Abstract
Cardiac myosin binding protein-C (cMyBP-C) plays a role in sarcomeric structure and stability, as well as modulating heart muscle contraction. The 150 kDa full-length (FL) cMyBP-C has been shown to undergo proteolytic cleavage during ischemia–reperfusion injury, producing an N-terminal 40 kDa fragment (mass 29 kDa) that is predominantly associated with post-ischemic contractile dysfunction. Thus far, the pathogenic properties of such truncated cMyBP-C proteins have not been elucidated. In the present study, we hypothesized that the presence of these 40 kDa fragments is toxic to cardiomyocytes, compared to the 110 kDa C-terminal fragment and FL cMyBP-C. To test this hypothesis, we infected neonatal rat ventricular cardiomyocytes and adult rabbit ventricular cardiomyocytes with adenoviruses expressing the FL, 110 and 40 kDa fragments of cMyBP-C, and measured cytotoxicity, Ca2+ transients, contractility, and protein–protein interactions. Here we show that expression of 40 kDa fragments in neonatal rat ventricular cardiomyocytes significantly increases LDH release and caspase 3 activity, significantly reduces cell viability, and impairs Ca2+ handling. Adult cardiomyocytes expressing 40 kDa fragments exhibited similar impairment of Ca2+ handling along with a significant reduction of sarcomere length shortening, relaxation velocity, and contraction velocity. Pull-down assays using recombinant proteins showed that the 40 kDa fragment binds significantly to sarcomeric actin, comparable to C0–C2 domains. In addition, we discovered several acetylation sites within the 40 kDa fragment that could potentially affect actomyosin function. Altogether, our data demonstrate that the 40 kDa cleavage fragments of cMyBP-C are toxic to cardiomyocytes and significantly impair contractility and Ca2+ handling via inhibition of actomyosin function. By elucidating the deleterious effects of endogenously expressed cMyBP-C N-terminal fragments on sarcomere function, these data contribute to the understanding of contractile dysfunction following myocardial injury.
Keywords: Proteolysis, Pathogenesis, Muscle contractility, Actin, Acetylation, Ca2+, Transients
Introduction
Cardiac myosin binding protein-C (cMyBP-C) has specific roles in both the structural assembly and stability of the sarcomere as well as in modulating muscle contractility (Harris et al. 2002). Much of our knowledge about cMyBP-C has emerged in the last decade based on the discovery that mutations in cMyBP-C cause familial hypertrophic cardiomyopathy (Barefield and Sadayappan 2010). The various regions of cMyBP-C appear to serve different interactions and functions (Winegrad 1999). Its N-terminal (N′) region mediates the most important regulatory role of cMyBP-C with thin and thick filament proteins (Sadayappan and de Tombe 2012). The N′ region consists of immunoglobulin (Ig)-like domains, such as C0, followed by C1, M and C2 domains. The C0 and C1 domains are connected by a proline-alanine linker (P/A linker), which is thought to bind actin and may play a role in modulating contraction (Squire et al. 2003). Unique differences in the P/A linker may, in turn, account for species-specific differences in actomyosin interactions (Shaffer et al. 2009, 2010). In addition, cMyBP-C contains four sites, Ser-273, Ser-282, Ser-302 and Ser-307, which are substrates for protein kinase A (PKA), C (PKC), D (PKD), ribosomal S6 kinases and Ca2+-calmodulin-dependent protein kinase phosphorylation that are known to modulate cardiac contractility (Sadayappan et al. 2011). All four phosphorylation sites reside in the myosin (M) binding domain that is located between the C1 and C2 domains. This region is proposed to be involved in conformational changes central to modulating myosin interaction (Gruen et al. 1999). In addition, contractility is modulated through phosphorylation of the M domain, which determines the binding property of myosin and actin (Shaffer et al. 2009, 2010; Howarth et al. 2012). cMyBP-C is highly sensitive to proteolytic cleavage following ischemia–reperfusion (I–R) and phosphorylation of the M domain has been shown to be cardioprotective against (I–R)-induced proteolysis (Sadayappan et al. 2006, 2009). Thus, understanding the molecular and cellular consequences of interactions within the N′ regions of cMyBP-C is critical to the design of new treatment strategies to protect the heart and improve cardiac function after I–R injury.
cMyBP-C is extensively phosphorylated under basal conditions (Sadayappan et al. 2005; Copeland et al. 2010); however, the level of cMyBP-C phosphorylation decreases in animal models during development of I–R injury (Sadayappan et al. 2005, 2006). The reduced phosphorylation is accompanied by contractile dysfunction and increased cleavage of cMyBP-C (Decker et al. 2005; Sadayappan et al. 2006). During I–R injury in mouse, rat and human, we previously identified the predominantly released N′ (40 kDa) fragment of cMyBP-C using N′-specific anti-cMyBP-C antibodies (Sadayappan et al. 2008) and determined that the increased level of such fragments in the circulatory system was well correlated with post-ischemic contractile dysfunction (Govindan et al. 2012). Importantly, the 40 kDa fragment is predominantly present in the ischemic region of the myocardium (Govindan et al. 2012). Mass spectrometry analyses revealed that the 40 kDa peptide is released by cleavage within the conserved M domain at 272-TSLAGAGRR-280 (Sadayappan et al. 2008; Govindan et al. 2012). The 40 kDa fragment contains the entire C0 and C1 domains along with the first 17 residues of the M domain (1–271 residues). Because the C1–M–C2 domains of cMyBP-C bind to the S2 region of myosin and actin, partial expression of this region in the 40 kDa fragment (named previously C0–C1f or 29 kDa fragment) may alter the regulation of actomyosin interaction and thus may have detrimental consequences on sarcomeric function (Sadayappan et al. 2008; Mun et al. 2011; Weith et al. 2012).
In the present study, our aim was to define the pathogenic role of the 40 kDa fragment of cMyBP-C in sarcomere function in vitro. Our data show that the 40 kDa fragment of cMyBP-C acts as an inhibitory peptide by strongly interacting with actin, thereby potentially inhibiting actomyosin function, resulting in the impairment of contractility and Ca2+ handling, as well as an increase in cell death. The present study, which focuses on restricted proteolytic modification of cMyBP-C, represents a novel area of research that will provide valuable information to better understand the functional and pathogenic role(s) of cMyBP-C on post-translational regulation of cardiac muscle function in health and diseases.
Materials and methods
I–R injury
Cardiac I–R injury was performed in mice (FVB/N) that were 8–10 weeks of age to determine the degradation profile of cMyBP-C. The left anterior descending coronary artery was occluded for 1 h, after which the myocardium was reperfused for 24 h as described previously (Sadayappan et al. 2006, 2009). Upon completion of the reperfusion period, mice were sacrificed by CO2 asphyxiation, and the hearts were removed for analysis of cMyBP-C degradation as described previously (Sadayappan et al. 2006, 2009). This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health, using protocols approved by the Loyola University Chicago Institutional Animal Care and Use Committee.
Culture of neonatal rat ventricular cardiomyocytes (NRVCMs)
NRVCMs were isolated from 1-day-old Sprague–Dawley rats as previously described (Sarkey et al. 2011), plated on gelatin-coated 60 mm2 dishes (2.5–3.0 × 106 cells/plate) in serum-free PC-1 medium (Lonza, Walkersville, MD), and allowed to attach for 18 h. NRVCMs were subsequently cultured in (4:1) DMEM/medium 199 (HyClone Laboratories Inc., Logan, UT) containing (100 U/mL) penicillin/(100 μg/mL) streptomycin (Invitrogen, Carlsbad, CA).
Culture of adult rabbit ventricular cardiomyocytes and analysis of contractility
Adult rabbit ventricular cardiomyocytes were prepared as previously described (Sarkey et al. 2011), plated on laminin coated 25 mm2 coverslips (3.0–5.0 × 104 cells/coverslip) in serum-free PC-1 medium (Lonza, Walkersville, MD), and allowed to attach for 2 h prior to infection with adenovirus using a multiplicity of infection (MOI) of 50. Cardiomyocytes were cultured for 24 h while pacing with 10 V, 5 ms pulse duration, at 0.1 Hz. For contractility measurements, cultured cardiomyocytes were perfused with Tyrode solution (1 mM Ca2+), and stimulated with 20 V, 6 ms pulse duration, at 0.5 Hz. Sarcomere lengths (SLs), relaxation velocities, and contraction velocities were measured simultaneously with Ca2+ transients and analyzed using IonOptix software (IonOptix, LLC, Milton, MA).
