percentage of ACIII-positive cilia showing D1-ciliary localization. All data are expressed as mean ± standard error of the mean (SEM). Statistical significance was tested using Student’s t test.
Yeast two-hybrid analysis
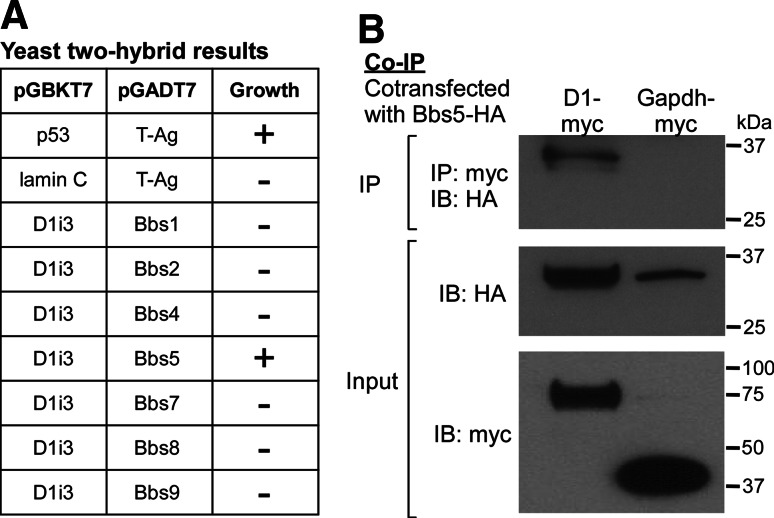
The analysis utilized the Yeastmaker Yeast Transformation System 2 (Clontech). The i3 loop of mouse D1 (nucleotides 652–819) was cloned into the yeast bait vector pGBKT7 and transformed into the yeast mating strain AH109. The prey vector pGADT7 containing each BBSome subunit was individually transformed into the yeast mating strain Y187. AH109 and Y187 yeast expressing the respective constructs were mated and plated. Growth was assessed on -Leu/-Trp and -Ade/-His/-Leu/-Trp (X-α-gal) selective media.
Protein isolation
The protein isolation protocol was adapted from Gomes et al. [18]. HEK293T cells (ATCC) were maintained in DMEM supplemented with 10% FBS and 1.5 g/L of sodium bicarbonate (Invitrogen). Bbs5-HA and D1-myc or Gapdh-myc were cotransfected by electroporation into HEK293T cells. After 48 h, cells were lysed in solubilization buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1% NP-40) supplemented with sodium orthovanadate and protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Following a 1 h incubation at 4°C with rotation, cell debris and unsoluble material were cleared by centrifugation for 20 min at 15,000×g at 4°C. The resulting supernatant containing soluble protein was collected, and the concentration was determined by the Bradford assay (Bio-Rad, Richmond, CA, USA).
Co-immunoprecipitation
Soluble protein was precleared at 4°C with rotation for 1 h with protein A-Sepharose beads (GE Healthcare, Piscataway, NJ, USA) pre-equilibrated in detergent buffer (1 mM Tris pH 7.5, 5 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1% NP-40) supplemented with protease inhibitor cocktail (Roche). Precleared protein was incubated overnight at 4°C with anti-myc antibody (9E10; Santa Cruz Biotechnology) immobilized to protein A-Sepharose beads. Samples were centrifuged and washed three times with 1× PBS supplemented with protease inhibitor cocktail. Immunoprecipitated proteins were subjected to 60°C heat for 15 min in SDS sample buffer to elute purified proteins. Purified proteins were then analyzed by immunoblotting.
Immunoblotting
Protein samples were run on a denaturing 4–15% gradient polyacrylamide gel (Bio-Rad) and transferred to a PVDF membrane (Millipore, Billerica, MA, USA). Membranes were blocked in TBS-T [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween 20] with 5% milk and incubated overnight at 4°C with appropriate antibodies diluted in TBS-T with 5% milk. Membranes were probed with horseradish peroxidase-conjugated secondary antibodies diluted in TBS-T with 5% milk for 1 h at room temperature. The secondary antibody was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL, USA), and bands were visualized using Blue Ultra Autorad Film (ISC Bioexpress, Kaysville, UT, USA). Immunoprecipitated proteins were probed with anti-HA (3F10; Roche) and cotransfected cell-lysate was probed with either anti-HA or anti-myc.
Prediction of human GPCR intracellular loop sequences
A database of the predicted intracellular sequences for all human GPCR proteins was generated as previously described [8]. The resulting database was searched using the ambiguous regular expression “AXXXQ” and “VXP/LXP” to identify GPCRs predicted to contain these motifs within their i3 loop and C-tail sequences, respectively.
