Abstract
The transmissible agent of prion disease consists of prion protein (PrP) in β-sheet-rich state (PrPSc) that can replicate its conformation according to a template-assisted mechanism. This mechanism postulates that the folding pattern of a newly recruited polypeptide accurately reproduces that of the PrPSc template. Here, three conformationally distinct amyloid states were prepared in vitro using Syrian hamster recombinant PrP (rPrP) in the absence of cellular cofactors. Surprisingly, no signs of prion infection were found in Syrian hamsters inoculated with rPrP fibrils that resembled PrPSc, whereas an alternative amyloid state, with a folding pattern different from that of PrPSc, induced a pathogenic process that led to transmissible prion disease. An atypical proteinase K-resistant, transmissible PrP form that resembled the structure of the amyloid seeds was observed during a clinically silent stage before authentic PrPSc emerged. The dynamics between the two forms suggest that atypical proteinase K-resistant PrP (PrPres) gave rise to PrPSc. While no PrPSc was found in preparations of fibrils using protein misfolding cyclic amplification with beads (PMCAb), rPrP fibrils gave rise to atypical PrPres in modified PMCAb, suggesting that atypical PrPres was the first product of PrPC misfolding triggered by fibrils. The current work demonstrates that a new mechanism responsible for prion diseases different from the PrPSc-templated or spontaneous conversion of PrPC into PrPSc exists. This study provides compelling evidence that noninfectious amyloids with a structure different from that of PrPSc could lead to transmissible prion disease. This work has numerous implications for understanding the etiology of prion and other neurodegenerative diseases.
Introduction
Prion diseases, or transmissible spongiform encephalopathies, are fatal neurodegenerative disorders that can arise spontaneously and be inherited or acquired through transmission (Prusiner, 1997). Three general mechanisms have been put forward to explain the diversity in etiology of prion diseases. Spontaneous misfolding and aggregation of the normal cellular isoform of the prion protein (PrP), PrPC, into the disease-related infectious isoform, PrPSc, is believed to underlie the sporadic forms of prion diseases (Cohen and Prusiner, 1998). Inherited prion diseases have been linked to a number of single point mutations, truncation, or octarepeat expansion mutations in the PRNP gene, with more than 20 disease-inducing mutations identified (Prusiner and Scott, 1997; Kong et al., 2004; Jackson et al., 2009; Sigurdson et al., 2009). In prion diseases acquired through the transmission of the infectious form, PrPSc, the abnormal conformation replicates itself in an autocatalytic manner by recruiting and converting the PrPC of the host. According to this template-assisted mechanism, the folding pattern of a newly recruited polypeptide chain accurately reproduces that of a PrPSc template (Cohen and Prusiner, 1998).
The current work suggests that a new mechanism responsible for the etiology of transmissible prion diseases exists that is different from those of template-assisted conversion and spontaneous conversion of PrPC into PrPSc. In the current work, three conformationally distinct and well defined amyloid states were prepared in vitro in the absence of any cellular cofactors using only highly purified full-length Syrian hamster recombinant PrP (rPrP). Surprisingly, no signs of prion infection were found in animals inoculated with rPrP amyloid state that resembled PrPSc, whereas an alternative rPrP amyloid state, with a folding pattern significantly different from that of PrPSc, induced a pathogenic process that eventually led to transmissible prion disease. Accumulation of atypical, proteinase K (PK)-resistant PrP (atypical PrPres) with a PK-resistant core that closely resembled that of the rPrP fibrils preceded authentic PrPSc. While no PrPSc particles were found in preparations of the rPrP fibrils in serial protein misfolding cyclic amplification with beads (PMCAb), the rPrP fibrils produced atypical PrPres in modified PMCAb, proving their capability to seed misfolding of PrPC and demonstrating their structural difference with authentic PrPSc. Moreover, amplification of both brain-derived and fibril-triggered PMCAb-derived atypical PrPres was found to be RNA independent, whereas amplification of PrPSc was RNA dependent. Taken together, these results indicate that atypical PrPres was the first product of PrPC misfolding triggered by fibrils and that the RNA-independent structure of atypical PrPres gave rise to structurally different, RNA-dependent PrPSc.
The new mechanism of prion disease introduced here postulates that structures substantially different from that of PrPSc can induce transmissible disease. The current study raises great concern that structures which are considered noninfectious could lead to a transmissible disease. Because pathological changes associated with diverse neurodegenerative diseases can be induced or transmitted in a prion-like manner through aggregated forms of nonprion proteins (for review, see Aguzzi and Rajendran, 2009; Miller, 2009; Frost and Diamond, 2010), the current work has far-reaching implications.
Materials and Methods
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Maryland, Baltimore, MD (assurance number A32000-01; permit number: 0309001).
Expression and purification of rPrP and formation of rPrP fibrils.
Syrian hamster full-length recombinant PrP encompassing residues 23–231 was expressed and purified according to a described previously procedure (Bocharova et al., 2005a) with minor modifications (Makarava et al., 2010). Lyophilized rPrP was dissolved in 5 mm HEPES, pH 7.0, immediately before use. To form fibrils for inoculations, the rPrP stock solution was supplemented with 50 mm MES, pH 6.0, and 0.5 or 2.0 m guanidine hydrochloride (GdnHCl) and incubated at 37°C under continuous agitation. Fibrillation in 0.5 m GdnHCl was performed with 0.5 mg/ml rPrP under 600 rpm horizontal shaking using a DELFIA plate shaker (Wallac), while fibrillation in 2 m GdnHCl was performed with 0.25 mg/ml rPrP under 24 rpm rotation using a Clay Adams Nutator (model 1105). S-fibrils were formed in 2 m GndCl with 0.5 mg/ml rPrP under shaking as described previously (Makarava and Baskakov, 2008). Amyloid formation was confirmed by thioflavin T fluorescence assay, epifluorescent microscopy, and electron microscopy as described previously (Bocharova et al., 2005a). Fibrils were dialyzed into 10 mm sodium acetate, pH 5.0, for analysis or into PBS, pH 7.4, for inoculations. As a control, a rPrP stock solution was diluted in PBS, pH 7.4, for a final protein concentration of 0.5 mg/ml and used for inoculation as normally folded α-rPrP. Conformation of α-rPrP was confirmed by circular dichroism.