Adenovirus preparation and infection
Mouse FL cMyBP-C (sequence ID O70468 UniPort, 150 kDa, residues 1–1270), 110 kDa (residues 272–1270) and 40 kDa (1–271 residues) were constructed using Ad-Easy Adenoviral Vector System (Stratagene, La Jolla, CA). Both FL and 110 kDa cDNA were cloned with a human Myc-tag (EQKLISEEDL) to differentiate them from endogenous cMyBP-C. The 40 kDa was generated with and without the Myc-tag. We used the adenoviruses expressing 40 kDa with Myc tag for the immunofluorescent staining. The rest of the experiments, we used the adenoviruses expressing 40 kDa without Myc tag since it is a small peptide, when compared to FL and 110 kDa, as the inclusion of the Myc-tag may well have influenced the pathogenic properties. NRVCMs were infected with these viruses after 24 h of culture at a multiplicity of viral infection (MOI) of 50 and 100 and then allowed 48 h for protein expression. We used adenovirus expressing green fluorescence protein (GFP), an empty viral vector, and uninfected NRVCMs as controls.
Western blot analysis
Transgenic protein expression was determined in NRVCMs by western blot analyses using antibodies against cMyBP-C 2–14 residues (EPGKKPVSAFSKK, C0 domain), 1200–1212 residues (AVRGSPKPKISWFK, C10 domain) and Myc-tag (Roche) (Govindan et al. 2012). Antibodies against sarcomeric α-actin (Sigma, St. Louis, MO) were used as a loading control (Sadayappan et al. 2006, 2009).
Cardiomyocyte cytotoxicity
To determine the cytotoxicity of NRVCMs that express the FL, 110 kDa and 40 kDa fragments, we measured lactate dehydrogenase (LDH) release (Roche, Indianapolis, IN) at two different MOI. LDH release into the NRVCMs culture medium, which is an indicator of the presence of membrane damage, was measured spectrophotometrically according to the manufacturer’s instructions. Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltet-razolium bromide (MTT). The assay measured the reduction in MTT (0.5 mg/mL) with a microplate spectro-photometric reader at 570 nm (Molecular Probes, Eugene, OR). Caspase-3 is an intracellular protease that becomes activated during the development of apoptosis. The enzymatic activity of caspase-3 by colorimetric reaction at a wavelength of 405 nm (R&D Systems, Minneapolis, MN).
Immunofluorescent analysis
Integration and localization of transgenic exogenous expression of the 40 kDa fragment into the sarcomere, compared to endogenous cMyBP-C, was determined by confocal microscopy. Cardiomyocytes were washed in phosphate buffered saline (PBS) and fixed in 4 % paraformaldehyde/cardioplegic buffer (50 mmol/L KCl, 5 % dextrose in PBS) for 30 min. The slides were washed with PBS (3 times, 5 min each), incubated in blocking solution (1 % BSA, 0.1 % cold water fish skin gelatin, 0.05 % sodium azide and 0.1 % Tween-20 in PBS) for 1 h at room temperature and probed with 1:1300 diluted polyclonal antibody against cMyBP-C and Myc tag in PBS for overnight at 4 °C. The slides were then washed with PBS 3 times for 5 min each, incubated with blocking solution for 20 min, and incubated with secondary goat anti-rabbit IgG antibody (1:100) and goat anti-mouse IgG antibody (1:100) conjugated with Alexa-488 (green fluorescence) and Alexa-565 in PBS. Finally, the slides were washed with PBS 3 times for 5 min each, and mounted with Vecta Shield (Vector Laboratories, Inc.). Specimens were examined using confocal microscopy (Zeiss LSM 510) and software (LSM Image Browser).
Measurement of Ca2+ transients
NRVCMs and adult rabbit cardiomyocytes were plated (5.0 × 105 cells/cover slip and 5.0 × 104 cells/cover slip respectively), infected with adenovirus expressing the FL, 110 kDa, 40 kDa fragments (50 MOI) or empty vector control, and cultured for 48 h. Cardiomyocytes were loaded with 2 μM Indo-1 AM calcium sensor dye for 20 min at room temperature, perfused with Tyrode solution containing 1 mM Ca2+. NRVCMs were stimulated with 40 V, 6 ms pulse duration, at 1 Hz. Adult rabbit cardiomyocytes were stimulated with 20 V, 6 ms pulse duration, at 0.5 Hz. Sarcoplasmic reticulum (SR) Ca2+ load was determined by rapid exposure to 10 mM caffeine. Ca2+ transient recordings were collected and analyzed using IonOptix software (IonOptix, LLC, Milton, MA) as previously described (Sarkey et al. 2011).
Pull-down assay
Pull-down assays were performed to determine the interaction of the 40 kDa fragment with myosin or actin, as described previously (Sadayappan et al. 2006). The soluble His-tagged C0–C1, 40 kDa or C0–C2 recombinant peptides were generated by using the pET expression system (Novagen, San Diego, CA). Total ventricular tissues of normal mouse hearts were homogenized in RIPA buffer (25 mM Tris–HCl, pH 7.6, 1 % NP-40, 1 % sodium desoxycholate, 0.1 % SDS) with 500 mM NaCl and protease/phosphatase inhibitors (Cocktail I and II, Sigma, St. Louis, MO). To pre-clear nonspecific proteins, 200 μg of proteins were incubated with 25 μL of Ni–NTA agarose beads (Qiagen, Valencia, CA) at 4 °C for 2 h. A brief spin at 2000 rpm was used to remove the beads. Precleared supernatant were incubated with 10 μg of recombinant peptides and 20 μL of Ni–NTA agarose beads for 2 h at 4 °C and washed with Tris buffer (50 mM) as described previously (Sadayappan et al. 2006). Proteins bound to the beads were eluted in Laemmli sample buffer (Bio-Rad, Hercules, CA) and subjected to western blot analysis with polyclonal anti-cMyBP-C2–14, monoclonal anti-actin (Sigma, St. Louis, MO) or monoclonal anti-α-myosin heavy chain (clone BA-G5, ATCC, Rockville, MD) antibodies.
Immunoprecipitation and in vitro acetylation
Two hundred micrograms of total ventricular proteins from sham and I–R injured mouse hearts in 200 μL high-salt RIPA buffer were pre-cleared with 25 μL Protein G Magnetic Beads (New England Biolabs, Inc., Ipswich, MA) at 4 °C for 2 h. Using a magnetic field, the beads were pulled down to the side of a 1.5 mL tube. The pre-cleared supernatant was then incubated with 5 μg of goat anti-cMyBP-C2–14 antibody at 4 °C. After 1 h of incubation, 25 μL of Protein G Magnetic Beads were added and pulled-down using a magnetic field. After extensive washing with high-salt RIPA buffer, the beads were boiled with 100 μL of 2× SDS-PAGE loading buffer (Bio-Rad, Hercules, CA), and western blots were carried out with rabbit anti-cMyBP-C2–14 antibody for cMyBP-C and anti-acetylated-lysine antibody (Cell Signaling, Danvers, MA). To in vitro acetylate, 5 μg of the recombinant 40 kDa fragments were incubated with histone acetyltransferases (HAT, p300, Millipore, Billerica, MA), assay buffer (50 mM ammonium bicarbonate, 1 mM DTT, 10 % glycerol, 0.5 mM EDTA), 0.5 mM acetyl-CoA, 50 mM Nicotinamide (Sigma, St. Louis, MO) and 10 μM Trichostatin A (Cell Signaling, Danvers, MA) at 37 °C for 2 h. For the deacetylation assay, five μg of recombinant 40 kDa peptides were incubated with Histone Deacetylase Assay buffer (25 mM Tris–HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2 and 0.1 mg/mL BSA). One hundred nanograms of proteins were resolved by SDS-PAGE and analyzed by western blotting using anti-acetylated-lysine antibody (Gupta et al. 2008).