Results
Ciliary GPCRs contain localization sequences in the i3 loop and C-tail
The ciliary GPCR rhodopsin contains a putative i3 loop ciliary localization consensus sequence [8] as well as a sequence (VxP) at the terminus of the carboxy terminal tail (C-tail) that is required for its trafficking across the photoreceptor-connecting cilium [19, 20]. We reasoned that other ciliary GPCRs may also have a combination of these sequences. Examination of the C-tail sequences from human SSTR3, HTR6, and MCHR1 revealed that all of them contain a VxP sequence, suggesting that the presence of sequences in both the i3 loop and C-tail may be important for ciliary localization of certain GPCRs. Ciliary localization sequences should be conserved across species in which the receptor is ciliary. Therefore, we asked whether the C-tail VxP sequence is conserved in mouse Sstr3, Htr6, and Mchr1, which all localize to cilia [8]. The C-tail VxP sequence is conserved in mouse Htr6 and Mchr1 but is not conserved in mouse Sstr3. However, the C-tail of mouse Sstr3 contains two LxP sequences that are conserved in human SSTR3. Since valine and leucine are structurally similar, we considered that VxP or LxP sequences in the C-tail in combination with the conserved i3 loop sequence (AxxxQ) might be a good predictor of novel ciliary GPCRs. Searching a database of the sequences of the predicted i3 loops and C-tails of all human GPCRs and narrowing the results to those GPCRs in which the i3 loop and C-tail sequences were conserved in mouse revealed eight candidate ciliary GPCRs (Table 1). One of these, dopamine receptor 1 (D1), was considered an especially good candidate for ciliary localization as it is involved in a variety of functions in the brain that are disrupted in ciliopathies, including cognition and food intake [21].
Table 1.
Human G protein-coupled receptors (GPCRs) with putative ciliary localization sequences in the predicted i3 and C-tail domains that are conserved in mouse. Known ciliary GPCRs are indicated
| G protein-coupled receptor | i3 sequence(s) | C-tail sequence(s) | Localization |
|---|---|---|---|
| Beta-1-adrenergic receptor | ALREQ | LEP | ? |
| Cadherin EGF LAG seven-pass G-type receptor 2 | AAQRQ | LNP | ? |
| Dopamine receptor 1 | AKNCQ | LCP/LIP/LSP | ? |
| GPR15 | AHYQQ | LCP | ? |
| GPR54 | ALQGQ | VCP | ? |
| GPR75 | ASRPQ/ASRLQ | LSP | ? |
| GPR92 | ATQSQ | LRP | ? |
| GPR123 | ASVLQ | LSP | ? |
| Melanin-concentrating hormone receptor 1 | APASQ | VKP | Ciliary |
| Opsin 1 (short-wave sensitive) | AVAAQ | VGP | Ciliary |
| Opsin 1 (medium-wave sensitive 2) | AVAKQ | VSP | Ciliary |
| Opsin 1 (long-wave sensitive) | AVAKQ | VSP | Ciliary |
| Rhodopsin | AAAQQ | VAP | Ciliary |
| Serotonin receptor 6 | AARKQ | VLP/LPP/LLP | Ciliary |
| Somatostatin receptor 3 | APSCQ | LRP/LLP | Ciliary |
Dopamine receptor 1 localizes to cilia