Fourier transform infrared spectroscopy.
Fourier transform infrared (FTIR) spectroscopy was performed as described previously (Ostapchenko et al., 2010). Briefly, rPrP fibrils were dialyzed in 10 mm sodium acetate, pH 5.0, loaded into a BioATR II cell of a Bruker Tensor 27 FTIR spectrometer (Bruker Optics), and their infrared spectra were then collected at 2 cm−1 resolution. Five hundred twelve scans were averaged for each fibril species and treated with Opus software (Bruker Optics) to obtain FTIR second derivative spectra.
Bioassay.
For the first passage, weanling Golden Syrian hamsters (all males) were inoculated intracerebrally with three different rPrP amyloid states or α-rPrP described above (see Expression and purification of rPrP and formation of rPrP fibrils). Each animal was anesthetized with 2% O2/4 minimum alveolar concentration (MAC) isoflurane before receiving 50 μl of inoculum. Starting from the third month postinoculation, hamsters were observed daily for disease. They were killed at 664–723 d postinoculation without any signs of clinical disease and their brains were removed aseptically and saved for analysis and second passage. For the second passage, 10% brain homogenates (BHs) prepared by sonication in PBS, pH 7.4 (Makarava et al., 2010), were dispersed by an additional 30 s of sonication immediately before inoculation. Each hamster received 50 μl of 10% BH inoculum intracerebrally under 2% O2/4 MAC isoflurane anesthesia. After inoculation, hamsters were observed daily for disease using a ‘blind’ scoring protocol.
Proteinase K digestion.
Brains were collected aseptically and cut in half with disposable scalpels. One half was used to prepare 10% BHs in PBS or conversion buffer as described previously (Makarava et al., 2010), while the second half was stored at −80°C for future analysis or fixed in formalin for histopathology. For the PK digestion of BH in sarcosyl, an aliquot of 10% BH was mixed with an equal volume of 4% sarcosyl in PBS supplemented with 50 mm Tris, pH 7.5, and digested with 20 μg/ml PK for 30 min at 37°C with 1000 rpm shaking using a DELFIA plate shaker (Wallac) placed in 37°C incubator. PK digestion was stopped by adding SDS sample buffer and heating the samples for 10 min in a boiling water bath. For PK digestion of recombinant fibrils, fibrils were diluted to 10 μg/ml in 1% normal BH (NBH) in conversion buffer supplemented with 0.25% SDS and digested with 10 or 50 μg/ml PK for 1 h at 37°C. After loading onto NuPAGE 12% Bis-Tris gels and transfer to PVDF membrane, PrP was detected with 3F4 (epitope 109–112), D18 (epitope 133–157), or SAF-84 (epitope 160–170) antibody, as indicated. Density profiles for Western blots were generated using WCIF ImageJ software (National Institutes of Health).
Protein misfolding cyclic amplification with beads.
Ten percent NBH from healthy hamsters was prepared as described previously (Makarava et al., 2011) and used as a substrate for PMCA with beads (Gonzalez-Montalban et al., 2011a). For the first round, 10 μl of 10% BH from inoculated animals was added to 90 μl of NBH. The standard sonication program consisted of 30 s sonication pulses delivered at 50% power efficiency applied every 30 min during a 24 h period. For each subsequent round, 10 μl of the reaction from the previous round were added to 90 μl of fresh substrate. Each PMCAb reaction was carried out in the presence of three 3/32” Teflon beads (McMaster Carr). To analyze production of PK-resistant PrP material in PMCAb, 10 μl of sample were supplemented with 5 μl of SDS and 5 μl of PK to a final concentration of SDS and PK of 0.25% and 50 μg/ml respectively, followed by incubation at 37°C for 1 h. The digestion was terminated by addition of SDS-sample buffer and heating the samples for 10 min in a boiling water bath. Samples were loaded onto NuPAGE 12% Bis-Tris gels, transferred to PVDF membrane, and probed with 3F4 or SAF-84 antibodies.
Protein misfolding cyclic amplification with partially deglycosylated substrate.
To produce substrate for protein misfolding cyclic amplification with partially deglycosylated substrate (dgPMCAb), 10% NBH from healthy hamsters prepared for PMCAb (see Protein misfolding cyclic amplification with beads) was treated with peptide N-glycosidase F (PNGase F) (New England BioLabs, glycerol-free) as follows. After preclearance of NBH at 500 × g for 2 min, 1500 U/ml PNGase F was added to the supernatant, and the reaction was incubated on a rotator at 37°C for 5 h. The resulting substrate was used in dgPMCAb using sonication conditions as described for PMCAb. To prepare RNA-depleted normal brain homogenate, NBH was precleared at 500 × g for 2 min, and then 100 μ/ml RNase A (Sigma, catalog no. R4875) was added to the supernatant and the reaction was incubated on a rotator at 37°C for 1 h as described previously (Gonzalez-Montalban et al., 2011b). Lack of RNA in RNA-depleted NBH was confirmed by RNA analysis in agarose gel.
Histopathological studies.
Formalin-fixed brain halves divided at the midline (right hemisphere), spinal cord, and spleen were processed for hematoxylin-eosin (H&E) stain as well as for immunohistochemistry for PrP, using the mouse monoclonal anti-PrP antibody 3F4 (1:1000, Covance) and anti-glial fibrillar acidic protein (GFAP; 1:3000, Dako). Blocks were treated in formic acid (96%) before being embedded in paraffin. For detection of disease-associated PrP, we applied a pretreatment of 30 min hydrated autoclaving at 121°C followed by 5 min in 96% formic acid. We evaluated all tissues for the presence of inflammation and PrP immunoreactivity, and the brain for the presence of spongiform change and degree of gliosis. Degree of spongiform change, neuronal loss, and gliosis and intensity of PrP immunostaining were semiquantitatively evaluated (0, none; 1, mild; 2, moderate; 3, severe) in the following anatomical regions as described previously: frontal cortex, hippocampus, caudate-putamen, thalamus, brainstem, and cerebellum (Makarava et al., 2010). Lesion profiles were obtained by averaging scores of spongiform change, neuronal loss, and gliosis for each anatomical region and animal group.
Conformational stability assay.