Mass spectrometry to determine acetylation
FL and N′ peptides of cMyBP-C were resolved by SDS-PAGE, and the band to be analyzed was excised from the gel. Gel pieces were destained using 100 mM ammonium bicarbonate pH 8.9 in 50 % acetonitrile and then treated with 100 μL of 50 mM ammonium bicarbonate, pH 8.0, and 10 μL of 10 mM TCEP [Tris (2-carboxyethyl) phosphine HCl] at 37 °C for 30 min. Protein digestion was carried out using 1:50 sequencing grade trypsin in 50 mM ammonium bicarbonate, pH 7.5. Digested peptide samples were desalted with C8 OptiPak column (Optimize Technologies) and analyzed by liquid chromatography electro-spray tandem mass spectrometry (LC–MS/MS) on a Thermo LTQ Orbitrap Hybrid FT Mass Spectrometer (Kim et al. 2006). Positive ion mass spectra were acquired in the reflectron mode. Ions selected for MS/MS were subsequently placed on an exclusion list using an isolation width of 1.6 Da, low mass exclusion of 0.8 Da, and high mass exclusion of 0.8 Da. Tandem mass spectra were extracted by Readw.exe version 3.0. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK) data explorer software.
Statistical analysis
Data were expressed as mean ± standard error of mean. Statistical analyses between the experimental groups were performed by one-way or two-way analysis of variance (ANOVA) with Tukey’s post hoc analysis. The threshold of statistical significance for all tests was defined at P < 0.05.
Results
Forty kDa fragment is the predominant N′ peptide of cMyBP-C released during I–R injury
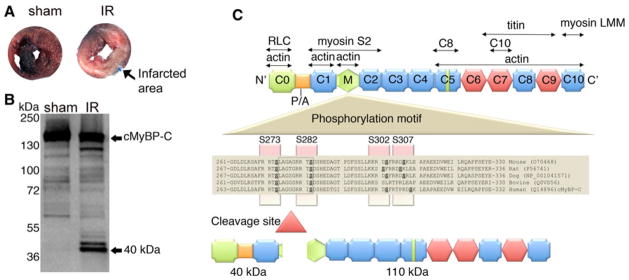
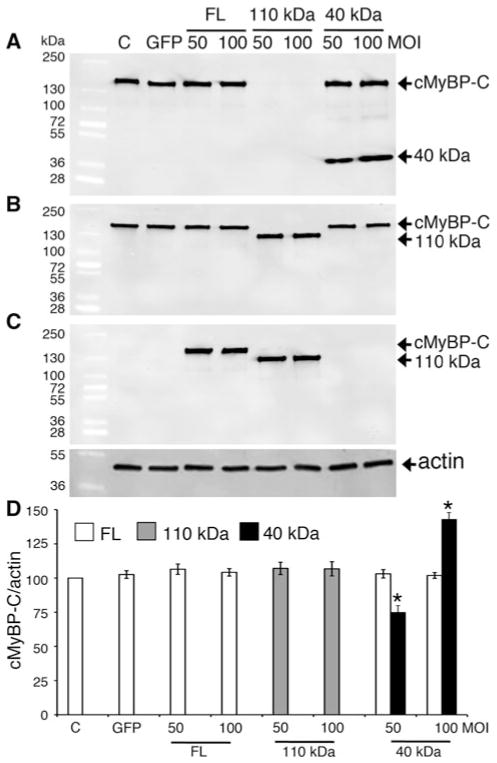
I–R injury refers to myocardial dysfunction that is induced by the restoration of blood supply to ischemic cardiomyocytes, resulting in cardiomyocyte damage and infarction (Fig. 1a). I–R injury leads to dramatic distortions in myocyte architecture and physiology, including sarcomere, sarcolemmal and mitochondrial injury, and alterations in intracellular Ca2+ handling. While the initial restoration of blood supply during reperfusion period is necessary to maintain cardiomyocyte structure and function, oxidative stress and protein proteolysis occurring subsequently to reperfusion may result in irreversible damage and cell death. We have previously shown that cMyBP-C is readily cleaved post-MI (Govindan et al. 2012), resulting in the significant release of 40 kDa N′ fragments associated with the impairment of cardiac function. Compared to sham-operated mouse hearts, when mice were subjected to I–R injury (Fig. 1a), cMyBP-C was extensively degraded and the 40 kDa fragment was the predominant peptide released (Fig. 1b). cMyBP-C was cleaved approximately at residue 271, which generates the release of the 40 kDa N′-fragment (Fig. 1c) (Sadayappan et al. 2008; Govindan et al. 2012). Based on these data, we hypothesized that the presence of 40 kDa is deleterious to the sarcomeres post-MI injury. To determine whether the presence of the 40 kDa N′ fragment alters sarcomere function in vitro, the 40 and 110 kDa and FL cMyBP-C were overexpressed in NRVCMs using adenoviral constructs. Western blot analyses show that expression of FL cMyBP-C at two different MOI did not significantly increase the total FL cMyBP-C protein content, consistent with the fact that sarcomeric proteins can never be overexpressed in the sarcomere beyond 100 % stoichiometry (Fig. 2a–d). Likewise, expression of the 110 kDa protein completely replaced the FL cMyBP-C, maintained stoichiometry, and did not increase in an MOI-dependent manner (McConnell et al. 1999). Strikingly, expression of the 40 kDa fragment did not replace endogenous FL cMyBP-C, but increased in an MOI-dependent manner, suggesting that 40 kDa N′ regions can be overexpressed several fold without competing with FL cMyBP-C in terms of stoichiometry. Based on these data, we hypothesize that the cleaved N′ 40 kDa fragment of cMyBP-C may interfere with normal cardiomyocyte function.
Fig. 1.
cMyBP-C interacting partners and generation of the 40 kDa cleavage fragment. Ischemia–reperfusion injury induces myocardial infarction, resulting in tissue damage and necrosis. Three month-old mice were induced with 60-min ischemia and 24-h reperfusion, as described previously (Sadayappan et al. 2009). Following I–R injury, hearts were fixed with 2 % Evan’s Blue in PBS, sectioned and staining with 2 % triphenyl tetrazolium chloride. Compared to sham operated mouse heart, I–R injured heart shows severe infarct region (a). The 40 kDa fragment is the predominant N′ fragment of cMyBP-C that is released in the sarcomere during I–R injury (b). Ten micrograms of total myofilament proteins were used to perform western blot analysis with anti-cMyBP-C2–14 antibodies. The antibody detects all the N′ fragments of cMyBP-C in which the 40 kDa is predominant. Although the 40 kDa fragment runs at the 40 kDa position in the gel, MS/MS sequence analysis identified the site of cleavage and molecular weight as 29 kDa (Sadayappan et al. 2008; Govindan et al. 2012). (c) cMyBP-C consists of 11 modules labeled C0–C10 from the N- to the C-terminus and belongs to the intracellular Ig super-family which is composed of repeating domains of Ig (Plaque) and fibronectin type-3 (Hexagon). The interacting regions of cMyBP-C with titin, myosin, actin and regulatory light chain are shown. The C8 and C10 domains that interact with C5 and C7 of cMyBP-C are marked. The cardiac-specific regions (C0 and M domain, and an insert in C5 domain) are marked in light green. A proline-alanine (P/A)-rich linker sequence located between the C0 and C1 domains is shown. The four-phosphorylation sites, Ser-273, Ser-282, Ser-302 and Ser-307, are highlighted across mouse to human species. The calpain cleavage occurs at 272–280 residues, resulting in both 40 (residues 1–271) and 110 kDa (residues 272–1270) fragments
Fig. 2.
