Abstract
Glioma remains incurable despite great advancements in medicine. Targeting the cell of origin for gliomas could bring great hope for patients. However, as a collection of diverse diseases, each subtype of glioma could derive from a distinct cell of origin. To resolve such a complex problem, one must use multiple research approaches to gain deep insights. Here we review current evidence regarding the cell of origin from clinical observations, whole-genome molecular pathology and glioma animal models. We conclude that neural stem cells, glial progenitors (including oligodendrocyte progenitor cells) and astrocytes could all serve as cells of origin for gliomas, and that cells incurring initial mutations (cells of mutation) might not transform, while their progeny cells could instead transform and act as cells of origin. Further studies with multidisciplinary approaches are needed to link each subtype to a particular cell of origin, and to develop effective therapies that target the signaling network within these cells.
Keywords: animal model, astrocyte, cell of mutation, cell of origin, cell-targeted therapy, malignant glioma, molecular pathology, neural stem cell, oligodendrocyte progenitor cell, tumor subtype
A thorough understanding of the cellular origin of glioma relies on multidisciplinary approaches
Malignant glioma is the most common primary brain tumor. Despite its relatively low occurrence among all cancer types (~1 out of 10,000), the diagnosis of glioma is devastating news for patients and their families. Even with great advancement of surgical procedures and treatment regimens with radiation and chemotherapy, malignant gliomas remain incurable [1]. This is due to their resistance to all conventional therapies and the diffuse infiltrative nature of the tumor cells, which makes complete tumor resection impossible. The most aggressive form, glioblastoma multiforme (GBM; WHO grade IV), has a 12–15-month medium survival rate. Even the lower-grade gliomas (WHO grade II and III gliomas) that appear less aggressive at the time of diagnosis eventually progress into the malignant phase within 5–10 years. In addition to the risk of fatality, the destructive effect of such tumors on brain function often leads to devastating cognitive and emotional deficits for patients. Therefore, accurate diagnosis and curative therapeutic strategies that target the cell of origin for gliomas are urgently needed.
One of the major difficulties for the identification of the cell of origin for gliomas is the complex cellular composition of this disease, or more precisely, these diseases. The histopathological classification of gliomas relies on morphological resemblance of tumor cells to normal cell types in the brain; tumors with astrocytic features are classified as astrocytomas, while those with oligodendroglial features are termed oligodendrogliomas. However, gliomas can also show mixed cellular features and even markedly anaplastic cells; such tumors are classified as oligoastrocytomas and glioblastomas. The source of this heterogeneity is not yet understood: do different subtypes of gliomas originate from different cell types, or do they originate from the same cell type but later give rise to various pathological manifestations due to the effects of distinct genetic mutations or the local microenvironment?
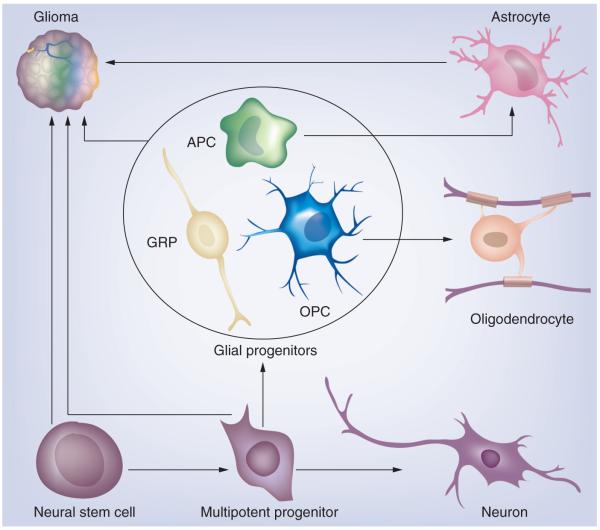
To fully address the cell of origin issue in gliomas, it is important to briefly introduce our current understanding of normal glial development (Figure 1). During brain development, neural stem cells (NSCs) and multipotent progenitor cells that reside in proliferative zones surrounding the ventricles give rise to lineage-restricted progenitor cells, which in turn generate neurons and glial cells (astrocytes and oligodendrocytes). Glial progenitors are heterogeneous; while some of them appear to be bipotent for both astrocyte and oligodendrocyte lineages [2-5], others appear to have relatively restricted potential, such as oligodendrocyte progenitor cells (OPCs), also known as NG2 cells [6]. NSCs and glial progenitors persist into adulthood, continue to self-renew and generate progeny cells throughout a person’s life, and thus are prime suspects as the cell of origin for gliomas [7,8]. However, there are notable differences in the abundance and distribution of these cell populations, as well as in their intrinsic potential to proliferate and differentiate, which may affect the type of tumors these cells can generate. For example, adult multipotent NSCs that express markers such as Nestin and GFAP are generally believed to be relatively small populations that are restricted to discrete brain regions, such as the subventricular zone (SVZ) of the lateral ventricles. By contrast, OPCs that express markers such as OlIG2, NG2 and PDGFRα are abundant and widely distributed throughout the brain, but their potential is normally restricted to the oligodendrocyte lineage. However, it is important to note that glial progenitors can display remarkable phenotypic plasticity under certain conditions, and cells that are normally restricted to one lineage can be induced to express markers of another lineage [8]. Furthermore, OPCs can even be reprogrammed to acquire a multipotent NSC-like phenotype in culture when treated with the appropriate growth factors [9]. Therefore, we must keep in mind possible dedifferentiation during gliomagenesis while interpreting data from studies concerning at the glioma cell of origin.
Figure 1. The gliomagenic potential of the different neural lineages.
Neural stem cells and multipotent progenitors in the subventricular zone can give rise to cells of neuronal and glial lineage. Glial progenitors, which include a heterogeneous population of immature cycling cells, can give rise to astrocytes and oligodendrocytes. Glioma is a heterogeneous disease that can be divided into distinct subtypes. We propose that the different glioma subtypes arise from different cells of origin.
APC: Astrocyte progenitor cell; GRP: Glial-restricted progenitor; OPC: Oligodendrocyte progenitor cell.