Ten percent BHs were sonicated for 30 s at 50% efficiency within Misonix-4000 microplate horn filled with 350 ml of water. Then the samples were diluted 2-fold with conversion buffer and incubated with various concentrations of GdnHCl in PBS, pH 7.4, for 1 h at room temperature. After incubation, all samples were diluted with nine volumes of 2% sarcosyl in PBS and incubated for additional 1 h at room temperature. Then, 20 μg/ml PK were added to each sample followed by incubation for 1 h at 37°C with 1000 rpm shaking using a DELFIA plate shaker (Wallac). The PK digestion was stopped by 2 mm PMSF, and the samples were precipitated by addition of 4 volumes of ice-cold acetone and overnight incubation at −20°C. The following day, samples were centrifuged for 30 min at 13,000 rpm in an AccuSpin Micro centrifuge (Fisher Scientific), the supernatants were aspirated, and the pellets were dried for 30 min at room temperature, dissolved in 1× SDS-loading buffer, denatured by incubation in boiling water bath for 10 min, and loaded onto NuPAGE 12% Bis-Tris gels. After transfer to PVDF, PrP was detected with 3F4 antibody.
To analyze the conformational stability of rPrP fibrils, dialyzed preparations of fibrils were supplemented with various concentrations of GdnHCl in 50 mm MES, pH 6.0, incubated for 1 h at room temperature, and then diluted out of GdnHCl with 1% NBH in conversion buffer before PK digestion with 2 μg/ml PK for 1 h at 37°C. The reaction was stopped by PMSF, precipitated by acetone, and analyzed by Western blot with SAF-84 antibody. Signal intensity of Western blots was measured using WCIF ImageJ software (National Institute of Health).
Deglycosylation of PrPres.
Removal of N-linked glycans was performed as described previously (Deleault et al., 2008) using glycerol-free PNGase F and supplied buffers (New England BioLabs). Ten percent BH or dgPMCAb product were mixed with equal volume of 4% sarcosyl in PBS, pH 7.4, digested with 20 μg/ml or 50 μg/ml PK as described above, deglycosylated following the procedure described previously (Makarava et al., 2010), and assayed by Western blot with SAF-84 or D18 antibody.
Results
Generating three conformationally distinct amyloid states
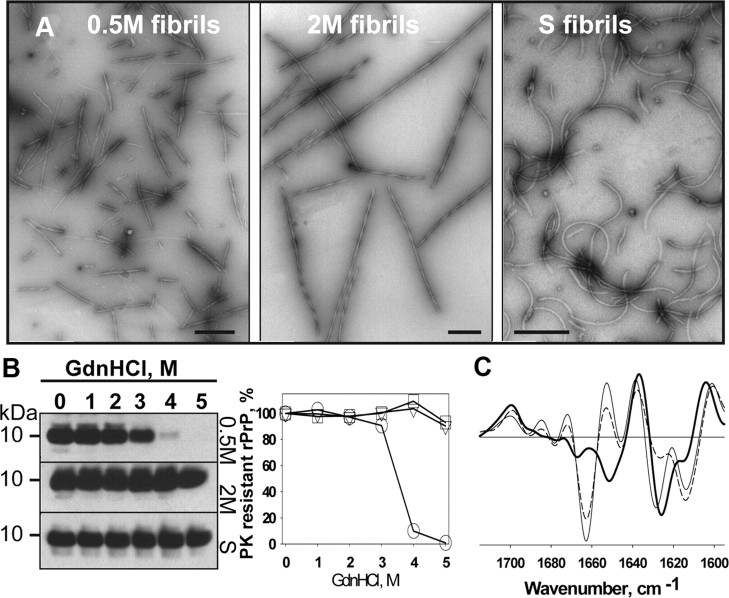
Three experimental protocols that produce three well defined and conformationally distinct amyloid states were employed as described previously (Makarava and Baskakov, 2008; Sun et al., 2008; Ostapchenko et al., 2010). Briefly, rPrP fibrils were formed in 0.5 m GdnHCl (referred to as 0.5 m fibrils) in 2 m GdnHCl under rotation (referred to as 2 m fibrils), or in 2 m GdnHCl under shaking (referred to as S-fibrils, Fig. 1A). rPrP expression, purification, and fibrillation procedures were conducted in a laboratory that was never exposed to transmissible spongiform encephalopathies (TSEs). In agreement with the previous studies, 0.5 m fibrils were substantially shorter in length than the 2 m fibrils (Sun et al., 2008), whereas S fibrils displayed a characteristic curvy shape (Fig. 1A) (Makarava and Baskakov, 2008; Ostapchenko et al., 2010). The differences in length between 0.5 m and 2 m fibrils were presumably due to higher intrinsic fragility of the 0.5 m fibrils (Sun et al., 2008). As judged from the GdnHCl-induced denaturation assay, 2 m or S fibrils were substantially more stable than the fibrils produced in 0.5 m GdnHCl (Fig. 1B). In agreement with previous studies that employed FTIR (Makarava and Baskakov, 2008; Ostapchenko et al., 2010), 0.5 m and 2 m but not S fibrils were characterized by a large peak at 1663 cm−1, which is typically assigned to β- or γ-turns (Fig. 1C). In S fibrils, the cross-β structure was represented by a peak at 1625 cm−1 with a shoulder at 1614 cm−1, whereas in 0.5 m or 2 m fibrils, the cross-β structure exhibited two peaks at 1628 cm−1 and 1614 cm−1, where the latter is believed to represent more rigid β-sheets than the former. As judged from electron microscopy, the interface surface between filaments was greater in 2 m than in 0.5 m or S fibrils (Fig. 1A), consistent with the larger fraction of rigid β-structure in 2 m fibrils as evident from a predominant peak at 1614 cm−1 (Fig. 1C).
Figure 1.
Analysis of rPrP fibrils produced in vitro. A, Negative staining electron microscopy of 0.5 m fibrils (left), 2 m fibrils (middle), or S fibrils (right). Scale bars, 0.1 μm. B, Analysis of fibril conformational stability. Western blots (left) and the conformational stability profiles (right) for fibrils subjected to GdnHCl-induced denaturation and treated with PK: 0.5 m fibrils (circles), 2 m fibrils (triangles), S fibrils (squares). C, Analysis of fibril secondary structure using FTIR spectroscopy. Second derivatives of FTIR spectra for 0.5 m fibrils (thin line), 2 m fibrils (dashed line), or S fibrils (bold line).