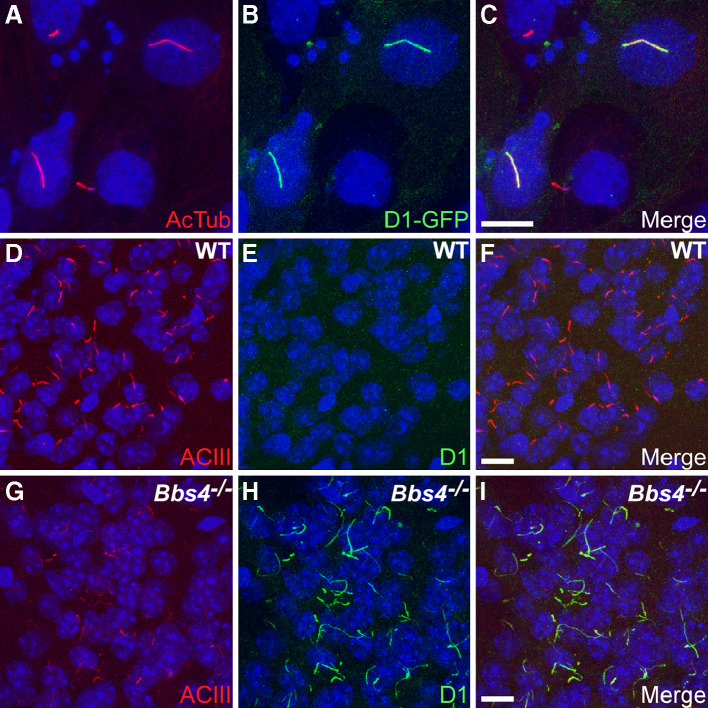
To test whether D1 localizes to cilia, we expressed a construct encoding mouse D1 with EGFP fused to the C-terminus in IMCD cells and assessed subcellular localization. Notably, D1 localized to cilia when expressed in IMCD cells (Fig. 1a–c). Recently, human D1 was also found to localize to cilia when heterologously expressed in NIH3T3 cells and rat striatal neurons [22]. We then tested whether D1 localizes to neuronal cilia in tissue and if that localization requires BBS proteins, similarly to Sstr3 and Mchr1. Brain sections from adult wild-type (WT) and Bbs4-null (Bbs4 −/−) littermates were colabeled with an antibody to type III adenylyl cyclase (ACIII), a prominent marker of primary cilia throughout the mouse brain [23], and an antibody to D1, the specificity of which was confirmed by immunofluorescence and immunoblotting (Suppl. Figs. 1, 2). Remarkably, D1-immunoreactive cilia were rarely detected in any region of WT brains but were abundant in several regions of Bbs4 −/− brains, including the striatum, olfactory tubercle, and amygdala (Fig. 1d–i). Labeling of sections from Bbs2-null (Bbs2 −/−) mice revealed a similar abundance of D1-immunoreactive cilia compared to WT littermates (Suppl. Fig. 3). To test whether the differences in abundance of D1-immunoreactive cilia were due to differences in D1 expression levels, we immunoblotted lysates from WT and Bbs4 −/− brains. Quantification of D1 protein levels indicated the receptor is expressed at equivalent levels in WT and Bbs4 −/− tissues (Suppl. Fig. 4). These results indicate that D1 localizes to neuronal cilia in vivo and suggest that BBS proteins are required for proper regulation of D1 ciliary localization.
Fig. 1a–i.
Dopamine receptor 1 (D1) localizes to cilia, and ciliary localization is increased in the brains of Bbs4 −/− mice. a–c Representative image of transiently transfected inner medullary collecting duct (IMCD) cells expressing D1 fused at the C-terminus to EGFP. a Acetylated α-tubulin (AcTub; red) marks the cilia; b EGFP fluorescence (green) shows expression of the D1 receptor; c merged images. Representative images of the basolateral amygdala in adult WT (d–f) and Bbs4 −/− (g–i) mice (n = 3 animals for each genotype) showing labeling for type III adenylyl cyclase (ACIII; red) and D1 (green). Nuclei are stained with DRAQ5 (blue). The appearance and distribution of ACIII-positive cilia is similar between WT (d) and Bbs4 −/− (g) sections. The identical fields showing labeling for D1 (green) reveal a lack of D1-positive cilia in the WT (e) section but abundant D1-positive cilia in the Bbs4 −/− (h) section. Merged images showing no D1 labeling of cilia in the WT (f) section and colocalization of ACIII and D1 to cilia in the Bbs4 −/− (i) section. Scale bars represent 10 μm
D1 ciliary localization is dynamic