In addition to the issue of cellular plasticity, the interpretation of the complex cellular composition of gliomas should also take into account the fact that these tumors are diffusely infiltrative. Therefore, neoplastic cells intermingle with non-neoplastic brain cells, including reactive and entrapped astrocytes, oligodendrocytes, neurons, microglia and blood vessels. While the heterogeneous mass of proliferating cells could arise from the clonal evolution of a transformed cell of origin, recent animal model studies have revealed that gliomas can recruit non-neoplastic glia progenitors to proliferate extensively [10], which in turn acquire their own transforming genetic alterations [11]. The extent to which recruitment and transformation of bystander cells contributes to growth and progression in human disease still awaits further investigation. Nevertheless, such an intriguing prospect has important implications with regards to the question of glioma cellular origin.
We want to acknowledge that the complexity of malignant gliomas has led to many interpretations, one of which is the cancer stem cell hypothesis. The central idea of this hypothesis is that very few renewable and multipotent cancer stem cells propagate and sustain the full composition of cell types in the tumor mass. While this is a tantalizing concept, cancer stem cell and cell of origin are distinct concepts; the former is about a subset of highly malignant tumor cells while the later is about what type of normal cells could be transformed into glioma through genetic mutations. Therefore, cancer stem cell research is beyond the scope of this review, and excellent reviews on this topic can be found elsewhere [12,13].
We believe that a clear understanding of the cell of origin problem for malignant glioma must come from a multidisciplinary approach. In the next three sections, we will summarize the current understanding of cell of origin from three areas: clinical observations; whole-genome molecular/ genetic phenotyping; and glioma animal models. In the last section, we will synthesize the information from all three areas to formulate our current understanding, raise further question and discuss potential clinical implications of the knowledge of the cellular origin for malignant gliomas.
Clues from clinical data
Glioma is a diverse disease that can present with a variety of clinical and histopathological features, suggesting that these tumors may arise from a variety of different cell types. While it is not possible to definitively resolve this question by studying end-stage human tumors, clinical data provides important clues as to the possible origins of gliomas.
What do gliomas look like?
Historically, gliomas have been classified on the basis of histopathological features, with reference to their proposed relationship to specific glial lineages. Some gliomas (called astrocytomas) show morphological similarities to normal and reactive astrocytes, and express astrocytic lineage markers, such GFAP, YKL-40 and ApoE [14]. Other gliomas (called oligodendrogliomas) show similarities to cells of the oligodendrocyte lineage, specifically OPCs, which often express OPC markers including Olig2, NG2 and PDGFRα [14-16]. By contrast, gliomas usually do not express markers of mature oligodendrocytes, such as myelin basic protein, proteolipid protein and myelin-associated glycoprotein [17]. Gliomas are further classified on the basis of tumor grade, with less aggressive astrocytomas and oligodendrogliomas designated as WHO grade II, more aggressive forms designated as WHO grade III and the most aggressive ones as GBM (WHO grade IV). As the name implies, GBM is predominantly composed of cells that resemble immature glia, which display varying degrees of similarity to different glial lineages. Like their lower-grade counterparts, some GBMs show predominantly astrocytic features while others show features of the oligodendroglial lineage. It has long been suspected that these apparent lineage-specific features reflect the developmental origins of gliomas, and suggest that gliomas arise from cells with a glial-restricted potential. However, there can be intratumoral heterogeneity with regards to the expression of these lineage markers, and it is not uncommon for gliomas to show a mixed oligodendroglial–astrocytic phenotype. This suggests that some gliomas may arise from bipotent glial progenitors with the capacity to differentiate along both astrocyte and oligodendrocyte lineages. It is also possible that gliomas arise from multipotent progenitor/stem cells, or from more differentiated glial cells that acquire an immature glial phenotype during the process of transformation.
Where do gliomas occur?
While most gliomas occur in the cerebral hemispheres, gliomas can arise anywhere in the CNS, suggesting that cells with the capacity to form gliomas are widely distributed throughout the brain and spinal cord. OPCs represent the most abundant and widely distributed population of cycling cells in the adult brain [18-21], making OPCs a compelling candidate for the potential cell of origin for gliomas. However, considering the migratory potential of glioma cells, it is also possible that some gliomas arise from more discretely localized populations of progenitors or stem cells, which disperse throughout the brain during the process of gliomagenesis. Radiographic data suggest that some GBMs arise in, or near, the SVZ adjacent to the lateral ventricles, while other GBMs arise in the more superficial subcortical white matter [22,23]. Furthermore, these studies suggest that tumors arising adjacent to the lateral ventricles may have distinct growth characteristics and clinical outcomes from tumors arising in the more superficial white matter. In addition, a recent study has shown that proneural gliomas with IDH1 mutations are significantly more likely to occur in frontal lobes, suggesting that these tumors may arise from a distinct population of progenitors that reside in this region of the brain [24]. While it is possible that variation of tumor phenotypes could be caused by region-specific differences in the brain microenvironment, it could also be affected by the intrinsic properties of the local progenitor population, as has been shown for medulloblastoma and ependymoma [25,26]. Together, these studies suggest that tumors arising in different regions of the brain may arise from distinct progenitor populations. More work is needed to elucidate how region-specific differences in progenitor populations may affect gliomagenesis.
When do gliomas occur?
Gliomas can occur at any age. However, the majority of malignant gliomas occur in adults, suggesting that the cells that give rise to these tumors reside in the adult brain, either as genetically normal cells, or as partially transformed cells that have not yet formed clinically detectable lesions. Gliomas that occur at different ages are clinically and molecularly distinct, suggesting that the tumorigenic potential of the cells that give rise to them is different. For example, primary GBMs tend to occur in older patients, whereas lower-grade gliomas and secondary GBMs (which progress from lower-grade gliomas) tend to occur in younger patients. Furthermore, age at diagnosis is one of the most important determinants of outcome, with younger patients having a significantly better prognosis [27,28]. While better survival may be explained, in part, by the overall better health status of younger patients, there is also good reason to suspect that tumors in younger patients are biologically distinct. The tumors from young and old patients tend to harbor different types of molecular/genetic alterations, suggesting that the progenitor populations that give rise to these different types of gliomas have distinct sensitivity to certain genetic mutations. In support of this idea, several studies have found that the proliferation and differentiation potentials of progenitor populations change during aging [4,29,30]. Furthermore, studies have suggested that these age-related changes of progenitor properties may result from alterations in growth factor responsiveness and tumor suppressor expression [31-33]. It is also possible that developmentally related changes in the brain environment, such as age-related changes in immune function and progenitor recruitment, could have effects on the patterns of gliomagenesis [34,35]. Additional studies are needed to tease these possibilities apart.