Fibrils (0.5 m) seeded formation of atypical PrPres in animals
The three rPrP fibrillar types or rPrP folded into normal α-helical conformation (α-rPrP) were inoculated into Syrian hamsters. No clinical signs were observed in any animal groups until old age (664–723 d postinoculation), when all animals were killed. However, six of seven animals inoculated with 0.5 m fibrils showed atypical, PK-resistant, C-terminal bands of ∼13, 16, and 23 kDa (designated as atypical PrPres) (Fig. 2B). Atypical PrPres was immunoreactive with SAF-84 antibody (epitope 160–170), but not with 3F4 antibody (epitope 109–112) (Fig. 2 A,B). Upon treatment with PNGase F, the three PK-resistant bands of 23, 16, and 13 kDa in atypical PrPres merged into a single band of ∼13 kDa, illustrating that these three bands represent diglycosylated, monoglycosylated, and unglycosylated forms of a single PK-resistant fragment, respectively (Fig. 2D). This fragment was immunoreactive with SAF-84 (epitope 160–170) antibody, but not with D18 (epitope 133–157) antibody (Fig. 2D). The PK-resistant core of 0.5 m rPrP fibrils consisted of two major fragments of 12 and 10 kDa (Fig. 2D). As judged from mass spectroscopy and epitope mapping, the 12 and 10 kDa fragments were previously found to encompass residues ∼138–231 and ∼152–231, respectively (Bocharova et al., 2005b). Consistent with these studies, both PK-resistant fragments were immunoreactive with SAF-84 antibody, but only the 12 kDa fragment reacted with D18 antibody (Fig. 2D). In summary, as judged from the epitope mapping, the PK-resistant core of the atypical PrPres matched the 10 kDa band of rPrP fibrils and presumably encompassed residues ∼152–231. The difference between PAGE-mobility of the atypical PrPres and the 10 kDa rPrP fragment was likely due to a glycosylphosphatidylinositol (GPI) anchor in the first one.
Figure 2.
Bioassay of rPrP fibrils. A, B, Western blots of brain homogenates from the animals inoculated with 0.5 m fibrils, 2 m fibrils, S fibrils, or α-rPrP and stained with 3F4 (A) or SAF-84 (B) antibody. In A, films were overexposed to emphasize a variation in the amounts of PrPSc. Undigested BHs are provided as references. BHs from the animals with numbers in rectangles were used for the second passage. C, Western blots of products of sPMCAb reactions seeded with BHs from the animals inoculated with three fibril types or α-rPrP. sPMCAb consisted of three rounds; 3F4 antibody was used for staining. D, Western blots of brain material containing atypical PrPres (lanes 1–3) or 0.5 m fibrils produced in vitro (lanes 4–6) and stained with SAF-84 or D18 antibodies. BH in lanes 3 was treated with PNGase F. The 0.5 m rPrP fibrils in lanes 5 were treated with 10 μg/ml and in lanes 4 with 50 μg/ml PK. Undigested BH and 0.5 m fibrils are provided as references. E, Mean incubation time to disease ±SD (light gray bars) and mean duration of clinical disease ± SD (dark gray bars) in four animal groups inoculated with BHs from the animals #3, #4, #5, and #7. Numbers within light bars show the attack rates within individual groups. At 665 d postinoculation (marked by dashed line) all remaining animals were killed due to old age and regardless of the disease stage.
In addition to atypical PrPres, four of six animals inoculated with 0.5 m fibrils showed variable amounts of PK-resistant material with a band-shift typical for authentic PrPSc that could be detected by 3F4 and the C-terminal antibodies (Fig. 2A). After three serial PMCAb (sPMCAb) rounds, brain homogenates from all six animals that were positive for atypical PrPres showed PrPSc immunoreactive with 3F4 (Fig. 2C). In contrast, BH from one animal that lacked atypical PrPres did not show any detectible PrPSc even after three sPMCAb rounds.
No animals from the groups inoculated with 2 m fibrils, S-fibrils, or α-rPrP showed atypical PrPres or PrPSc in their brain material (Fig. 2A,B). Furthermore, no PK-resistant materials were detected in any animals from these three groups after three PMCAb rounds (Fig. 2C). In summary, we concluded that six of seven animals were infected upon inoculation of 0.5 m fibrils, whereas no signs of prion infection could be detected upon inoculation of 2 m fibrils, S-fibrils, or α-rPrP.
Transition from atypical PrPres to PrPSc
To test whether the pathogenic process triggered by 0.5 m fibrils leads to transmissible prion disease, brain material from animals #3, #4, #5, and #7 was selected for the second passage. BHs from these animals contained large (animal #3), medium (#7), small (#5), or undetectable (#4) amounts of PrPSc, but approximately equal amounts of atypical PrPres. Animals from all four groups developed clinical signs of prion disease (Fig. 2E). All four groups showed similar clinical symptoms, including startle response, difficulty in righting themselves, substantially reduced activity, and obesity. After the first signs, the clinical disease progressed very slowly, which resembled the slow progression of disease reported for two other synthetic strains, SSLOW and LOTSS (Makarava et al., 2010, 2011).
The incubation time to clinical disease correlated well with the amounts of PrPSc in the inoculums (Fig. 2A,E). The shortest incubation time (347 ± 7 d) was found for the animal group inoculated with BH #3, whereas the longest incubation time (512 ± 82 d) and incomplete attack rate (5 out 7 animals showed clinical signs) were observed for the group inoculated with BH #4 (Fig. 2A,E). Because the amounts of atypical PrPres in four BHs were approximately the same, these results suggest that the differences in the incubation time and the attack rate were likely attributed to the differences in the amounts of PrPSc.
The brain materials from four animal groups were analyzed by Western blot using SAF-84 antibody that detects both atypical PrPres and PrPSc (Fig. 3). Because of the overlaps between the diglycosylated and monoglycosylated atypical PrPres with the monoglycosylated and unglycosylated PrPSc, respectively (Fig. 3C, presented as a schematic diagram), the PK-resistant profiles of atypical PrPres and PrPSc mixtures consisted of four bands (Fig. 3A,B). Nevertheless, the relative amounts of atypical PrPres versus PrPSc could be estimated based on the peak ratios (Fig. 3B). After careful analysis of the individual PK resistant profiles, several important observations can be made.