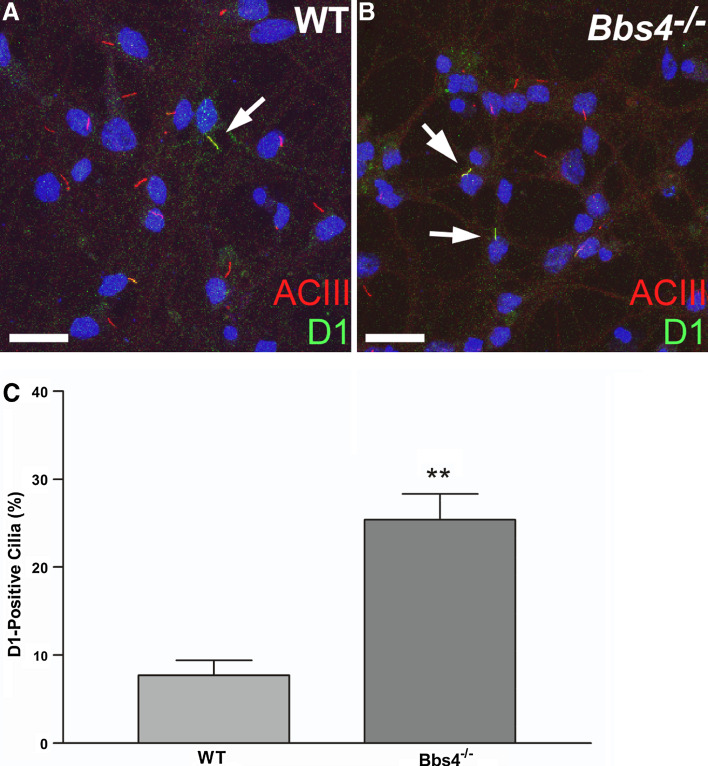
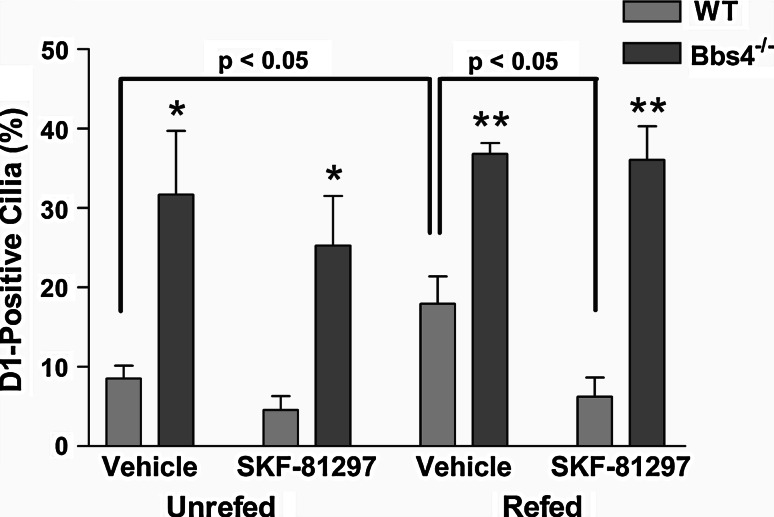
To confirm our in vivo findings of D1 ciliary localization, we generated serum-free primary cultures enriched for amygdala neurons from newborn WT and Bbs4 −/− mice. After 7 days in culture, the cells were colabeled with antibodies to D1 and ACIII (Fig. 2a, b) and the percentage of D1-positive cilia was quantified (Fig. 2c). Similar to our observations in vivo, the percentage of D1-positive cilia was significantly higher in Bbs4 −/− cultures (25.4%) than WT cultures (7.7%). To further confirm that the increase in D1 ciliary localization in Bbs4 −/− neurons was not due to differences in D1 expression levels, we performed real-time PCR analysis of RNA from day 7 WT and Bbs4 −/− amygdala cultures and found D1 was expressed at equivalent levels in WT and Bbs4 −/− neurons (Suppl. Fig. 4). Interestingly, the percentage of D1-positive cilia in WT cultures was much greater than expected given how rarely we detected them in brain sections. We reasoned the relative abundance of D1-positive cilia on cultured WT neurons might indicate that D1 ciliary localization is dynamic and regulated by signaling in the brain. To test this hypothesis, we treated cultures enriched for amygdala neurons from WT and Bbs4 −/− mice on day 7 with the D1 agonist SKF-81297 or vehicle. The WT and Bbs4 −/− cultures were split into two experimental groups; one group in which the medium was changed 30 min prior to treatment and a second group in which the medium was not changed prior to treatment. After 15 min of treatment the cells were fixed, colabeled with antibodies to D1 and ACIII, and the percentage of D1-positive cilia was quantified (Fig. 3). The percentage of D1-positive cilia in unrefed WT cultures treated with vehicle (8.5%) was similar to our previous result. Remarkably, when the WT cultures were refed prior to vehicle treatment the percentage of D1-positive cilia (17.9%) was significantly increased compared to unrefed WT cultures (Fig. 3). Moreover, agonist treatment of refed WT neurons led to a significant decrease in the percentage of D1-positive cilia (6.2%) (Fig. 3). Notably, the percentage of D1-positive cilia in Bbs4 −/− cultures was significantly greater than WT cultures under all conditions and did not change significantly in response to refeeding or agonist treatment (Fig. 3). In WT cultures, pretreatment with a D1 antagonist prevented the agonist-mediated decrease in D1-positive cilia (Suppl. Fig. 5), suggesting the agonist is acting through D1. Thus, these results suggest D1 ciliary localization is dynamic and the receptor can be recruited to the ciliary membrane in response to refeeding and translocated from cilia in response to receptor agonist binding. Furthermore, our data suggest the translocation of D1 out of cilia requires BBS proteins.