When do the initiating genetic mutations occur?
Inherited germline mutations associated with familial cancer syndromes, such as Li–Fraumeni syndrome [36], are responsible for a small fraction of glioma cases. These rare familial cases provide important ‘proof of principle’ as to the potential mechanisms of gliomagenesis. Even though the genetic alteration is inherited by every cell in the body, and is present throughout embryonic and postnatal development, the resulting tumors often do not manifest until the patient is in their second or third decade of life. Thus, in some cases gliomagenesis can be a prolonged multistep process that begins with a predisposing genetic alteration in an early progenitor and then takes years to evolve into clinical disease. Conversely, there is evidence from patients that have had MRI for unrelated reasons prior to developing clinical symptoms of brain tumor that gliomas can evolve from being radiographically undetectable to full-blown GBM in a matter of months. These rare cases provide insight into the dynamics of gliomagenesis, and further suggest that GBM is a heterogeneous disease that cannot be attributed to a single cell of origin or a single pattern of transformation.
Clues from whole-genome molecular/genetic phenotyping
Cancer is a genetic disease and some of the most dramatic improvements in the treatment of tumors seen in the past two decades have been the result of associations between genomic abnormalities and targeted therapies [37-39]. However, such success has not yet been seen in the treatment of gliomas. Unbiased surveys of genomic profiles are required to identify novel recurrent alterations that may be therapeutically targetable. The Cancer Genome Atlas (TCGA) project is an ambitious initiative founded by the National Cancer Institute in 2005, which aims to accelerate understanding of the molecular basis of cancer through cataloging the genomic changes present in more than 3% of cases. In an initial report, TCGA described that the majority of GBM samples contain alterations in three critical pathways: the RB1 pathway, to remove cell cycle checkpoints; the TP53 pathway, to evade apoptosis and senescence responses; and RTK pathways, resulting in increased proliferation [40]. Interestingly, these findings were corroborated in a set of mouse models in which glioma was induced and the genetic alterations were extensively characterized [41]. TCGA findings were based on the sequencing of a set of 600 genes in 91 tumor samples, in addition to genome-wide DNA copy number profiling on 206 GBMs. TCGA has expanded its sample collection to over 500 cases and is currently undertaking genome-wide analysis of its entire cohort, which may indicate that defects or activating mutations outside these three key pathways are necessary to induce tumorigenesis. Given the variability in histological and clinical presentation of GBM, it appears unlikely that the full spectrum of the disease can be explained by a single unifying pattern of molecular/genetic alterations. Rather, we propose that gliomas originating from different progenitor cell types are subject to distinct mechanisms of malignant transformation.
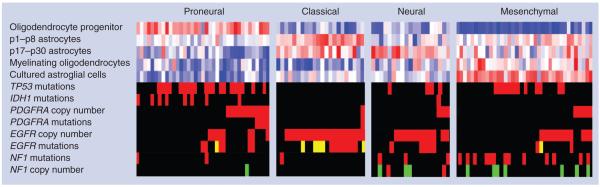
Recently, molecular subtypes of GBM have been described based on different patterns of expression profiles and methylation profiles [42-51]. The presence of four expression subtypes were reported in TCGA GBM expression data set, referred to as proneural, neural, classical and mesenchymal, which were confirmed in an independent set of published GBM expression profiles [50]. Interestingly, specific associations between expression subtypes, genomic abnormalities and relationships to glial lineages were found. For instance, EGFR amplification, mutations and vIII intragenic deletions (known as EGFRvIII variant) were found in 97% of cases expressing the classical signature; the subtype that showed astrocyte gene expression signatures (Figure 2). Interestingly, EGF receptor (EGFR) signaling plays an important role in maintaining pluripotency of NSCs and EGF stimulation can induce progenitor/stem cells in the SVZ to proliferate extensively and form glioma-like lesions [52]. In contrast, amplification and mutations of PDGFRA are most commonly seen in proneural GBM, and proteomic studies revealed that activation of PDGF signaling is associated with this phenotype [53]. Since PDGFRα is selectively expressed in OPCs in the normal brain and PDGF signaling is critical to stimulate the proliferation and migration of OPCs [54-56], OPCs could be the candidate cell of origin for proneural gliomas. The proneural subtype is further characterized by the presence of mutations in IDH1 that co-occur with the glioma-CpG island methylator phenotype (G-CIMP) hypermethylation signature [46]. Although unsupervised expression cluster analysis did not separate G-CIMP/IDH1 mutant tumor samples into an individual cluster, supervised expression analysis could predict the presence of G-CIMP and the proneural subtype may thus be further separated into G-CIMP and non-G-CIMP tumor samples [46]. Patients whose tumor harbors an IDH1 mutation form a unique class of GBM patients with a prolonged survival and a younger age. IDH1 mutations are found in the majority of lower-grade gliomas and secondary GBMs [57]. Although IDH1 is thought to play a role in cellular metabolism, recent reports showed that mutant, but not wild-type, IDH1 is able to remodel the methylome of primary human astrocytes [58].
Figure 2. Transcriptomic and genomic associations of glioblastoma multiforme gene expression subtypes.
Proneural, neural, classical and mesenchymal gene expression subtypes were established and associated with genomic abnormalities in a set of 116 glioblastoma multiforme (GBM) samples. The first five rows represent the overall level of expression of a number of developmental neural lineage gene sets in each of the 116 GBMs, with each column representing one GBM sample and each row representing a gene set. Blue indicates a lack of gene set activation, and red denotes a high level of expression. Gene set activation was assessed using single sample gene set enrichment analysis. Sample gene set enrichment analysis scores were calculated for five developmental mouse neural lineages. The next eight rows visualize the relationship between GBM expression subtypes and genomic associations. In rows TP53 mutations, IDH1 mutations, PDGFRA mutations, EGFR mutations and NF1 mutations: red indicates presence of a point mutation; yellow indicates presence of the vIII variant/intragenic deletion; and black indicates wild-type. In rows PDGFRA copy number, EGFR copy number and NF1 copy number: red indicates presence of a DNA copy number gain; green indicates presence of DNA copy number deletion; and black indicates diploid DNA copy number.
There are at least two possible explanations for the association between gene signature and genomic abnormality:
The ability of genomic alterations to stimulate tumor growth is determined by the tumor cell transcriptome and these changes are therefore more likely to be found in cells with the optimal expression context;
The expression pattern is the result of the set of genomic alterations found in the tumor.