Figure 3.
Analysis of brain materials from the second passage. A, Western blots of brain material from the animals of second passage inoculated with BHs #3, #4, #5, and #7 and stained with SAF-84. Two animals marked by asterisks in the group #4 were asymptomatic up to 665 d postinoculation. The animal marked with ampersand (&) was euthanized at 534 d postinoculation due to an unrelated health problem. B, PK resistance profiles for individual animals from two groups: those inoculated with BH #4 (top panel) or BH #3 (bottom panel). Four peaks at 33, 23, 16, and 13 kDa represent six PK-resistant bands. C, Schematic representation of the PK resistance profile showing overlap between atypical PrPres and PrPSc, where atypical PrPres and PrPSc are represented by gray and black boxes, respectively. D, Hypothetical mechanism illustrating genesis of PrPSc in animals inoculated with rPrP fibrils. During the first passage, rPrP fibrils seeded atypical PrPres, a new transmissible form of PrP capable of self-replicating without detectible clinical signs. Replication of atypical PrPres occasionally produces PrPSc in seeding events that appeared to be relatively rare and stochastic, and are described by a deformed templating mechanism (Makarava et al., 2009, 2011). PrPSc replicates faster than atypical PrPres and eventually replaces its ancestor.
First, substantial amounts of atypical PrPres observed in animals from the first and second passages for up to 665 d postinoculation indicate that this form is self-replicating and its replication does not require the assistance of PrPSc (Figs. 2A,B, 3A). Second, the observations that (1) atypical PrPres appeared before PrPSc, (2) PrPSc was never detected in animals negative for atypical PrPres, and (3) brain material from animals positive for atypical PrPres always gave rise to PrPSc during serial transmission suggested that atypical PrPres was a precursor of PrPSc (Fig. 2A–C). Third, the group inoculated with BH #3 (with the largest amounts of PrPSc) showed the highest PrPSc/atypical PrPres ratio when compared to the other three groups. The group inoculated with BH #4 (with undetectable amounts of PrPSc) showed the lowest PrPSc/atypical PrPres ratio (Fig. 3A,B). Furthermore, the PrPSc/atypical PrPres ratio was variable not only between the four groups but also within individual groups (Fig. 3A). The greatest variation was observed for the group inoculated with BH #4. Significant variations in PrPSc/atypical PrPres ratios at the end of the first and second passages suggest that the seeding of PrPSc by atypical PrPres appeared to be a stochastic event. Fourth, PrPSc was predominantly diglycosylated, whereas atypical PrPres was predominantly monoglycosylated (Figs. 2A,D). This observation illustrates that these two forms had different preferences in recruiting the three PrPC glycoforms and supports the notion that the two forms were structurally different. Fifth, accumulation of atypical PrPres alone was not sufficient for development of clinical disease. In support of this notion, two animals from the group inoculated with BH #4 that showed large amounts of atypical PrPres but very little if any PrPSc were asymptomatic (Fig. 3A). Sixth, because atypical PrPres was eventually replaced by PrPSc (Fig. 3A), PrPSc appears to replicate faster and outcompete the atypical PrPres.
Serial transmission of 0.5 m fibrils leads to a new disease phenotype
Histopathological studies of animals from the second passage revealed characteristic signs of TSE infection, including spongiform degeneration, neuronal loss, reactive astrogliosis, and deposition of disease-associated PrP in the brains without signs of inflammatory infiltration. Spongiform change, neuronal loss, and reactive astrogliosis were predominantly found in the thalamus and the brainstem followed by caudate-putamen (Fig. 4 A,B). However, they were noted also in the frontal cortex, hippocampus, and, to a lesser extent, in the cerebellum. A diffuse/synaptic type of immunoreactivity was observed in all examined subregions (Fig. 4B). In addition, perineuronal deposits were found predominantly in the thalamus, basal ganglia, and deeper layers of the cortex (Fig. 4C). Prominent large plaques and amorphous deposits were present in the subpial, periventricular, and periaqueductal subependymal regions (Fig. 4C). Furthermore, small PrP plaques were observed around vessels, mainly in the white matter of the cerebellum (Fig. 4C). The new synthetic strain that emerged upon inoculation of 0.5 m fibrils will be designated as S05.
Figure 4.

Histopathological analysis. A, Lesion profile (left) and PrP immunopositivity score (right) in hamsters inoculated with BH #3 (squares, red lines). The histopathological profiles for the second passage of LOTSS-inoculated hamsters (circles, blue lines) are provided as a reference (Makarava et al., 2011). B, Comparison of spongiform changes in the hippocampus, frontal cortex, thalamus, and cerebellum stained with H&E (top) or anti-PrP 3F4 antibody (bottom) (scale bar, 100 μm for all panels). C, Plaques in the brain subependymal region stained with H&E and 3F4 antibody (scale bar, 30 μm), perineuronal PrP immunoreactivity (indicated by a white arrow) in the thalamus (scale bar, 10 μm), and perivascular PrP plaques (indicated by black arrowheads) in the cerebellar white matter (scale bar, 10 μm).