Overexpression of the 40 kDa fragments in NRVCMs does not affect expression of endogenous cMyBP-C. Representative western blot analyses show the expression of the transgenic FL, 110 and 40 kDa fragments. Fifteen micrograms of total lysates from infected NRVCMs for 48 h were used for SDS-PAGE, followed by western blot analyses with respective antibodies. FL of both endogenous and transgenic cMyBP-C and 40 kDa fragments were recognized with anti-cMyBP-C2–14 antibodies (N′-specific, (a)). FL of both endogenous and transgenic cMyBP-C and 110 kDa proteins were recognized with anti-cMyBP-CC10 antibodies (C-terminal-specific, (b)). FL and 110 kDa cMyBP-C were transgenically tagged with Myc and recognized with anti-Myc antibodies (c). Data are summarized for respective antibodies (n = 4, (d)). Sarcomeric α-actin was used as a loading control
Expression of 40 kDa peptides in NRVCMs is pathogenic
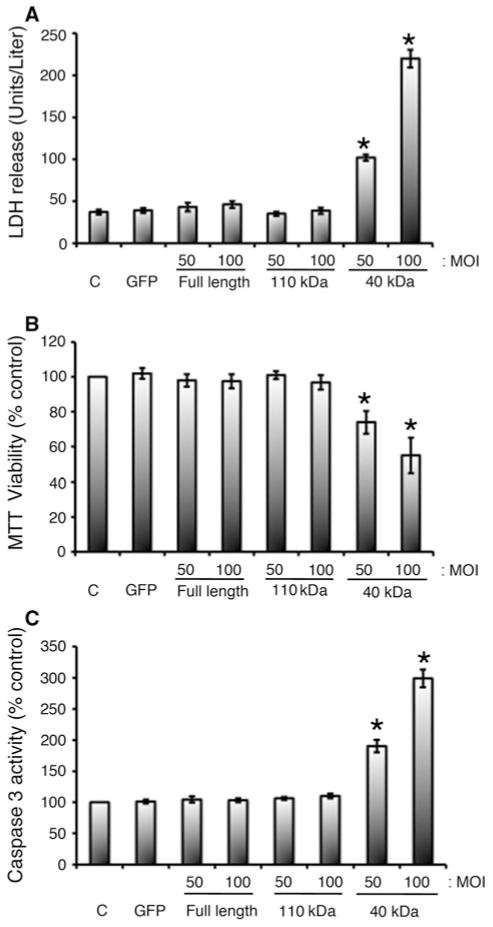
Next, we determined whether the expression of the 40 kDa fragment in NRVCMs induces cytotoxicity compared with the 110 kDa fragment and FL cMyBP-C controls by measuring LDH release, cell viability and apoptotic markers. The release of LDH into the medium is widely used as an indicator of cell injury. NRVCMs that were infected with the 40 kDa fragment in two different MOI concentrations showed significant increase of LDH release into the cell culture medium (102 ± 4 and 220 ± 11 units/liter at 50 and 100 MOI, respectively, as compared to the control values, *P < 0.001) (Fig. 3a). In contrast, expression of the 110 kDa and FL cMyBP-C had no effect on the release of LDH in NRVCMs. Next, we measured cell viability using a standard MTT assay. Expression of the 40 kDa fragment at 50 and 100 MOI resulted in a significant reduction in cell viability of with 26 ± 7 and 51 ± 10 % reduction respectively, compared to the controls (*P < 0.001, Fig. 3b). Conversely, expression of the 110 kDa and FL cMyBP-C had no effect on cell survival. Increased caspase-3 activity is an indication of apoptosis and has been directly linked with apoptotic process in the myocardium of end-stage HF. Expression of the 40 kDa fragment at 50 and 100 MOI significantly increased relative caspase-3 activity (190 ± 10 and 298 ± 14 respectively, *P < 0.001), compared to the controls (Fig. 3c). In contrast, expression of the FL and 110 kDa had no effect on cell viability (MTT assay), damage (LDH release) and cell death (caspase-3 activity). These data indicate that the exogenous expression of the 40 kDa fragment of cMyBP-C is toxic to cardiomyocytes.
Fig. 3.
Overexpression of 40 kDa fragments is cytotoxic to neonatal cardiomyocytes. Cardiomyocytes were infected with 50 and 100 MOI of respective adenoviruses for 48 h. LDH release in medium (a), MTT conversion to formazan for cell viability (b), and caspase-3 activity (c) were assessed as described in “Materials and methods” section. *P < 0.001 * versus control (n = 4). Data for LDH activity were shown as activities, units/liter, and as absorption units at 540 nm for MTT assay. For caspase-3 activity, absorption values were expressed as % of control, which was set as 100 %
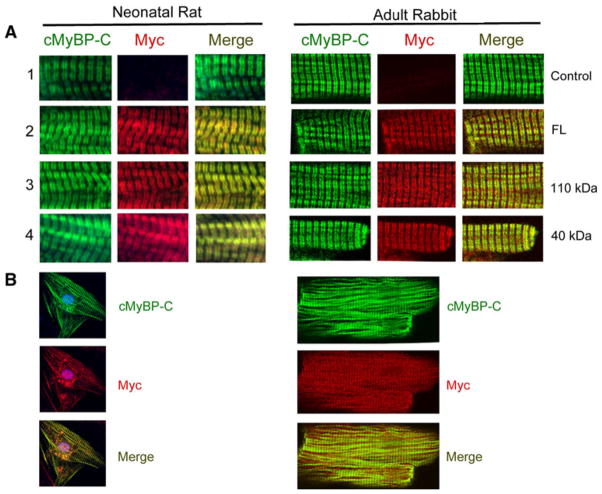
Exogenous 40 kDa protein incorporates into the sarcomere
In order to determine whether the 40 kDa fragment incorporates into the sarcomere and co-localizes with endogenous cMyBP-C within the C-zone, we performed immunofluorescent analyses using NRVCMs and adult rabbit ventricular cardiomyocytes that were infected with adenoviruses (50 MOI). Antibodies against cMyBP-C and Myc tag were used to determine co-localization of the 40 kDa with cMyBP-C. Results show that the exogenous 40 kDa fragments were normally incorporated within sarcomeres and co-localized with cMyBP-C both in NRVCMs and adult rabbit ventricular cardiomyocytes (Fig. 4a, b). These data demonstrate that N′-fragments of cMyBP-C are able to incorporate into the sarcomere where they could potentially alter sarcomere function.
Fig. 4.