Fig. 2a–c.
D1 ciliary localization is increased in amygdala-enriched neuronal cultures. Co-immunolabeling of day 7 amygdala neurons from WT (a) and Bbs4 −/− (b) mice with antibodies to ACIII (red) and D1 (green) shows the presence of numerous ACIII-positive cilia in both genotypes, a subset of which are also positive for D1 (arrows). Nuclei are stained with DRAQ5 (blue). Scale bars represent 20 µm. c Percentage of ACIII-positive cilia in WT and Bbs4 −/− cultures (n = 3 animals for each genotype) that are positive for D1. D1 localizes to 7.7 ± 1.7% (n = 324) of ACIII-positive cilia in WT cultures and 25.4 ± 2.9% (n = 289) of ACIII-positive cilia in Bbs4 −/− cultures. Note the percentage of D1-positive cilia is significantly higher in Bbs4 −/− cultures compared to WT cultures. Values are expressed as mean ± SEM. **Significantly different from WT percentage (p < 0.01)
Fig. 3.
In amygdala neuronal cultures, agonist treatment causes a decrease in D1 ciliary localization in WT but not Bbs4 −/− cultures. Percentage of ACIII-positive cilia in WT and Bbs4 −/− cultures (n = 3–4 animals for each genotype and condition) that are positive for D1. In unrefed WT cultures (left side of graph), D1 localizes to 8.5 ± 1.6% (n = 216) of ACIII-positive cilia in cultures treated with vehicle and 4.5 ± 1.8% (n = 297) of ACIII-positive cilia in cultures treated with the D1 agonist SKF-81297. In unrefed Bbs4 −/− cultures, D1 localizes to 31.7 ± 8% (n = 157) and 25.3 ± 6.3% (n = 140) of ACIII-positive cilia in cultures treated with vehicle and agonist, respectively. In refed WT cultures (right side of graph), D1 localizes to 17.9 ± 3.4% (n = 185) of ACIII-positive cilia in cultures treated with vehicle and 6.2 ± 2.4% (n = 189) of ACIII-positive cilia in cultures treated with agonist. Note the percentage of D1-positive cilia is significantly higher in refed vehicle-treated WT cultures compared to both unrefed vehicle-treated WT cultures (p < 0.05) and refed agonist-treated WT cultures (p < 0.05). In refed Bbs4 −/− cultures, D1 localizes to 36.8 ± 1.4% (n = 165) and 36.1 ± 4.3% (n = 144) of ACIII-positive cilia in cultures treated with vehicle and agonist, respectively. Note the percentage of D1-positive cilia in Bbs4 −/− cultures is significantly greater than WT cultures under all conditions and does not change significantly in response to refeeding or agonist treatment. Values are expressed as mean ± SEM. *Significantly different from WT percentage (p < 0.05). **Significantly different from WT percentage (p < 0.01)
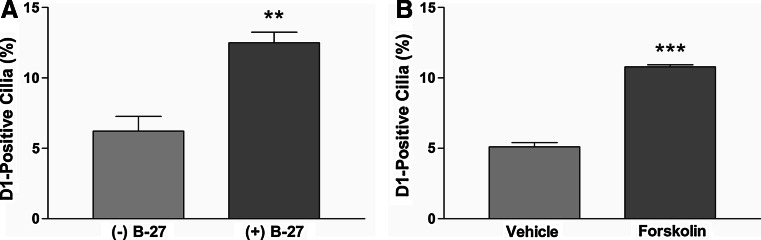
To further investigate the increase in D1-ciliary localization induced by refeeding, we refed WT cultures with neurobasal-A medium with or without B-27 supplement. We focused on the B-27 supplement for two reasons. First, we noticed that the baseline percentage of D1-positive cilia in our neuronal cultures differed between lots of the B-27 supplement. Second, we reasoned that if the effect was due to the addition of some factor(s), it was likely contained in the B-27 supplement, which is comprised of over 20 different components. After 45 min the cells were fixed, colabeled with antibodies to D1 and ACIII, and the percentage of D1-positive cilia was quantified (Fig. 4a). Interestingly, the percentage of D1-positive cilia in WT cultures refed medium with B-27 (12.5%) was significantly increased compared to WT cultures refed medium without B-27 (6.2%) (Fig. 4a). This result suggests the increase in D1-ciliary localization induced by fresh medium is due to the addition of some factor(s) present within B-27, rather than removal of a factor(s). In renal cells, forskolin-stimulated increases in intracellular cAMP levels stimulate D1 recruitment from intracellular vesicles to the plasma membrane [24, 25]. In addition, the ciliary GPCR Smoothened translocates to cilia in response to forskolin treatment [26, 27]. Thus, we hypothesized that forskolin treatment might stimulate D1 translocation to cilia. Indeed, treatment of WT neurons with forskolin led to a significant increase in D1-positive cilia (10.8%) compared to vehicle-treated WT neurons (5.2%) (Fig. 4b). Together, these results suggest D1 rapidly translocates to cilia in response to signals, such as increased cAMP levels, and out of cilia in response to agonist binding.