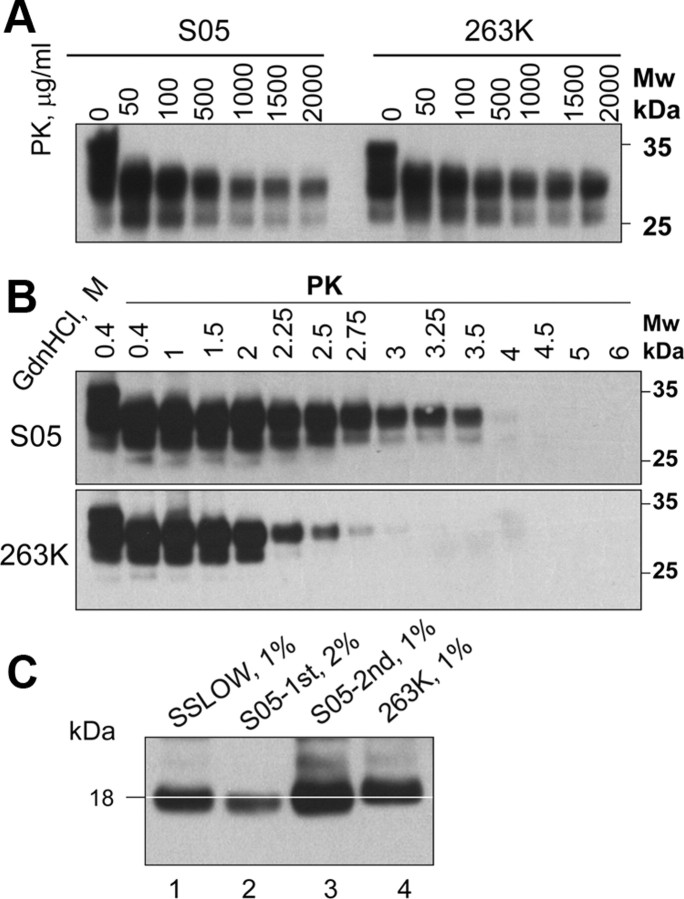
Biochemical properties of S05 PrPSc were assessed using several assays. While S05 PrPSc and 263K PrPSc showed very similar PK resistance (Fig. 5A), S05 PrPSc was conformationally more stable than 263K PrPSc (Fig. 5B). PNGase treatment revealed that the size of the PK-resistant core of S05 PrPSc was slightly shorter than that of 263K PrPSc (Fig. 5C). Overall, taking aside strain-specific differences, biochemical assays confirmed that S05 PrPSc exhibits physical features typical for authentic PrPSc.
Figure 5.
Analysis of S05 PrPSc biochemical features. A, Analysis of PK-resistance. BHs from S05- or 263K-inoculated animals were treated with increasing concentrations of glycerol-free proteinase K (Sigma, catalog no. P6556) in the presence of 0.25% SDS for 1 h at 37°C. BH from the second passage of S05 was used. B, Analysis of PrPSc conformational stability. One percent BHs from animals inoculated with S05 or 263K were incubated with increasing concentrations of GdnHCl from 0.4 to 6 m for 1 h, as indicated, then diluted out of GdnHCl, equilibrated for 1 h at room temperature, and digested with 20 μg/ml PK. Undigested brain material exposed to 0.4 m GdnHCl is provided as a reference. BH from the second passage of S05 was used. C, Analysis of a size of PrPSc PK-resistant core. Two or one percent BHs from the first (lane 2) or second (lane 3) passages of S05, respectively, treated with PK and PNGase F and analyzed by Western blot. BHs from the SSLOW-inoculated (lane 1) or 263K-inoculated animals (lane 4) are shown as references. White line indicates the center of S05 unglycosylated PrPSc. Western blots were stained with 3F4 in all experiments.
Preparation of 0.5 m fibrils lacks any detectible PrPSc
An alternative to the hypothesis that noninfectious rPrP fibrils gave rise to PrPSc is the hypothesis that the preparation of 0.5 m fibrils contains small amounts of PrPSc. To test whether small amounts of PrPSc were present in the preparations of 0.5 m fibrils, we employed sPMCAb protocol capable of detecting single PrPSc particles. First, we demonstrated that miniscule amounts of S05 PrPSc can be efficiently detected by sPMCAb. Ten percent BHs prepared from clinically ill animals from the second passage of S05 were diluted in 10-fold serial steps, and then aliquots from each dilution were used to seed sPMCAb reactions. In 100 μl of PMCAb reaction volume, 1010- and 1011-fold diluted S05 brain material was detected with 100% and 66% success rate, respectively (Fig. 6A). The reactions seeded with 1012-fold or higher dilutions were all negative. Five sPMCAb rounds were sufficient for amplification of the highest dilutions of S05 brain material to the detectible level. An increase in the number of sPMCAb rounds did not improve the success rate in the reactions seeded with highly diluted brain material (Fig. 6A). These results illustrate that the limiting dilution was reached at 1011-fold dilution. At 1011-fold dilution there is a 66% chance of finding at least one PMCAb-active PrPSc particle per 100 μl of reaction volume. This experiment demonstrated that sPMCAb can effectively detect as little as a single S05 PrPSc particle.
Figure 6.
Preparations of 0.5 m fibrils have no detectible PrPSc. A, S05 brain material was serially diluted to up to 1013-fold, and each dilution was subjected to five (top) or six rounds (bottom) of sPMCAb. Representative results are shown on Western blot stained with 3F4 antibody. B, sPMCAb reactions were seeded with 0.5 m rPrP fibrils (lanes 8–13), 1010-fold diluted S05 brain material (lanes 2–4), or 0.5 m fibrils and 1010-fold diluted S05 brain material (lanes 5–7); then six rounds of sPMCAb were conducted for each condition. The final concentration of 0.5 m fibrils in sPMCAb reaction was 2.5 μg/ml. Brains from clinically ill animals from S05 second passage were used for all experiments. Undigested 10% NBH is provided as a reference. Western blots were stained with 3F4.
In the next series of experiments, sPMCAb was employed to detect PrPSc in the preparations of 0.5 m rPrP fibrils. Multiple sPMCAb reactions were seeded with 0.5 m fibrils from two independent amyloid preparations. No positive signals were detected in any reaction after six sPMCAb rounds (Fig. 6B). As judged from the previous experiment, five sPMCAb rounds were sufficient to detect S05 PrPSc at the level of a single particle in 100 μl of reaction volume. To rule out the possibility that the negative results in detecting PrPSc in the preparations of 0.5 m fibrils were due to an inhibitory effect of fibrils on PrPSc amplification, sPMCAb reactions were seeded with 0.5 m fibrils mixed with 1010-fold diluted S05 brain material. This dilution was the highest at which 100% sPMCAb reactions were positive. Three independent sPMCAb reactions were conducted, and all were positive (Fig. 6B). This experiment confirmed that the presence of rPrP fibrils did not diminish the sensitivity of detection of S05 PrPSc by sPMCAb. These experiments revealed that the preparations of rPrP amyloid fibrils did not contain any PrPSc particles that could be detected by sPMCAb. In our experience and consistent with the previously published data (Saa et al., 2006; Makarava et al., 2012), sPMCAb is up to ∼4000-fold more sensitive than bioassay. These experiments also showed that 0.5 m fibrils failed to convert PrPC into PrPSc in sPMCAb if used as a source of seeds.