Exogenously expressed 40 kDa peptides incorporate into the sarcomere and co-localize with endogenous cMyBP-C. Immunofluorescent staining of cMyBP-C with anti-cMyBP-C2–14 (1, 2 and 4) and anti-cMyBP-CC10 polyclonal antibodies (3) with anti-Myc monoclonal antibodies (a, 60×). Green and red identify cMyBP-C and Myc tag, respectively. Immunofluorescent staining of cMyBP-C and 40 kDa proteins in neonatal cardiomyocytes and adult rabbit cardiomyocytes are shown, respectively (b, 20×). Data demonstrate that exogenously expressed 40 kDa fragments properly localize to the sarcomere and co-localized with cMyBP-C
Expression of 40 kDa fragments impairs intracellular Ca2+ handling and contractility in cardiomyocytes in vitro
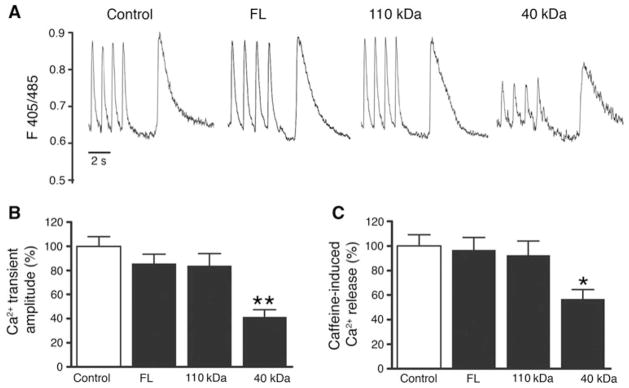
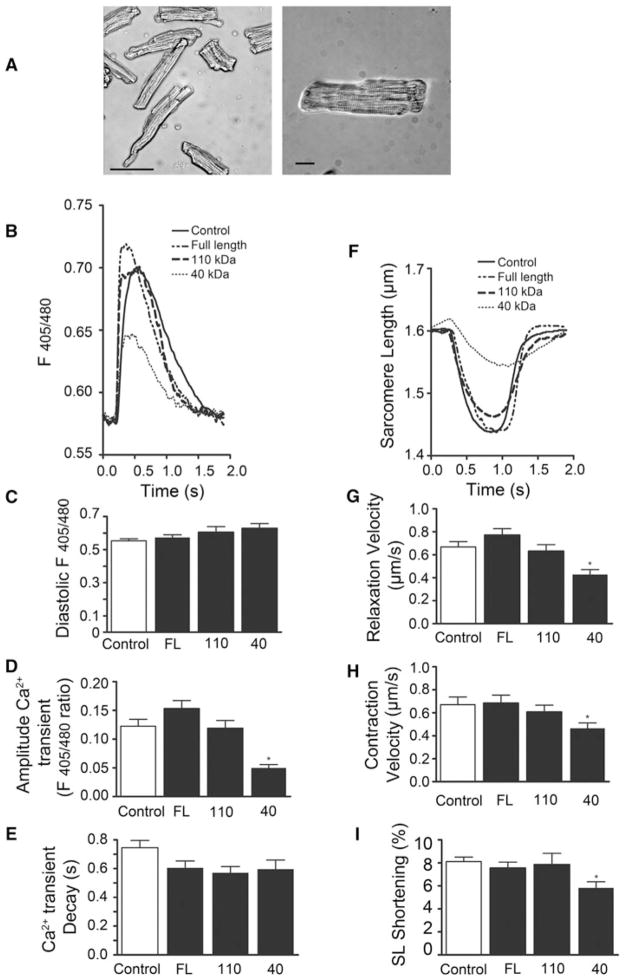
cMyBP-C has been previously shown to bridge thick and thin filaments in striated muscle, implying a potential role of cMyBP-C in the modulation of Ca2+-dependent regulation of actin-myosin interaction (Luther et al. 2011). To determine whether the presence of cleaved fragments of cMyBP-C affects intracellular Ca2+ handling, NRVCMs were infected at 50 MOI with either FL cMyBP-C, 110 or 40 kDa fragments, and Ca2+ transients were recorded from Indo 1-AM-loaded cells during electrical stimulation and caffeine-induced contractures. Compared to control, over-expression of either the 110 kDa fragment of cMyBP-C or FL cMyBP-C had no significant effect on Ca2+ handling or Ca2+ content within the SR. However, overexpressing the 40 kDa fragment of cMyBP-C significantly decreased the amplitude of Ca2+ release evoked by electrical stimulation (Fig. 5), without affecting diastolic Ca2+ levels, the time constant of Ca2+ decay (tau), or the time of peak calcium release (data not shown). In addition, 40 kDa fragment overexpression significantly reduced Ca2+ release from the SR (SR load) evoked by brief application of caffeine without affecting the time constant of Ca2+ decay of the caffeine-induced Ca2+ peak. These findings indicate that the 40 kDa fragment of cMyBP-C reduces Ca2+ release from the SR during twitches and SR Ca2+ load, possibly by affecting sodium-calcium exchanger or SERCA2 activity. However, since NRVCMs and adult cardiomyocytes differ with respect to their structure and Ca2+ homeostatic mechanisms, we next utilized adult rabbit ventricular cardiomyocytes, which are very close to human in terms of Ca2+ homeostasis, and contractile structure and function. To determine whether the presence of cleaved fragments of cMyBP-C affects Ca2+ handling and contractility, adult rabbit cardiomyocytes were infected with either FL cMyBP-C, 110 or 40 kDa fragments, and Ca2+ handling and cell shortening was recorded from cells during electrical stimulation (Fig. 6a). Compared to control, overexpression of either the 110 kDa fragment of cMyBP-C or FL cMyBP-C had no significant effect on Ca2+ handling or contractile function. However, overexpressing the 40 kDa fragment of cMyBP-C significantly decreased the amplitude of Ca2+ release evoked by electrical stimulation (Fig. 6b, d) without affecting diastolic Ca2+ levels (Fig. 6c) or the time constant of Ca2+ decay (tau) (Fig. 6e). In addition, overexpression of the 40 kDa fragment significantly reduced SL shortening (Fig. 6f, i), relaxation velocity (Fig. 6g), and contraction velocity (Fig. 6h). These data suggest that the presence of 40 kDa fragments in the sarcomere of both neonatal and adult cardiomyocytes alters Ca2+ homeostasis and contractile function.
Fig. 5.
N-terminal 40 kDa fragments of cMyBP-C induce abnormal Ca2+ handling in neonatal cardiomyocytes. Representative Ca2+ transients and caffeine (10 mM)-releasable SR Ca2+ content (a). Averaged Ca2+ transient amplitudes (b) and caffeine-releasable SR Ca2+ content (c) normalized to control. *P < 0.05; **P <0.001 (n = 3; 10 NRVCMs/time). One-way ANOVA with Tukey’s post hoc analysis
Fig. 6.
N-terminal 40 kDa fragments of cMyBP-C induce abnormal Ca2+ handling and contractile dysfunction. Isolated adult rabbit cardiomyocytes cultured for 24 h. Scale bar 100 μM (panel 1); scale bar 20 μM (panel 2) (a). Representative averaged Ca2+ transients in adult rabbit cardiomyocytes infected with empty control vector, FL cMyBP-C, 110 kDa fragment cMyBP-C, or 40 kDa fragment cMyBP-C (b). Averaged diastolic calcium levels (c), Ca2+ transient amplitudes (d), and transient decay (tau) (e). Representative SL shortening (f) and averaged relaxation velocities (g), contraction velocities (h), and SL shortening (i). *P < 0.001; **P <0.01; ***P < 0.05 (n = 3 experiments; ≥10 cells/experiment). One-way ANOVA with Tukey’s post hoc analysis
Strong interaction of 40 kDa region with sarcomeric actin
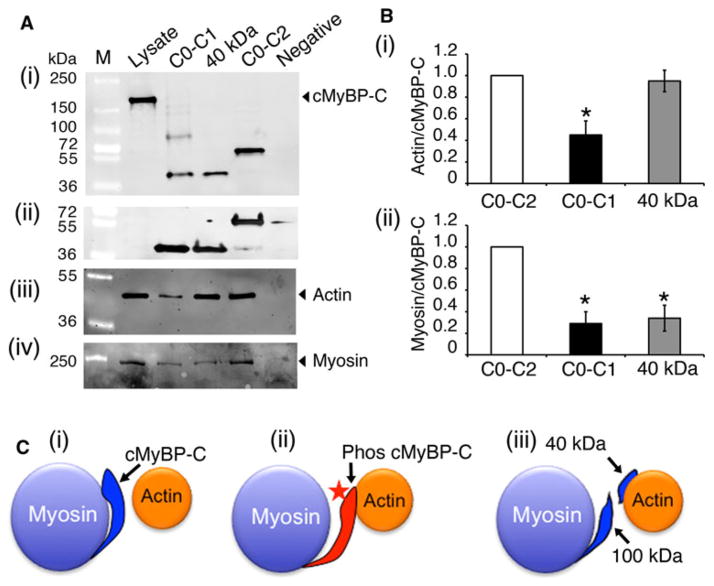
It has been shown that C1, M and C2 domains directly interact with myosin S2 region (Gruen et al. 1999; Sadayappan et al. 2006). Recent studies showed that C1 and M domains also interact with actin (Shaffer et al. 2009). In this interaction, phosphorylation regulates M domain interaction, whereas C1 interaction is independent of phosphorylation. In order to determine whether the 40 kDa fragment interacts with either myosin or actin, a pull-down assay was performed using the recombinant soluble proteins of C0–C1, 40 kDa fragment (C0–C1–17 residues of the M domain) and C0–C2 domains. Results show that binding between 40 kDa fragments and actin is proportionately similar to binding between actin and C0–C2, whereas C0–C1 had a 50 % significant reduction its binding properties with actin (Fig. 7a, b). In contrast, the 40 kDa fragment showed a significant 70 % reduction in its binding with myosin, compared to C0–C2 fragments, suggesting that 40 kDa fragment lost its interaction with myosin significantly, but could still interact with actin. Considering that the 40 kDa fragment does not contain the phosphorylatable residues of the M domain, these data suggest that the cleaved 40 kDa fragment can not perform the phosphorylation-dependent on/off interaction with myosin and actin (Fig. 7c), as does the FL cMyBP-C (Barefield and Sadayappan 2010).