Fig. 4a, b.
D1 ciliary localization is increased in WT amygdala neuronal cultures in response to media containing B-27 supplement or forskolin treatment. a Percentage of ACIII-positive cilia in WT cultures (n = 3 animals for each condition) that are positive for D1. D1 localizes to 6.2 ± 1.0% (n = 322) of ACIII-positive cilia and 12.5 ± 0.8% (n = 327) of ACIII-positive cilia in cultures refed medium without B-27 and with B-27, respectively. Note the percentage of D1-positive cilia is significantly higher in WT cultures refed medium containing B-27 compared to medium lacking B-27. Values are expressed as mean ± SEM. **Significantly different from vehicle-treated percentage (p < 0.01). b Percentage of ACIII-positive cilia in WT cultures (n = 3 animals for each condition) that are positive for D1. D1 localizes to 5.1 ± 0.3% (n = 310) of ACIII-positive cilia in cultures treated with vehicle and 10.8 ± 0.2% (n = 269) of ACIII-positive cilia in cultures treated with forskolin. Note the percentage of D1-positive cilia is significantly higher in forskolin-treated WT cultures compared to vehicle-treated WT cultures. Values are expressed as mean ± SEM. ***Significantly different from vehicle-treated percentage (p < 0.0001)
Bbs5 interacts with D1
As BBS proteins are involved in D1 ciliary distribution and the BBSome is known to localize to cilia, we tested the hypothesis that subunits of the BBSome interact with D1. Since the i3 loop of certain GPCRs is important for ciliary localization, we first screened the i3 loop of D1 against the seven BBSome subunits using yeast two-hybrid analysis and found that Bbs5 specifically interacts with the i3 loop of D1 (Fig. 5a). We then confirmed the interaction of D1 and Bbs5 by co-immunoprecipitation of transiently transfected epitope-tagged full-length proteins. Lysates of HEK293T cells expressing HA-tagged Bbs5 and Myc-tagged D1 or Myc-tagged Gapdh, as a negative control, were immunoprecipitated with an anti-Myc monoclonal antibody followed by immunoblotting with anti-HA. Bbs5 was detected in the D1 immunoprecipitate but not the Gapdh immunoprecipitate, indicating that D1 and Bbs5 interact (Fig. 5b). These results suggest the BBSome regulates D1 ciliary localization through an interaction that may be mediated by Bbs5.
Fig. 5a, b.
D1 and Bardet-Biedl syndrome 5 (Bbs5) proteins interact. a Results of yeast matings between cells expressing the i3 loop of mouse D1, p53 (positive control), or lamin C (negative control) in the bait vector (pGBKT7) and cells expressing each of the seven BBSome subunits or large T antigen (T-Ag) in the prey vector pGADT7. Note that only D1 i3 loop plus Bbs5 mating and the positive control p53 plus T-Ag mating show yeast growth (indicated as +) on selective media. b HA-tagged Bbs5 (Bbs5-HA) was co-expressed with myc-tagged D1 (D1-myc) or myc-tagged glyceraldehyde-3-phosphate dehydrogenase (Gapdh-myc) in HEK293T cells. Cell extracts were immunoprecipitated (IP) with an anti-myc antibody. Immunoprecipitates were analyzed by Western blotting (IB) with an anti-HA antibody (upper). Note that Bbs5 is immunoprecipitated with D1 but not Gapdh. The input, confirming expression of each protein, is also shown (middle and bottom)
Discussion
Our results show D1 localizes to cilia on central neurons, thereby implicating neuronal cilia in dopamine signaling. Furthermore, D1 ciliary localization is dynamic and the receptor can be recruited to the ciliary membrane in response to environmental signals, such as increased cAMP levels, and translocated from cilia in response to receptor agonist binding. Interestingly, D1 ciliary localization is rarely observed in WT mouse brains but is commonly detected on cultured WT neurons. We hypothesize D1 is constantly transported to and from the cilium and the amount of ciliary D1 reflects an equilibrium between signals that recruit D1 to the cilium and signals that stimulate D1 translocation from the cilium. Thus, D1 ciliary localization may be rarely detected in vivo due to a higher relative level of signals stimulating D1 translocation from cilia and these signals are reduced or absent in vitro resulting in an increase in ciliary localization.