Atypical PrPres could be amplified in modified PMCAb using partially deglycosylated PrPC
While atypical PrPres appeared to represent one of the self-replicating PrP structures, our numerous attempts to amplify atypical PrPres in PMCAb failed (data not shown). We hypothesized that the PrPC glycoform ratio in NBH favors PrPSc amplification but is disadvantageous for atypical PrPres, which is predominantly monoglycosylated. Indeed, when NBH was pretreated with PNGase F (Fig. 7A), atypical PrPres could be readily amplified (Fig. 7B,C). Pretreatment of NBH with PNGase F did not fully remove glycosyls but shifted the ratio of PrPC glycoforms from predominantly diglycosylated to predominantly monoglycosylated (Fig. 7A). The PMCAb format that uses PNGase F-pretreated NBH as a substrate is designated as dgPMCAb. Notably, much lower amounts of PrPSc was amplified in dgPMCAb than in standard PMCAb seeded with the same brain material (Fig. 7B). Nevertheless, the amounts of dgPMCAb-produced PrPSc correlated well with the amounts of PrPSc seeds in brain material: the reactions seeded with material #3 showed the most abundant PrPSc signal, whereas the reactions seeded with material #4 produced the least abundant signal (Figs. 2A, 7B). Again, no signs of atypical PrPres were detected in PMCAb seeded with the same brain material containing atypical PrPres (Fig. 7B,C). These results suggest that (1) atypical PrPres and PrPSc are two self-replicating states, which compete for a substrate in dgPMCAb, and (2) dgPMCAb favors atypical PrPres, whereas standard PrPSc had a selective advantage over atypical PrPres in standard PMCAb (Fig. 7B,C).
Figure 7.
Amplification of atypical PrPres in modified PMCAb. A, Analysis of PrPC glycoform composition in NBH or NBH treated with PNGase F. B, PMCAb or dgPMCAb reactions were seeded with 102-fold diluted brain materials from animals #2, #3 or #4 (see Fig. 2A), 0.5 m fibrils (fibr.), S fibrils, or mouse rPrP fibrils and subjected to four serial rounds. The final concentration of rPrP fibril seeds in all reactions was 2.5 μg/ml. The fourth round of nonseeded PMCAb or dgPMCAb reactions is shown as control. The film was overexposed to show minor amounts of PrPSc amplified in dgPMCAb. C, Serial PMCAb or dgPMCAb reactions seeded with 102-fold diluted brain materials from the #4 animals. The reaction products were treated with 50 or 500 μg/ml PK as indicated. PMCAb amplified PrPSc, whereas dgPMCAb amplified atypical PrPres. D, Analysis of dgPMCAb-derived and brain-derived atypical PrPres fragments before and after PNGase F treatment. A minor amount of deglycosylated PrPSc marked by an asterisk appeared as a result of dgPMCAb amplification. E, Serial dgPMCAb reactions seeded with 0.5 m rPrP fibrils or serial dgPMCAb in the absence of seeds. The final concentration of 0.5 m fibril seeds in the reaction was 2.5 μg/ml. In B, D, and E, the reaction products were treated with 50 μg/ml PK. For all serial PMCAb or dgPMCAb reactions, 10-fold dilution between serial rounds was used. All Western blots were stained with SAF-84.
While both brain-derived PrPres and dgPMCAb-derived atypical PrPres were predominantly monoglycosylated, dgPMCAb-derived PrPres displayed two PK-resistant unglycosylated bands instead of the one in brain-derived PrPres (Fig. 7B,D). These two bands were produced as a result of PK cleavage at two closely located sites. When dgPMCAb-derived PrPres was treated with higher concentration of PK, only a single unglycosylated band similar in size to unglycosylated brain-derived PrPres was observed (Fig. 7C). In fact, 0.5 m fibrils, too, displayed two PK-digestion sites at a low PK concentration and a single site at high PK concentration (Fig. 2D).
Fibrils (0.5 m) triggered atypical PrPres in dgPMCAb
To test whether 0.5 m fibrils can trigger atypical PrPres in vitro, serial dgPMCAb was seeded with fibrils. Remarkably, the reactions seeded with 0.5 m fibrils produced the same PK-resistant band pattern as those observed in dgPMCAb seeded with brain material containing atypical PrPres (Fig. 7B,E). We performed numerous dgPMCA reactions using several independent preparations of 0.5 m fibrils and several preparations of PNGase F-treated NBH substrate. In all experiments, 0.5 m fibril-induced formation of atypical PrPres was robust. The PK-resistant band patterns produced by 0.5 m fibrils and brain-derived atypical PrPres in dgPMCAb were always remarkably similar if not identical. No PrPres bands were observed in nonseeded serial dgPMCAb reactions or serial dgPMCAb reactions seeded with hamster rPrP S-fibrils or fibrils produced from full-length mouse rPrP (Fig. 7B). These results illustrated that the process of inducing atypical PrPres in dgPMCAb was species specific and 0.5 m fibril structure specific. Most important, these results supported the hypothesis that 0.5 m fibrils gave rise to atypical PrPres in animals and that 0.5 m fibrils and atypical PrPres are structurally very similar.
The attempts to use 0.5 m fibril-induced atypical PrPres to seed PrPSc in standard sPMCAb failed, pointing out that authentic PrPSc was still absent in dgPMCA products of rPrP fibrils, nor was it generated in sPMCAb. The fact that PrPSc was observed in dgPMCAb products of amplification of brain-derived atypical PrPres (Fig. 7B) can be explained by the fact that PrPSc was already present in those brains in small amounts (Fig. 2A).
Amplification of atypical PrPres is RNA independent, whereas amplification of PrPSc is RNA dependent
Successful amplification of atypical PrPres in gdPMCA seeded with 0.5 m fibrils pointed to the fact that atypical PrPres was the first product of PrPC misfolding triggered by fibrils. Formation of atypical PrPres accomplishes an important step in triggering pathogenic process: switching from rPrP to PrPC, a GPI-anchored protein with complex glycosylation. The failure to produce PrPSc in PMCAb upon seeding with dgPMCAb-derived atypical PrPres supports the idea that atypical PrPres and PrPSc are structurally different.