As EGFR alterations are found in nonclassical GBMs, and PDGFRA alterations are seen in nonproneural samples, these genomic changes are unlikely to be the only or even the dominant determinant of the tumor cell transcriptome, deeming the latter option less likely. The set of genomic abnormalities found in a tumor sample is a combination of initiating genetic mutations and acquired alterations. The former are present in the tumor cell of origin and will be propagated to all clonally related tumor cells. Analysis of the percentage of tumor cells in which mutations and DNA copy number changes are found may shed light on the relation between expression signature and genomic abnormality. Analysis of the number of cytogenetically defined tumor clones using interphase FISH and assessment of histopathology features suggested that GBMs typically harbor multiple subclones [59,60]. In a survey of TP53 mutations in grade II–IV gliomas, distinct mutations were found in different sections of the same GBM [61]. Analysis of TP53 mutations in TCGA data set showed that approximately 25% of GBMs contain multiple TP53 mutations [40]. A small subset of GBM tumors were reported to contain mosaic amplification of multiple receptor tyrosine kinases, such as PDFRA and EGFR, which led to the conclusion that different subpopulations existed within each tumor that were all derived from the same clonal origin [62,63].
The frequent infiltration of nontumor cells in GBM samples affects the expression profiles by adding nontumoral mRNA to the transcriptomic mixture. This is reflected in the expression subtypes through the increased level of necrosis and vascular proliferation seen in samples expressing the mesenchymal signature [50]. However, it is unlikely that the mesenchymal class is exclusively the result of infiltrating vasculature and necrosis since glioma cell lines and glioma mouse models expressing the mesenchymal signature have been identified [41,50]. Furthermore, one study found that gene expression profiles from different regions of the same tumor showed a surprising similarity, even though there was histological variability with respect to the amount of vascular proliferation and necrosis seen in these different regions [43]. These results suggest that the global expression profiles are largely determined by inherent properties of the tumor cells, which we propose are derived from the cell of origin. However, most studies of intratumoral heterogeneity of GBM suffer from small sample size and more research is necessary before conclusive insights into the developmental mechanism of tumorigenesis are obtained. Notably, studies of the clonality of genomic aberrations are becoming increasingly feasible with the ability to perform deep sequencing of tumor DNA with a large series of GBM samples by TCGA, which should provide further insights into the clonality of GBM [64]. Interestingly, mutational analysis recently suggested that the proneural-associated mutations in IDH1 are an initiating and thus clonal event in the cell of origin [24].
Clues from animal models
While analysis of clinical data and molecular signatures has been providing critical information that implicates the cell of origin for gliomas, one cannot definitively identify the cell of origin by studying terminal-stage tumors. In this respect, studies with animal models have a unique advantage since one can introduce gliomagenic mutations into a specific cell type and directly test its transforming ability.
To create the mouse models, one needs to introduce genetic mutations that are relevant to human glioma into specific cell types. As stated above, gliomagenic mutations include the aberrant overexpression or activation of RTK pathways due to activating mutations or overexpression of PDGFRα, EGFR, loss of NF1 or PTEN, and the loss or decreased activity of cell cycle checkpoint genes such as p53, p16INK4a, p19Arf and Rb. To introduce these mutations into specific cell types in mouse models, at least three genetic methods are commonly used: the RCAS-TVA system; the retroviral system; and the Cre/loxP or tTA/TetO binary genetic system. The cellular specificity of mutations is achieved by expressing the TVA receptor, Cre recombinase or growth factor(s) and their constitutively active receptors, under the control of cell type-specific promoters.
A few pioneering mouse models successfully induced malignant gliomagenesis with combined defects in both RTK signaling and cell cycle regulating pathways, such as EGFR overexpression plus INK4a deficiency [65], the loss of p53 and NF1 [66], or inactivated Rb pathway and PTEN [67]. These studies functionally demonstrated the important roles of these pathways in gliomagenesis. Furthermore, the cell of origin problem was investigated using the RCAS-TVA system to introduce mutations into either nestin-expressing NSCs or GFAP-expressing astrocytes [65]. In this system it was found that gliomas formed with higher penetrance in nestin-TVA mice, suggesting that nestin-expressing progenitors or NSCs may have higher transforming potentials than GFAP-expressing astrocytes. However, recent studies have shown that NSCs in the SVZ also express GFAP [68]; thus one cannot distinguish NSCs from astrocytes on the basis of GFAP expression alone. Two additional studies have addressed the transforming potentials of GFAP astrocytes and NSC. In one study, it was shown that both GFAP-expressing astrocytes and NSCs could be transformed in culture with a combination of EGFR activation and loss of both p16INK4a and p19ARF tumor suppressors, and would give rise to a similar high-grade glioma phenotype when transplanted into the brains of SCID mice [69]. Another study used GFAP-CreER that is expressed in both NSCs and mature astrocytes to inactivate p53 and PTEN simultaneously [41]. Although many tumors were found at regions contiguous to NSC niches, one-fifth of tumors were located in brain parenchyma far away from the NSC proliferative zone. Furthermore, no aberrant proliferation was found in the SVZ of these brains, arguing against the possibility that these tumors arose from NSCs and then migrated out into brain parenchyma. Together, these studies suggest that both GFAP-expressing NSCs in the SVZ and GFAP-expressing astrocytes outside the SVZ have the capacity to form brain tumors. Interestingly, the tumors localized in the different brain regions had different phenotypes, with tumors involving the leptomeninges having a more mesenchymal expression profile, suggesting that the genetic alterations required to transform these cells and the phenotypes of the tumors they give rise to may be different.
The gliomagenic potential of NSCs was substantiated by a few elegant genetic models. The inactivation of p53 and PTEN in embryonic NSCs led to gliomagenesis [70]. The introduction of p53 and NF1 mutations into either embryonic NSCs with hGFAP-Cre [71] or adult NSCs with nestin-CreER [72] consistently generated malignant gliomas with many pathological features found in human tumors. The close proximity of early lesions to the SVZ, and the successful induction of glioma by stereotaxic injection of Cre into the SVZ (but not into white matter or cortex), suggest that NSCs gave rise to glioma in this model [72]. Taken together, these data suggest that both embryonic and adult NSCs could serve as the glioma cell of origin.