Fig. 7.
The 40 kDa fragment interacts with actin. Pull-down assay was performed to determine the direct interaction between N′ region of cMyBP-C with either actin or myosin (a). Ten micrograms of recombinant His-tagged C0–C1, 40 kDa and C0–C2 peptides were mixed with 200 μg of total mouse heart lysates (lysate). Following this, the protein complex was pulled down with Ni–NTA beads (see “Materials and methods” section), and the proteins were separated by SDS-PAGE and western blotted with anti-cMyBP-C2–14 (i), anti-His antibodies (ii), anti-sarcomeric actin (iii) and anti-α-myosin heavy chain (iv) antibodies. Two micrograms of total lysates were used for positive control, and Ni–NTA beads-alone (without peptides) was used as a negative control. Quantitation data show (b) that 40 kDa interacts with actin in the same ratio as C0–C2 (i), but has significantly reduced binding with myosin in the same ratio as C0–C1 fragments (ii). *P <0.001 versus C0–C2, n = 4. A schematic diagram illustrates the 40 kDa-actin interaction (c). In the absence of cMyBP-C phosphorylation (i), the position of the actin-binding site of cMyBP-C would lie about 3 nm from the thin filament and tightly interact with myosin S2. Phosphorylation (star) of cMyBP-C (ii) would extend the cross-bridge to the surface of the thin filament and lose the packing of the rod portion of the myosin molecule. The 40 kDa fragment may possibly constantly remain with actin (ii) to inhibit its movements during cross-bridge cycling
Acetylation of the 40 kDa fragments in vivo and in vitro
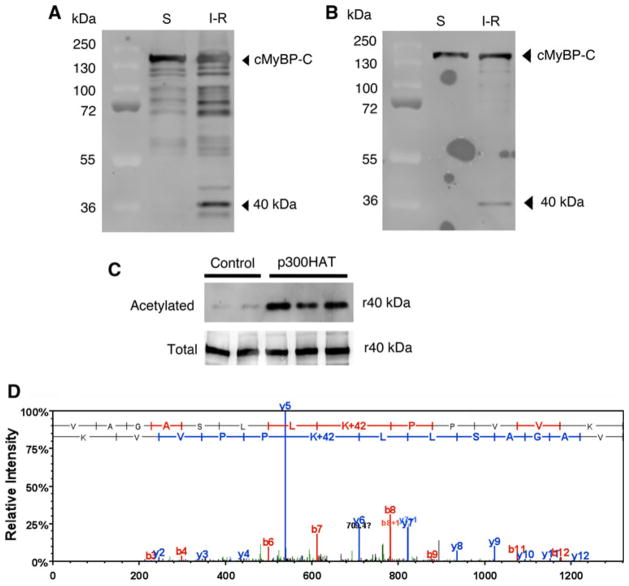
According to MS/MS data, the 40 kDa fragment was cleaved at Thr 272-Arg 280 residues in FL cMyBP-C resulting in a 1–271-residue fragment with a molecular weight calculated at 29 kDa (Sadayappan et al. 2008). However, it is interesting to note that these 29 kDa peptides migrated close to the 40 kDa position in SDS-PAGE (Figs. 1a, 8a, b). It has been previously demonstrated that cleaved cMyBP-C fragments are highly susceptible to lysine-acetylation, which causes a migration shift in SDS-PAGE (Ge et al. 2009). To determine whether 40 kDa fragments are acetylated in vivo, we immunoprecipitated cMyBP-C and its fragments from sham and I–R-injured mouse hearts (Fig. 8a), using anti-acetylated-lysine antibody for western blot analysis (Fig. 8b). Data show that cMyBP-C and the N′ fragments are heavily acetylated under basal conditions and post-I–R injury. Using recombinant 40 kDa proteins, we further demonstrated that the N′ region of cMyBP-C could also be acetylated in vitro with p300 HAT (Fig. 8c). The acetylated 40 kDa fragments and FL cMyBP-C were used for LC–MS/MS analysis to determine the site of acetylation. Data show that there are eight predominant acetylated sites in cMyBP-C (K7, K185, K190, K193, K202, K442, K935 and K962). Five of these sites are within the C0–C1 domains (K7, K185, K190, K193 and K202) (Fig. 8d) and four sites (K185, K190, K193 and K202) are located in the actin-binding region. These data suggest that following I–R injury, 40 kDa cleavage fragments of cMyBP-C are potentially acetylated and may thereby contribute to altered contractile function.
Fig. 8.
cMyBP-C is acetylated in vivo and in vitro. cMyBP-C and its fragments were immunoprecipitated using ventricular lysates from sham (S) and I–R injured mouse hearts with goat anti-cMyBP-C2–14 antibodies and western blotted with rabbit anti-cMyBP-C antibodies (a). A representative blot of the respective antibodies was shown in (a) and (b). To determine whether cMyBP-C and its fragments are acetylated in vivo, the immunoprecipitated samples were western blotted with rabbit anti-acetylated lysine antibodies (b). The His-tagged recombinant 40 kDa (r40) was acetylated in vitro (c) and western blotted with anti-acetylated lysine antibodies (top) and rabbit anti-cMyBP-C2–14 antibodies (bottom). A sample LC–MS/MS spectrum of “VAGASLLKacPPVVK” that identified K185 as one of the acetylation sites in the 40 kDa fragment (d). To determine the acetylation site, the immunoprecipitated FL cMyBP-C and 40 kDa fragments from the I–R injured hearts were subjected to mass spectrometry according to the method described previously (Kim et al. 2006). Annotation of representative tandem mass spectra of trypsin-digested 40 kDa fragments showing the K185 acetylation
Discussion
Pathogenesis of N-terminal region of cMyBP-C
cMyBP-C is sensitive to proteolysis and undergoes severe degradation during I–R injury (Sadayappan et al. 2006, 2009; Govindan et al. 2012). The 40 kDa fragment is a predominant small N′ peptide that is released post-I–R injury (Sadayappan 2012; Sadayappan and de Tombe 2012). Recently, we found that the 40 kDa fragments of cMyBP-C are released into the circulatory system after acute MI (Govindan et al. 2012). Moreover, it is interesting to note that cMyBP-C is severely degraded during muscle atrophy, accounting for alterations in contractile function (Cohen et al. 2009). Despite its diagnostic importance, no significant pathogenic properties have ever been ascribed to this phenomenon. Extensive release of cardiac troponin I and T fragments from the sarcomere also occurs during IR injury (O’Brien 2006), and these proteolytic fragments of cardiac troponin I and T have been found be capable of activating caspase-3 to induce apoptosis in cardiomyocytes (Jeong et al. 2009; Communal et al. 2002). Therefore, it is clinically important to determine whether cMyBP-C fragments released from cardiac myofilaments post-I–R injury likewise induce apoptosis and to elucidate a possible additional pathogenic role of these fragments in contractile dysfunction. In the present study, we investigated the potential cytotoxicity of 40 kDa fragments in NRVCMs utilizing three independent assays to determine cytotoxicity, cell viability and apoptosis. Due to the significant amount of 40 kDa released in the ischemic region of the myocardium post-MI (Govindan et al. 2012), we hypothesized that the presence of the 40 kDa fragments of cMyBP-C in the cardiomyocytes contributes to cytotoxicity and altered contractile function of the sarcomere. Our data demonstrate that expression of the 40 kDa fragments induces apoptosis-based cytotoxicity and impaired Ca2+ handling and contractility. In contrast, the C-terminal 110 kDa protein did not induce any cytotoxicity or impairment of Ca2+ handling and contractility in NRVCMs or adult rabbit ventricular cardiomyocytes. The N′ region of the cMyBP-C is highly conserved across species (Shaffer et al. 2010). As such, cytotoxicity of the 40 kDa fragment, as shown by our studies, indicates a unique and universal feature of the N′ region of the cMyBP-C (Howarth et al. 2012). The 40 kDa fragment contains the C0 domain (99 residues), a P/A rich region (51 residues), the C1 domain (104 residues), and 17 residues of the M domain. However, future studies are necessary to define the region of toxicity within the 40 kDa fragment, compared with other N′ domains. Because the C-terminal region of the 110 kDa fragment binds with titin and myosin, which are necessary for cMyBP-C integration and stability, it is plausible that these interactions could prevent structure-based toxic effects.