Interestingly, D1 ciliary localization is abundant in BBS mouse brains, is significantly increased on cultured BBS neurons compared to WT neurons, and does not decrease in response to treatment with a D1 agonist. These results indicate BBS proteins are required for D1 translocation from cilia and lack of BBS proteins leads to accumulation of D1 in cilia. This is contrary to previous results showing Sstr3 and Mchr1 require BBS proteins for ciliary localization and suggests a novel role for BBS proteins in trafficking of some GPCRs out of neuronal cilia. Indeed, our results are consistent with a recent study showing mutations of BBSome proteins in Chlamydomonas reinhardtii lead to the accumulation of signaling proteins in flagella [28]. Thus, BBS proteins appear to mediate the import and export of ciliary signaling proteins.
The transport of proteins within cilia is mediated by a highly conserved process called intraflagellar transport (IFT). IFT is a bidirectional microtubule-based transport process in which complexes of proteins called IFT particles are transported by molecular motors along the axonemal outer doublet microtubules from the base of the cilium to the distal tip and then back to the cell body [29–31]. IFT also mediates transport of transmembrane proteins, including TRPV channels in C. elegans and polycystin-2 in mammalian cells [32, 33]. Notably, in C. elegans BBS proteins coordinate IFT [34] and in mammalian cells the BBSome has been observed moving within cilia in a manner consistent with IFT [13]. Combined with our current results, it is tempting to speculate the BBSome acts as an adaptor between ciliary signaling proteins, such as GPCRs, and IFT particles to mediate trafficking into and out of cilia. This is supported by our finding that D1 and Bbs5 interact. In addition, our results suggest that the process of BBSome-mediated transport from cilia is regulated by environmental cues (i.e. agonist-mediated activation of D1). Perhaps agonist binding results in a structural change in D1 that allows it to bind to the BBSome and be transported from the cilium.
It is intriguing that Sstr3 and Mchr1 require BBS proteins for ciliary localization whereas D1 does not. This indicates D1 transport into cilia is BBSome-independent and suggests GPCRs are transported into cilia by more than one mechanism. One difference between these receptors is D1 preferentially couples to Gs, whereas Sstr3 and Mchr1 preferentially couple to Gi/q. However, it is unknown whether G protein coupling affects GPCR ciliary localization.
One question raised by our findings is how trafficking of D1 into and out of cilia affects D1-mediated signaling. D1 translocation to and from cilia is reminiscent of Hedgehog (Hh) signaling. The Hh receptor Patched localizes to the ciliary membrane and upon activation traffics out of the cilium, allowing Smoothened (Smo) to enter the cilium [35, 36]. Smo then activates signaling at the distal tip of the cilium where transcription factors are localized [37]. Perhaps ciliary localization of D1 allows interactions or coupling with signaling partners that are specifically located within the cilium. Interestingly, different types of D1-mediated signaling have been detected in different brain regions [38]. Although D1 dimerization with D2 has been proposed to explain these findings [38], it is also possible that different types of D1-mediated signaling could be due to localization of the receptor on the ciliary membrane versus the cell membrane. Nevertheless, we hypothesize that D1 signaling is disrupted in BBS.
It is intriguing that D1 ciliary localization is particularly abundant in the amygdala of BBS animals. It is well established that the amygdala plays an important role in fear and other emotional processes [39, 40]. BBS children display increased levels of anxiety and depression [41]. Whether this is related to disruption of dopamine signaling in BBS patients is not known. Similarly, Bbs4 −/− mice show increases in anxiety-related behavioral tests such as the open-field and light-dark box test [42]. However, BBS mice have additional phenotypes, such as obesity, retinal degeneration, and anosmia that can confound such analyses. Utilization of conditional knockout mice with a disruption in a BBS protein or cilia assembly protein specifically in the amygdala will be important for resolving the precise roles of D1 ciliary signaling in fear-related behaviors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Joshua Stowell for technical assistance and David Sibley (NINDS) for providing D1 WT and KO brains. This work was supported in part by the Systems and Integrative Biology Training Grant T32 GM068412(J.G.) and R01 GM083120 from the NIH/National Institute of General Medical Sciences (K.M.).