In addition to astrocytes and NSCs, an extensive body of work has provided evidence that glial progenitors widely distributed throughout the brain can also serve as the cell of origin for malignant gliomas. Several studies have described murine glioma models in which PDGF is overexpressed in various populations of glial and neural progenitors in the neonatal brains [73-75]. Moreover, infecting glial progenitors that reside in the subcortical white matter of adult rats and mice with PDGF-expressing retrovirus induces these cells to form tumors with close resemblance to human glioblastoma [10,76]. More recently, it was shown that selectively deleting tumor suppressors (PTEN and p53) in adult white matter progenitors cooperates with PDGF signaling to facilitate the transformation of these cells. Cross-species comparisons of global expression patterns showed that these PDGF-driven mouse gliomas most closely resemble proneural GBM [77]. Furthermore, this study showed that both the mouse and human proneural gliomas are highly enriched for OPC genes. In addition to the PDGF-overexpression models, a genetic mouse model that overexpresses an activated allele of EGFR (v-erbB) in a p53-mutant background demonstrated that OPCs can also give rise to tumors with the histological features of malignant oligodendrogliomas [78]. Furthermore, NG2-expressing OPC-like cells isolated from these tumors, but not CD15-expressing NSC-like cells, were able to form secondary tumors when transplanted into naive mice. Therefore, the notion of OPC as glioma cell of origin is also well supported by a wealth of evidence from animal models with diverse gene mutations.
Recently, a mouse model based on a genetic mosaic system, called mosaic analysis with double markers [79], added a new twist to our understanding of the cell of origin for gliomas [80]. Mosaic analysis with double markers generates sparse mutant cells that are unambiguously labeled with GFP and their sibling wild-type cells with RFP, allowing cellular resolution analysis of tumorigenic processes prior to malignancy. After introducing p53 and NF1 mutations into NSCs, cellular expansion (mutant to wild-type cell ratio) was quantified in each cell lineage derived from NSCs at a pretransforming stage. Surprisingly, there was no or limited expansions in NSCs, astrocytic and neuronal lineages but dramatic overgrowth of OPCs. At a later age, GFP-positive tumors with features of malignant gliomas formed at high penetrance. OPC features of GFP-positive tumor cells were confirmed by both marker staining and transcriptome profiling. Lastly, direct introduction of p53 and NF1 mutations into OPCs also resulted in glioma formation, suggesting that the initial genetic alterations may occur in either NSCs or OPCs; however, OPCs serve as the transforming origin in this model (Figure 3) [80] .
Figure 3. The distinction between cell of mutation and cell of origin for gliomas.
In genetically engineered animal glioma models, tumorigenic mutations could be introduced into a neural stem cell or glial progenitor population. It should be noted that NSCs, the cell type in which the mutations are introduced, might not have the signaling context for malignant transformation, and hence should be called the ‘cell of mutation‘. On the other hand, OPCs, a progeny cell type derived from NSCs, actively respond to the genetic mutations, overexpand and eventually lead to the formation of malignant glioma. In this case, OPCs, as the transforming cell type, should be considered the ‘cell of origin‘, a critical target for effective therapies.
NSC: Neural stem cell; OPC: Oligodendrocyte progenitor cell. Adapted with permission from [80].
While animal studies have been informative, there have also been controversies. These debates could be partly attributed to the intrinsic complexity of gliomas, and partly to technical caveats. Here we want to emphasize a few important considerations while using animal models to address the cell of origin issue. First, it is absolutely critical to validate transgenic lines for their faithful expression in intended cell types. For example, both nestin and GFAP promoters are dynamically regulated, and can drive expression in more than one cell type. Although nestin has been widely used as a NSC marker, its expression was also well documented in reactive astrocyte and in perinatal OPCs [81]. A similar situation applies to GFAP: despite being widely accepted as an astrocyte marker, recent studies demonstrated that GFAP is also expressed in NSCs [68], and the human GFAP promoter, but not the mouse GFAP promoter, is also active in OPCs [82]. Second, most studies using transgenic mice introduce genetic alterations at early stages of brain development and to a broad population of cells. This approach may recapitulate familial cancer syndromes, such as Li–Fraumeni syndrome, but clinical evidence suggests that the vast majority of gliomas occur in elderly individuals that do not harbor germline mutations. Therefore, it would be important to use inducible genetic tools to introduce genetic changes in adult animals to validate earlier findings. The fact that gliomas most likely start as clonal transformations also calls for the use of retroviral delivery or genetic mosaic models such as the mosaic analysis with double markers system mentioned above. Third, whenever mutations are introduced into multipotent progenitor cells (such as NSCs), it is critical to analyze the transforming ability of all subsequent lineages before claiming the cell of origin, as illustrated by a study mentioned above (Figure 3) [80]. Finally, one must consider the possibility of species–specific differences. The progenitor populations found in humans and mice might have different proliferative capacities and could rely on different mechanisms to safeguard against transformation. Nevertheless, cross-species comparisons are highly valuable to unveil the cellular origin of human gliomas.
In conclusion, based on the data from all these animal studies, we can now conclude (Figure 1) . We postulate that distinct combinations of genetic mutations could preferentially transform a particular cell of origin. The underlying molecular mechanisms for such a synergistic effect should be explored for developing effective treatments.
Summary
Current understanding of the cellular origin of glioma
In the sections above, we illustrated the importance of using multiple approaches to understand the cellular origin of gliomas. Clinical observations help us formulate highly relevant hypotheses because the marker expression by tumor cells, the location and onset timing of tumors all provide important clues toward the cell of origin of gliomas. Recent molecular phenotyping at a genome-wide level not only greatly advanced our categorization of gliomas by dividing morphologically indistinguishable tumors into four subtypes based on gene expression signatures and unique genomic aberrations, but also shed light on potential cell types that are responsible for particular subtypes of glioma. Finally, animal model studies have allowed us to directly introduce gene mutations commonly found in human glioma patients into specific progenitor cell types, providing the most direct way to test hypotheses formulated from the other two methods. These highly relevant mouse models can also be used for preclinical testing of drug candidates prior to human trials. Combining currently available evidence from multiple approaches, we conclude that NSCs, glial progenitors (including OPCs) and astrocytes could all serve as the cell of origin for gliomas. The relationship between the four subtypes and the cell of origin awaits further elucidation. It is possible that a distinct cell of origin could give rise to each subtype, or distinct genetic mutations might transform the same cell of origin into different pathological manifestations, or both could be true. Lastly, we would also like to point out that our interest in the cell of origin problem is not for the teleological argument since in principle every cell is derived from a fertilized egg but claiming it as the tumor cell of origin has no real meaning. Rather, we propose that defining the specific cellular context in which the process of transformation takes place will lead to a deeper understanding of the mechanisms of gliomagenesis and to the development of more effective methods to treat this disease.