The cytotoxicity of the 40 kDa fragments, which leads to apoptosis, particularly impacts cardiomyocytes. Specifically, expression data show that transgenic FL cMyBP-C, 110 and 40 kDa fragments were able to incorporate into the sarcomeres. Strikingly, however, the 40 kDa did not replace the FL protein (Fig. 2). Although the 110 kDa replaced the endogenous FL cMyBP-C without changing the overall stoichiometry of the cMyBP-C content, the 40 kDa fragment, was instead overexpressed. Previously it has been shown that C1–C2 peptides significantly alter myofilament Ca2+ sensitivity and cross bridge cycling kinetics (Harris et al. 2004), while C0–C2 peptides alter force development, Ca2+ sensitivity, and sarcomere-length-tension relationships (Herron et al. 2006). Since overexpression of 40 kDa does not alter expression or localization of endogenous cMyBP-C, the pathogenesis of 40 kDa may result from the direct disruption of actomyosin function. Previous studies have shown that modification of contractile proteins, such as cardiac troponin T, alters Ca2+ transients and contractile function (Haim et al. 2007; Knollmann et al. 2003). In agreement with these studies, our data demonstrate that expression of the 40 kDa N′ region is toxic to cardiomyocytes and alters intracellular Ca2+ handling and contractility. However, it is unclear whether such an alteration in the Ca2+ handling is a primary or secondary effect. Importantly, replacing endogenous cMyBP-C with the 110 kDa protein had no effect on cytotoxicity, Ca2+ handling, or contractility. As we demonstrated recently (Fig. 4 in Govindan et al. 2012), cMyBP-C undergoes significant cleavage within cardiomyocytes post-MI, thereby generating readily detectable levels of the 40 kDa fragment in the ischemia region. We also further showed that dephosphorylation of cMyBP-C at Ser-273 and Ser-282 is directly associated with the generation of the 40 kDa fragments. Therefore, we hypothesize that ischemic cardiomyocytes will contain a sufficient amount of 40 kDa proteins to cause detrimental effects on contractility. In the present study, we demonstrated that overexpression of the 40 kDa peptide in healthy cardiomyocytes causes toxic effects and that it impairs contractility. We therefore propose that the contractile dysfunction observed post-MI is due in part to the presence of 40 kDa fragments. In contrast, cardiomyocytes expressing of the 110 kDa (C′ region) exhibit normal function without any toxic effects, suggesting that the presence of N′ region of the truncated fragment is toxic to cardiomyocytes. Therefore, the downstream effects of I–R injury are associated with the degradation of sarcomeric proteins, including cMyBP-C. Thus, we hypothesize that maintenance of normal contractile function is dependent on the presence of intact sarcomeric proteins, as even partial degradation of cMyBP-C would affect sarcomeric structure and function. Taken together, these studies suggest that cleaved-off N′ region of cMyBP-C (40 kDa) is deleterious and contributes to the pathogenesis of cardiomyocytes.
Constant state of 40 kDa binding with actin is detrimental
The precise arrangement of actin-myosin filaments in the sarcomere depends on the ability of cMyBP-C to modulate myosin assembly and stabilize thick filaments. cMyBP-C consists of 11 modules labeled C0–C10 from the N- to the C-terminus (Fig. 1b) and belongs to the intracellular immunoglobulin superfamily which is composed of repeating domains of Ig and fibronectin type-3. cMyBP-C interacts with the S2 fragment of myosin via phosphorylation of the M domain in a phosphorylation-dependent manner (Gruen and Gautel 1999; Gruen et al. 1999; Sadayappan et al. 2006) and is strongly anchored to light meromyosin (LMM) via the C10 domain (Gilbert et al. 1999) and titin via the C8–C10 domains (Freiburg and Gautel 1996). In addition, a potential actin-binding sequence exists in the Pro-Ala-rich linker sequence located between the C0 and C1 domains (Shaffer et al. 2009), and recent data provide evidence for in vitro interaction between cMyBP-C and actin via the C0 domain (Kuli-kovskaya et al. 2003), C1 and M domain (Shaffer et al. 2009), and C5 domain (Rybakova et al. 2011). These studies indicate that the N′-region of cMyBP-C is critical for sarcomere stability. Strikingly, N′-regions are severely degraded during myocardial injury (Decker et al. 2005; Sadayappan et al. 2006, 2009). Dephosphorylation of cMyBP-C is directly associated with its degradation and the generation of 40 kDa cleavage fragments. Consistent with previous studies, we found that the 40 kDa fragments retain strong interaction with actin (Weith et al. 2012) and exhibit weaker interaction with myosin compared to C0–C2 domain. However, others studies have determined that the N′-fragments C0–C1 and C0–C4 do not bind to actin in a saturable manner (Rybakova et al. 2011) and have suggested alternative binding sites for the C0 domain, such as regulatory light chain (Ratti et al. 2011). We hypothesized here that the 17 residues of the M domain in the 40 kDa fragment are sufficient to interact with actin (Fig. 7c: i–iii), and that the constant binding state of the 40 kDa fragment with actin could potentially alter myosin-actin interaction, thereby impairing force generation and Ca2+ homeostasis (Weith et al. 2012). It is interesting to note, however, that immunofluorescent analysis demonstrated localization of the 40 kDa fragments at the C-zone within the sarcomere and not the I-zone, which would be expected if the 40 kDa fragment exhibited exclusive binding affinity for actin. While pull-down assays suggest the 40 kDa fragments exhibit lower myosin binding ability compared to C0–C2 domains, further studies are required to conclusively determine whether the 40 kDa fragments bind with higher affinity to myosin or actin. Ultimately, inhibiting actin function may lead to cytotoxicity against cardiomyocytes and promote apoptosis. Therefore, the role of cMyBP-C cleaved fragments in prompting cell death in I–R injury is sufficiently significant to warrant further studies in vivo.
Post-translational modification of cMyBP-C
Reduced phosphorylation is accompanied by contractile dysfunction and increased degradation of cMyBP-C, followed by the generation of 40 kDa cleavage fragments (Decker et al. 2005; Sadayappan et al. 2006). Studies show that histone deacetylases are activated to deacetylate lysine residues during HF (Kee et al. 2006) and cMyBP-C is a substrate of acetylation at lysine residues (Ge et al. 2009). In the present study, we identified eight acetylation sites that are predominantly acetylated in vivo. The acetylation site K190 is highly conserved across the species and, thus, may play an important role in myosin and actin interactions. One possible mechanism of pathogenesis is that acetylation of 40 kDa fragments in vivo may influence its binding with actin and accelerate contractile dysfunction, however, the functional and physiological consequences of the acetylation of these sites have yet to be characterized.
Conclusion
In summary, we demonstrated novel evidence that the N′ region of cMyBP-C is toxic to cardiomyocytes and able to induce cell death. The presence of abundant N′fragments of cMyBP-C in myocardial injuries could contribute to altered protein–protein interaction, impaired Ca2+ handling, and contractile dysfunction that may ultimately lead to the apoptosis. The underlying mechanism by which cMyBP-C fragments induce cytotoxicity remains to be elucidated. In the future, elucidation of molecular determinants responsible for allosteric modulation of cardiac contraction by the N-terminus of cMyBP-C and the mechanisms by which charge-induced modifications alter conformational states may provide novel approaches for controlling heart function post-I–R injury.