Abbreviations
- BBS
Bardet-Biedl syndrome
- GPCR
G protein-coupled receptor
- D1
Dopamine receptor 1
- Mchr1
Melanin-concentrating hormone receptor 1
- Sstr3
Somatostatin receptor 3
- ACIII
Type III adenylyl cyclase
- IMCD
Inner medullary collecting duct
- i3
Third intracellular
- C-tail
Carboxy terminal tail
References
- 1.Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8:97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 3.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han YG, Alvarez-Buylla A. Role of primary cilia in brain development and cancer. Curr Opin Neurobiol. 2010;20:58–67. doi: 10.1016/j.conb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Handel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Hollt V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/S0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- 6.Hamon M, Doucet E, Lefevre K, Miquel MC, Lanfumey L, Insausti R, Frechilla D, Del Rio J, Verge D. Antibodies and antisense oligonucleotide for probing the distribution and putative functions of central 5-HT6 receptors. Neuropsychopharmacology. 1999;21:68S–76S. doi: 10.1016/S0893-133X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 7.Brailov I, Bancila M, Brisorgueil MJ, Miquel MC, Hamon M, Verge D. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872:271–275. doi: 10.1016/S0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- 8.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin JL, Beales PL. Bardet-Biedl syndrome: beyond the cilium. Pediatr Nephrol. 2007;22:926–936. doi: 10.1007/s00467-007-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 14.Handlon AL, Zhou H. Melanin-concentrating hormone-1 receptor antagonists for the treatment of obesity. J Med Chem. 2006;49:4017–4022. doi: 10.1021/jm058239j. [DOI] [PubMed] [Google Scholar]
- 15.Mykytyn K, Mullins RF, Andrews M, Chiang AP, Swiderski RE, Yang B, Braun T, Casavant T, Stone EM, Sheffield VC. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura DY, Fath M, Mullins RF, Searby C, Andrews M, Davis R, Andorf JL, Mykytyn K, Swiderski RE, Yang B, Carmi R, Stone EM, Sheffield VC. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berbari NF, Bishop GA, Askwith CC, Lewis JS, Mykytyn K. Hippocampal neurons possess primary cilia in culture. J Neurosci Res. 2007;85:1095–1100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- 18.Gomes I, Filipovska J, Jordan BA, Devi LA. Oligomerization of opioid receptors. Methods. 2002;27:358–365. doi: 10.1016/S1046-2023(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 19.Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc Natl Acad Sci USA. 1998;95:10620–10625. doi: 10.1073/pnas.95.18.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green ES, Menz MD, LaVail MM, Flannery JG. Characterization of rhodopsin mis-sorting and constitutive activation in a transgenic rat model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:1546–1553. [PubMed] [Google Scholar]
- 21.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 22.Marley A, von Zastrow M. DISC1 regulates primary cilia that display specific dopamine receptors. PLoS One. 2010;5:e10902. doi: 10.1371/journal.pone.0010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop GA, Berbari NF, Lewis JS, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505:562–571. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 24.Brismar H, Asghar M, Carey RM, Greengard P, Aperia A. Dopamine-induced recruitment of dopamine D1 receptors to the plasma membrane. Proc Natl Acad Sci USA. 1998;95:5573–5578. doi: 10.1073/pnas.95.10.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi M, Narkar VA, Hussain T, Lokhandwala MF. Dopamine recruits D1A receptors to Na-K-ATPase-rich caveolar plasma membranes in rat renal proximal tubules. Am J Physiol Renal Physiol. 2004;287:F921–F931. doi: 10.1152/ajprenal.00023.2004. [DOI] [PubMed] [Google Scholar]
- 26.Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One. 2009;4:e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 31.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 32.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin H, Burnette DT, Bae YK, Forscher P, Barr MM, Rosenbaum JL. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 34.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 35.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 36.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates Hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 37.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rashid AJ, So CH, Kong MM, Furtak T, El-Ghundi M, Cheng R, O’Dowd BF, George SR. D1–D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc Natl Acad Sci USA. 2007;104:654–659. doi: 10.1073/pnas.0604049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeDoux J. The amygdala. Curr Biol. 2007;17:R868–R874. doi: 10.1016/j.cub.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Phelps EA, LeDoux JE. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron. 2005;48:175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 41.Barnett S, Reilly S, Carr L, Ojo I, Beales PL, Charman T. Behavioural phenotype of Bardet-Biedl syndrome. J Med Genet. 2002;39:e76. doi: 10.1136/jmg.39.12.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eichers ER, Abd-El-Barr MM, Paylor R, Lewis RA, Bi W, Lin X, Meehan TP, Stockton DW, Wu SM, Lindsay E, Justice MJ, Beales PL, Katsanis N, Lupski JR. Phenotypic characterization of Bbs4 null mice reveals age-dependent penetrance and variable expressivity. Hum Genet. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.