The prognostic significance of the cell of origin
Gliomas as a group, and GBM in particular, are highly aggressive tumors. The average survival is approximately 14 months after diagnosis and use of currently available therapy. However, there are exceptions to this general trend and a subset of patients with malignant gliomas can show robust response to therapy and live for many years. As a general rule, gliomas with a more astrocytic phenotype are associated with a worse prognosis; astrocytomas tend to behave more aggressively than do oligodendrogliomas of the same grade [83]. However, there is also marked variability in behavior within these general lineage-defined categories, and several studies have revealed molecular genetic traits associated with good prognosis, including loss of heterozygosity of 1p 19q [84], mutations in IDH1 [57] and G-CIMP [46]. Interestingly, these genetic alterations are more common in oligodendrogliomas and proneural GBMs, which show phenotypic similarities to OPC, suggesting that these prognostic features may be inherently linked to the specific cell types from which these tumors arise. These findings highlight the potential clinical relevance of categorizing gliomas from the perspective of the cell of origin.
Implications for diagnosis & treatment
The understanding of the cell of origin should also be very important for patient stratification with tailored treatment methods. Currently, the standard care for glioma patients is temozolomide with radiation therapy [1]. However, if we could devise delivery methods that target the specific type of cell of origin in each individual patient, we should be able to improve the effectiveness of treatment and reduce collateral damage. The identification of the cellular origin of gliomas could aid in drug development in two ways. First, we could target their surface molecules for drug delivery, thereby reducing toxicity to other cell types in the brain. Second, through the in-depth understanding of the intrinsic signaling network that allowed the glioma cellular origin to respond to particular genetic mutations, one could devise highly effective therapeutic strategies by disrupting critical nodes in these networks. The subtype-specific mutations or gene expression changes should be a great starting point to design these strategies.
Expert commentary
Current status of the field & recommendations for existing & new paradigm choices
Malignant glioma is the most common primary brain tumor and remains incurable. This is due to the resistance of tumors to all conventional therapies and the diffuse infiltrative nature of tumor cells, which makes complete tumor resection impossible. In addition to the high fatality rate, the destructive effect of such tumors on brain functions often leads to devastating cognitive and emotional deficits for patients. Therefore, accurate diagnosis and curative therapeutic strategies that target the cell of origin for gliomas are urgently needed. Unfortunately, one of major difficulties of identifying the cellular origin of gliomas is the complex cellular composition of the disease. Based on genome-wide expression analyses, gliomas can be divided into four distinct subtypes: proneural, neural, classical and mesenchymal. The source of this heterogeneity is not yet understood: these tumors could originate from different cell types, or they could originate from the same cell type but later give rise to various pathological manifestations due to the effects of distinct genetic mutations or influences of the local microenvironment. In addition to the complexity between different tumors, heterogeneity within the same tumor is another puzzle for understanding the cell of origin problem. Tumor cells display intratumoral heterogeneity in terms of gene mutations and chromosome aberrations, but more studies are required to identify the source of such a mixed cell population, derived either from surrounding cells attracted by tumor mass that go on a distinct mutational path, or from the continuous clonal evolution process of the original tumor cells. Overall, it is imperative to fully understand the cell of origin for gliomas for accurate diagnosis and effective treatment.
Five-year view
In the next 5 years, the field will quickly adopt a multidisciplinary approach to investigate the glioma cell of origin problem based on clinical data, molecular profiling and animal modeling. Effective communication and continuous revision of our current understanding will be key for the rapid progression of the field. With collective efforts, glioma could be reclassified based on cell of origin and corresponding genetic mutations. Research will intensify on the signaling context of each cell of origin, with the goal to identify the transforming susceptibility in their signaling network in response to tumorigenic mutations. While animal model research will help identify the cell of origin for each subtype of gliomas, cell biology and molecular genetic studies will provide critical information as to why the cell of origin is particularly susceptible to genetic mutations for malignant transformation. Cross-species comparisons will determine the clinical relevance of animal models to the human disease. Deep sequencing will allow studies of the clonality of mutations. Systems biology and biocomputation will be employed to resolve complex issues and identify nodes in the regulatory network as therapeutic targets. For the diagnostic process, molecular profiling will be much more frequently used to complement histopathology for patient stratification, prognostic prediction and treatment planning. For patient treatment, cell-targeted therapy will be explored in several ways: first, ‘smart missile‘ drugs that target cell surface molecules of the cell of origin; second, systemic intervention drugs that target signaling susceptibility of cell of origin; and third, differentiation therapy drugs that reduce the developmental potential of the glioma cell of origin and force the cells to differentiate into postmitotic mature cell types. Finally, the drug development process will be greatly informed by highly sophisticated animal models and drug efficacy will be assessed based on whole-genome molecular profiling in addition to simple measurement of tumor sizes. Overall, the field will grasp the cell of origin problem and move toward a deeper understanding of molecular and cellular changes in gliomas.
Key issues.
Glioma is a diverse disease that can present with a variety of clinical and histopathological features, for which accurate diagnosis and curative therapeutic strategies are urgently needed.
Understanding the cell of origin of glioma would allow for the design of targeted therapies. * A thorough understanding of the glioma cell of origin relies on multidisciplinary approaches including clinical observations, whole-genome molecular profiling and animal models.
Glioma cells resemble immature glial cells: some gliomas more closely resemble cells of the astrocyte lineage; others more closely resemble cells of the oligodendrocyte lineage; yet others show mixed cellular features.
Genome-wide expression profiling has identified four distinct subtypes of glioma, referred to as proneural, neural, classical and mesenchymal.
We propose that gliomas originating from different progenitor cell types are subject to distinct mechanisms of malignant transformation.
Animal model studies have shown that neural stem cells in the subventricular zone and glial progenitors, in particular oligodendrocyte progenitor cells, that are widely distributed throughout the brain can be induced to form gliomas.
Cross-species analyses have shown that certain animal models resemble specific subtypes of human glioma. More specifically, oligodendrocyte progenitor cell-originated gliomas in mouse models most closely resemble proneural gliomas, providing an experimental link between specific cells of origin and glioma subtype.