Acknowledgments
We thank our collaborators, Jody L. Martin, PhD, in the adenoviral core faculty for viral production and NRVCMs preparation, and Aleksey Zima, PhD, for providing us the rabbit adult ventricular cardiomyocytes at the Cell and Molecular Physiology, Loyola University Chicago, Stritch School of Medicine, Maywood, IL 60153, USA. We also thank, Jeffery D. Molkentin, PhD, Cincinnati Children’s Hospital, Cincinnati, OH 45229, USA, for providing us the mouse I–R injury surgery training.
Funding This study was funded by National Institutes of Health grants HL007692 (Dr. Sarkey), HL101297, HL62426 (Dr. de Tombe) and R01HL105826 (Dr. Sadayappan), American Heart Association—Post-Doctoral Training Grant (10POST4230040 to Dr. Sundaresan) and—Scientist Development Grant (0830311N to Dr. Sadayappan).
Abbreviations
- cMyBP-C
Cardiac myosin binding protein-C
- Ca2+
Calcium ions
- h
Hour(s)
- I–R
Ischemia–reperfusion
- Ig
Immunoglobulin
- kDa
Kilodaltons
- LDH
Lactate dehydrogenase
- MI
Myocardial infarction
- M domain
Myosin binding domain
- Min
Minutes
- MOI
Multiplicity of infection
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- N′
N-terminal
- NRVCM
Neonatal rat ventricular cardiomyocyte
- PBS
Phosphate buffered saline
- PKA
Protein kinase A
- SR
Sarcoplasmic reticulum
Contributor Information
Suresh Govindan, Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University Chicago, 2160 South First Ave., Maywood, IL 60153, USA.
Jason Sarkey, Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University Chicago, 2160 South First Ave., Maywood, IL 60153, USA.
Xiang Ji, Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University Chicago, 2160 South First Ave., Maywood, IL 60153, USA.
Nagalingam R. Sundaresan, Department of Surgery, University of Chicago, Chicago, IL 60637, USA
Mahesh P. Gupta, Department of Surgery, University of Chicago, Chicago, IL 60637, USA
Pieter P. de Tombe, Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University Chicago, 2160 South First Ave., Maywood, IL 60153, USA
Sakthivel Sadayappan, Email: ssadayappan@lumc.edu, Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University Chicago, 2160 South First Ave., Maywood, IL 60153, USA.
References
- Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Communal C, Sumandea M, de Tombe P, Narula J, Solaro RJ, Hajjar RJ. Functional consequences of caspase activation in cardiac myocytes. Proc Natl Acad Sci USA. 2002;99:6252–6256. doi: 10.1073/pnas.092022999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland O, Sadayappan S, Messer AE, Steinen GJ, van der Velden J, Marston SB. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J Mol Cell Cardiol. 2010;49:1003–1011. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Decker RS, Decker ML, Kulikovskaya I, Nakamura S, Lee DC, Harris K, Klocke FJ, Winegrad S. Myosin-binding protein C phosphorylation, myofibril structure, and contractile function during low-flow ischemia. Circulation. 2005;111:906–912. doi: 10.1161/01.CIR.0000155609.95618.75. [DOI] [PubMed] [Google Scholar]
- Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- Ge Y, Rybakova IN, Xu Q, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc Natl Acad Sci USA. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R, Cohen JA, Pardo S, Basu A, Fischman DA. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. J Cell Sci. 1999;112(Pt 1):69–79. doi: 10.1242/jcs.112.1.69. [DOI] [PubMed] [Google Scholar]
- Govindan S, McElligott A, Muthusamy S, Nair N, Barefield D, Martin JL, Gongora E, Greis KD, Luther PK, Winegrad S, Henderson KK, Sadayappan S. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J Mol Cell Cardiol. 2012;52:154–164. doi: 10.1016/j.yjmcc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- Gupta MP, Samant SA, Smith SH, Shroff SG. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J Biol Chem. 2008;283:10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim TE, Dowell C, Diamanti T, Scheuer J, Tardiff JC. Independent FHC-related cardiac troponin T mutations exhibit specific alterations in myocellular contractility and calcium kinetics. J Mol Cell Cardiol. 2007;42:1098–1110. doi: 10.1016/j.yjmcc.2007.03.906. [DOI] [PubMed] [Google Scholar]
- Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- Harris SP, Rostkova E, Gautel M, Moss RL. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ Res. 2004;95:930–936. doi: 10.1161/01.RES.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- Herron TJ, Rostkova E, Kunst G, Chaturvedi R, Gautel M, Kentish JC. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ Res. 2006;98:1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- Howarth JW, Ramisetti S, Nolan K, Sadayappan S, Rosevear PR. Structural insight into unique cardiac myosin-binding protein-C motif: a partially folded domain. J Biol Chem. 2012;287:8254–8262. doi: 10.1074/jbc.M111.309591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EM, Wang X, Xu K, Hossain MM, Jin JP. Nonmyofilament-associated troponin T fragments induce apoptosis. Am J Physiol Heart Circ Physiol. 2009;297:H283–H292. doi: 10.1152/ajpheart.01200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, Kim JK, Kim KK, Epstein JA, Kook H. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Knollmann BC, Kirchhof P, Sirenko SG, Degen H, Greene AE, Schober T, Mackow JC, Fabritz L, Potter JD, Morad M. Familial hypertrophic cardiomyopathy-linked mutant troponin T causes stress-induced ventricular tachycardia and Ca2+-dependent action potential remodeling. Circ Res. 2003;92:428–436. doi: 10.1161/01.RES.0000059562.91384.1A. [DOI] [PubMed] [Google Scholar]
- Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122:761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK, Winkler H, Taylor K, Zoghbi ME, Craig R, Padron R, Squire JM, Liu J. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci USA. 2011;108:11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JY, Gulick J, Robbins J, Woodhead J, Lehman W, Craig R. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410:214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien PJ. Blood cardiac troponin in toxic myocardial injury: archetype of a translational safety biomarker. Expert Rev Mol Diagn. 2006;6:685–702. doi: 10.1586/14737159.6.5.685. [DOI] [PubMed] [Google Scholar]
- Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286:12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. J Biol Chem. 2011;286:2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S. Cardiac myosin binding protein-C: a potential early-stage, cardiac-specific biomarker of ischemia–reperfusion injury. Biomark Med. 2012;6:69–72. doi: 10.2217/bmm.11.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, de Tombe P. Cardiac myosin binding protein-C: redefining its structure and function. Biophys Rev. 2012 doi: 10.1007/s12551-012-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein-C phosphorylation is cardioprotective. Proc Natl Acad Sci USA. 2006;103:16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, Greis KD, Robbins J. Phosphorylation-dependent proteolysis and pathogenesis of cardiac myosin binding protein-C. J Mol Cell Cardiol. 2008;44:S44. [Google Scholar]
- Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Robbins J. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation. 2009;119:1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, Gulick J, Osinska H, Barefield D, Cuello F, Avkiran M, Lasko VM, Lorenz JN, Maillet M, Martin JL, Brown JH, Bers DM, Molkentin JD, James J, Robbins J. A critical function for ser-282 in cardiac myosin binding protein-C phosphorylation and cardiac function. Circ Res. 2011;109:141–150. doi: 10.1161/CIRCRESAHA.111.242560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkey JP, Chu M, McShane M, Bovo E, Mou YA, Zima AV, de Tombe PP, Kartje GL, Martin JL. Nogo-A knockdown inhibits hypoxia/reoxygenation-induced activation of mitochondrial-dependent apoptosis in cardiomyocytes. J Mol Cell Cardiol. 2011;50:1044–1055. doi: 10.1016/j.yjmcc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JF, Kensler RW, Harris SP. The myosin binding protein-C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284:12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JF, Wong P, Bezold KL, Harris SP. Functional differences between the N-terminal domains of mouse and human myosin binding protein-C. J Biomed Biotechnol. 2010;2010:789798. doi: 10.1155/2010/789798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331:713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- Weith A, Sadayappan S, Gulick J, Previs MJ, Vanburen P, Robbins J, Warshaw DM. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winegrad S. Cardiac myosin binding protein C. Circ Res. 1999;84:1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]