Genetic mosaic mouse models demonstrated that the cell type that initially incurs mutations (cell of mutation) and the cell type that eventually transforms into malignancy (cell of origin) could be distinct, and that the latter should be the focus for therapeutic purposes.
Our in-depth understanding of the glioma cell of origin will provide critical information for clinical diagnoses and innovative ideas for drug development.
Acknowledgements
We thank Ben Barres for helpful discussions.
Footnotes
Financial & competing interests disclosure
H Zong would like to acknowledge grant support by National Cancer Institute (NIH/NCI) grant number R01CA136495 and the WM Keck Foundation. H Zong is a Pew Scholar in Biomedical Sciences, supported by The Pew Charitable Trusts. RGW Verhaak would like to acknowledge support by National Cancer Institute (NIH/NCI) grant number U24CA143883. P Canoll would like to acknowledge support by National Institute of Neurological Disease (NIH/NINDS) grant number R01NS066955. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH.The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Stupp R, Mason WP, Van Den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Raff MC, Abney ER, Fok-Seang J. Reconstitution of a developmental clock in vitro: a critical role for astrocytes in the timing of oligodendrocyte differentiation. Cell. 1985;42(1):61–69. doi: 10.1016/s0092-8674(85)80101-x. [DOI] [PubMed] [Google Scholar]
- 3.Raff MC, Williams BP, Miller RH. The in vitro differentiation of a bipotential glial progenitor cell. EMBO J. 1984;3(8):1857–1864. doi: 10.1002/j.1460-2075.1984.tb02059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolswijk G, Noble M. Identification of an adult-specific glial progenitor cell. Development. 1989;105(2):387–400. doi: 10.1242/dev.105.2.387. [DOI] [PubMed] [Google Scholar]
- 5.Rao MS, Mayer-Proschel M. Glial-restricted precursors are derived from multipotent neuroepithelial stem cells. Dev. Biol. 1997;188(1):48–63. doi: 10.1006/dbio.1997.8597. [DOI] [PubMed] [Google Scholar]
- 6.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 2009;10(1):9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 7.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005;353(8):811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 8.Canoll P, Goldman JE. The interface between glial progenitors and gliomas. Acta Neuropathol. 2008;116(5):465–477. doi: 10.1007/s00401-008-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289(5485):1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- 10.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 2006;26(25):6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. •• Showed that glial progenitors in adult white matter can be induced to form brain tumors that resemble glioblastoma multiforme (GBM) and that nontransformed glial progenitors are recruited by the tumor and stimulated to proliferate by paracrine growth factor stimulation.
- 11.Fomchenko EI, Dougherty JD, Helmy KY, et al. Recruited cells can become transformed and overtake PDGF-induced murine gliomas in vivo during tumor progression. PLoS One. 2011;6(7):e20605. doi: 10.1371/journal.pone.0020605. •• Showed that recruited glial progenitors accumulate genetic alterations and become neoplastic glia that contribute to tumor growth.
- 12.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58(6):832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 13.Park DM, Rich JN. Biology of glioma cancer stem cells. Mol. Cells. 2009;28(1):7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 14.Rousseau A, Nutt Cl, Betensky RA, et al. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2-2. J. Neuropathol. Exp. Neurol. 2006;65(12):1149–1156. doi: 10.1097/01.jnen.0000248543.90304.2b. [DOI] [PubMed] [Google Scholar]
- 15.Shoshan Y, Nishiyama A, Chang A, et al. Expression of oligodendrocyte progenitor cell antigens by gliomas: implications for the histogenesis of brain tumors. Proc. Natl Acad. Sci. USA. 1999;96(18):10361–10366. doi: 10.1073/pnas.96.18.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riemenschneider MJ, Koy TH, Reifenberger G. Expression of oligodendrocyte lineage genes in oligodendroglial and astrocytic gliomas. Acta Neuropathol. 2004;107(3):277–282. doi: 10.1007/s00401-003-0809-8. [DOI] [PubMed] [Google Scholar]
- 17.Sung CC, Collins R, Li J, et al. Glycolipids and myelin proteins in human oligodendrogliomas. Glycoconj. J. 1996;13(3):433–443. doi: 10.1007/BF00731476. [DOI] [PubMed] [Google Scholar]
- 18.Geha S, Pallud J, Junier MP, et al. NG2+/ Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 2010;20(2):399–411. doi: 10.1111/j.1750-3639.2009.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell Neurosci. 2003;24(2):476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 20.Rhee W, Ray S, Yokoo H, et al. Quantitative analysis of mitotic Olig2 cells in adult human brain and gliomas: implications for glioma histogenesis and biology. Glia. 2009;57(5):510–523. doi: 10.1002/glia.20780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy NS, Wang S, Harrison-Restelli C, et al. Identification, isolation, and promoter-defined separation of mitotic oligodendrocyte progenitor cells from the adult human subcortical white matter. J. Neurosci. 1999;19(22):9986–9995. doi: 10.1523/JNEUROSCI.19-22-09986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohman LE, Swanson KR, Moore JL, et al. Magnetic resonance imaging characteristics of glioblastoma multiforme: implications for understanding glioma ontogeny. Neurosurgery. 67(5):1319–1327. doi: 10.1227/NEU.0b013e3181f556ab. discussion 1327-1318 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim DA, Cha S, Mayo MC, et al. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro. Oncol. 2007;9(4):424–429. doi: 10.1215/15228517-2007-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J. Clin. Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. • Showed that proneural GBMs with IDH1 mutations are clinically and pathologically distinct, with features that suggest they arise from oligodendrocyte progenitor cells (OPCs).
- 25.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468(7327):1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curran WJ, Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three radiation therapy oncology group malignant glioma trials. J. Natl Cancer Inst. 1993;85(9):704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence YR, Mishra MV, Werner-Wasik M, et al. Improving prognosis of glioblastoma in the 21st century: Who has benefited most? Cancer. 2011 doi: 10.1002/cncr.26685. doi:10.1002/ cncr.26685. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 29.Stoll EA, Habibi BA, Mikheev AM, et al. Increased re-entry into cell cycle mitigates age-related neurogenic decline in the murine subventricular zone. Stem Cells. 2011;29(12):2005–2017. doi: 10.1002/stem.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gritti A, Dal Molin M, Foroni C, Bonfanti L. Effects of developmental age, brain region, and time in culture on long-term proliferation and multipotency of neural stem cell populations. J. Comp. Neurol. 2009;517(3):333–349. doi: 10.1002/cne.22153. [DOI] [PubMed] [Google Scholar]
- 31.Enwere E, Shingo T, Gregg C, Fujikawa H, Ohta S, Weiss S. Aging results in reduced epidermal growth factor receptor signaling, diminished olfactory neurogenesis, and deficits in fine olfactory discrimination. J. Neurosci. 2004;24(38):8354–8365. doi: 10.1523/JNEUROSCI.2751-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molofsky AV, Slutsky SG, Joseph NM, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol. Aging. 2009;30(3):483–497. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass R, Synowitz M, Kronenberg G, et al. Glioblastoma-induced attraction of endogenous neural precursor cells is associated with improved survival. J. Neurosci. 2005;25(10):2637–2646. doi: 10.1523/JNEUROSCI.5118-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler CJ, Black KL, Liu G, et al. Thymic CD8+ T cell production strongly influences tumor antigen recognition and age-dependent glioma mortality. J. Immunol. 2003;171(9):4927–4933. doi: 10.4049/jimmunol.171.9.4927. [DOI] [PubMed] [Google Scholar]
- 36.Kleihues P, Schauble B, Zur Hausen A, Esteve J, Ohgaki H. Tumors associated with p53 germline mutations: a synopsis of 91 families. Am. J. Pathol. 1997;150(1):1–13. [PMC free article] [PubMed] [Google Scholar]
- 37.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 38.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 39.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chow LM, Endersby R, Zhu X, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. •• Used transgenic mouse models to test the tumorigenic effects of specific combinations of initiating genetic hits and performed a genome-wide analysis of the copy number alterations that accumulated in the resulting tumors.
- 42.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc. Natl Acad. Sci. USA. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 45.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 46.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 48.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. • A pioneer paper using molecular profiling to divide gliomas into subclasses.
- 49.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. •• A comprehensive paper that use molecular profiling to divide gliomas into four subtypes, analyze distinct genetic mutations and chromosomal alterations and implicate the cell of origin of some subtypes of gliomas.
- 51.Zheng S, Chheda MG, Verhaak RG. Studying a complex tumor: potential and pitfalls. Cancer J. 2012;18(1):107–114. doi: 10.1097/PPO.0b013e3182431c57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36(6):1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 53.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pringle NP, Mudhar HS, Collarini EJ, Richardson WD. PDGF receptors in the rat CNS: during late neurogenesis, PDGFα-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development. 1992;115(2):535–551. doi: 10.1242/dev.115.2.535. [DOI] [PubMed] [Google Scholar]
- 55.Chojnacki AK, Mak GK, Weiss S. Identity crisis for adult periventricular neural stem cells: subventricular zone astrocytes, ependymal cells or both? Nat. Rev. Neurosci. 2009;10(2):153–163. doi: 10.1038/nrn2571. [DOI] [PubMed] [Google Scholar]
- 56.Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/ type-2 astrocyte progenitor cell. Nature. 1988;333(6173):560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- 57.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vital AL, Tabernero MD, Crespo I, et al. Intratumoral patterns of clonal evolution in gliomas. Neurogenetics. 2009;11(2):227–239. doi: 10.1007/s10048-009-0217-x. [DOI] [PubMed] [Google Scholar]
- 60.Paulus W, Peiffer J. Intratumoral histologic heterogeneity of gliomas. A quantitative study. Cancer. 1989;64(2):442–447. doi: 10.1002/1097-0142(19890715)64:2<442::aid-cncr2820640217>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 61.Ren ZP, Olofsson T, Qu M, et al. Molecular genetic analysis of p53 intratumoral heterogeneity in human astrocytic brain tumors. J. Neuropathol. Exp. Neurol. 2007;66(10):944–954. doi: 10.1097/nen.0b013e318156bc05. [DOI] [PubMed] [Google Scholar]
- 62.Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 63.Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. •• References [59-63] identified distinct pathological features, mutations or chromosome aberrations within the same tumor, demonstrating intratumoral heterogeneity in gliomas.
- 64.Durinck S, Ho C, Wang NJ, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1(2):137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holland EC, Hively WP, Depinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12(23):3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reilly KM, Loisel DA, Bronson RT, Mclaughlin ME, Jacks T. Nf1; Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat. Genet. 2000;26(1):109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 67.Xiao A, Wu H, Pandolfi PP, Louis DN, Van Dyke T. Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell. 2002;1(2):157–168. doi: 10.1016/s1535-6108(02)00029-6. [DOI] [PubMed] [Google Scholar]
- 68.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 69.Bachoo RM, Maher EA, Ligon KL, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1(3):269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 70.Zheng H, Ying H, Yan H, et al. p53 and PTEN control neural and glioma stem/ progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8(2):119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alcantara Llaguno S, Chen J, Kwon CH, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. •• References [70-72] demonstrated that neural stem cells (NSCs) could serve as the cell of origin for gliomas.
- 73.Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58(23):5275–5279. [PubMed] [Google Scholar]
- 74.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28(23):2266–2275. doi: 10.1038/onc.2009.76. •• Demonstrated that neonatal OPCs could serve as the cell of origin for tumors that resemble oligodendrogliomas.
- 76.Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling adult gliomas using RCAS/t-va technology. Transl. Oncol. 2009;2(2):89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lei L, Sonabend AM, Guarnieri P, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6(5):e20041. doi: 10.1371/journal.pone.0020041. •• Showed that adult glial progenitors give rise to proneural GBM and that both mouse and human proneural GBMs are enriched in OPC genes.
- 78.Persson AI, Petritsch C, Swartling FJ, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18(6):669–682. doi: 10.1016/j.ccr.2010.10.033. •• Demonstrated that mouse and human oligodendrogliomas resemble OPCs rather that NSCs.
- 79.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121(3):479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 80.Liu C, Sage JC, Miller MR, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. •• This paper reconciles the NSC and OPC hypothesis. In this model, NSC is the cell of mutation (nontransforming) while OPC serves as the cell of origin (transforming).
- 81.Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135(1):145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- 82.Casper KB, Mccarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol. Cell Neurosci. 2006;31(4):676–684. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 83.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005;64(6):479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 84.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin. Cancer Res. 2005;11(3):1119–1128. [PubMed] [Google Scholar]
- 85.Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]