Structural differences between atypical PrPres and PrPSc could be highlighted by the differences in RNA dependency of their amplification. Previous studies established that amplification of hamster-adapted scrapie strains in PMCA was RNA dependent (Deleault et al., 2003, 2007, 2010). Two experimental formats were used here to test the effect of RNA. In the first format, serially diluted S05 materials containing atypical PrPres and PrPSc were used to seed PMCAb or dgPMCAb reactions conducted in NBH or RNA-depleted NBH. In PMCAb (that selectively amplifies PrPSc), the amplification of S05 PrPSc was observed only in NBH but not in RNA-depleted NBH (Fig. 8A). The signal detected in RNA-depleted NBH was simply due to seed dilution (Fig. 8A). In dgPMCAb, amplification of atypical PrPres was observed in both NBH and RNA-depleted NBH, whereas robust amplification of PrPSc was seen only in NBH (Fig. 8B). RNA depletion substantially reduced the efficiency of PrPSc amplification.
Figure 8.
Analysis of RNA dependency of PrPSc and atypical PrPres amplification. A, B, Analysis of amplification rate of S05 PrPSc (A) or atypical PrPres (B) in NBH or RNA-depleted NBH [NBH(−RNA)]. A, S05 10% brain material from animal #3 was diluted 10-, 30-, 100-, 300-, or 1000-fold as indicated into 10% NBH or RNA-depleted NBH and amplified for one PMCAb round. Dilution of seeds without amplification is shown as control. B, Brain material from animal #3 was first amplified in dgPMCAb for four serial rounds, and then dgPMCAb products were diluted 10-, 30-, 100-, 300-, or 1000-fold, as indicated, into 10% NBH or RNA-depleted NBH and amplified for one dgPMCAb round. Dilution of seeds without amplification is shown as control. C, Serial dgPMCAb reactions seeded with S05-derived atypical PrPres and conducted in NBH or RNA-depleted NBH. To prepare S05-derived atypical PrPres, serial dgPMCAb was seeded with 103-fold diluted brain materials from animal #3 and subjected to four rounds. Small amounts of PrPSc could not be amplified after RNA depletion, whereas atypical PrPres amplifies well in both conditions. D, Serial dgPMCAb reactions seeded with 0.5 m fibril-derived atypical PrPres and conducted in NBH or RNA-depleted NBH. To prepare 0.5 m fibril-derived atypical PrPres, serial dgPMCAb was seeded with 0.5 m fibrils and subjected to four rounds as shown in Figure 7E. The 0.5 m fibril-derived atypical PrPres amplifies equally well in both NBH and RNA-depleted NBH. In all experiments the reaction products were treated with 50 μg/ml PK. All Western blots were stained with SAF-84.
In an alternative format, dgPMCAb reactions were seeded with S05 materials containing atypical PrPres and PrPSc, and then four serial rounds were performed in NBH or RNA-depleted NBH (Fig. 8C). While atypical PrPres was steadily amplified in both conditions, PrPSc could be only amplified in NBH containing RNA (Fig. 8C). Remarkably, just like brain-derived atypical PrPres, 0.5 m fibril-derived atypical PrPres could be also amplified equally well in the presence or absence of RNA (Fig. 8D). In summary, rPrP fibrils that were produced in the absence of RNA triggered in dgPMCA formation of PrPres, which was also able to amplify in the absence of RNA. Brain-derived atypical PrPres was RNA independent as well, while S05 PrPSc was RNA dependent, a feature typical for hamster prion strains.
Discussion
The current studies demonstrated that transmissible prion disease could be induced by rPrP fibrils with a structure different from that of PrPSc. A long, clinically silent stage accompanied by accumulation of atypical PrPres preceded development of clinical disease. Several lines of evidence suggested that the molecular mechanism leading to transmissible prion disease was fundamentally different from the previously known mechanisms, including the template-assisted conversion initiated by PrPSc or the spontaneous conversion of PrPC into PrPSc. According to the template-assisted conversion mechanism (Cohen and Prusiner, 1998), the folding pattern of the newly produced PrPSc accurately reproduces that of the PrPSc template or seeds. In contrast, in the current studies, prion infection and transmissible disease emerged upon the inoculation of 0.5 m fibrils with a structure different from that of PrPSc (Spassov et al., 2006; Wille et al., 2009; Ostapchenko et al., 2010; Piro et al., 2011).
In previous studies, cellular cofactors including RNA and lipids were required for generating authentic PrPSc structures in vitro from PrPC or rPrP (Deleault et al., 2007; Wang et al., 2010). Furthermore, cellular cofactors including RNAs are believed to be involved in PrPSc replication (Deleault et al., 2003, 2005). In the current work, rPrP fibrils were produced in the absence of any cofactors. As judged from x-ray and FTIR analyses, the PrP folding pattern within rPrP fibrils is significantly different from that of PrPSc (Spassov et al., 2006; Wille et al., 2009; Ostapchenko et al., 2010). Moreover, application of ultra-sensitive PMCAb, which is capable of detecting single PrPSc particles, demonstrated that preparations of 0.5 m rPrP fibrils had no detectible PrPSc. PMCAb has been very useful for amplification of minuscule amounts of PrPSc, but when seeded with recombinant fibrils, no product was generated, suggesting that authentic PrPSc was absent in the preparations of fibrils. On the other hand, 0.5 m fibrils produced atypical PrPres in dgPMCA, proving their capacity to seed misfolding of PrPC and demonstrating their structural difference with authentic PrPSc.
When triggered by structures different from that of PrPSc, genesis of authentic PrPSc appears to involve at least two major steps (Fig. 3D). The first step consisted of rPrP fibril-seeded formation of atypical PrPres. Similarities in size and position of the PK-resistant core suggest that atypical PrPres originated from that of rPrP fibrils (Fig. 2D). Moreover, 0.5 m fibrils triggered atypical PrPres in dgPMCAb with a PK resistance profile identical to that of brain-derived atypical PrPres. In opposite to PrPSc, amplification of both brain-derived and fibril-triggered atypical PrPres was RNA-independent.
The second step, the appearance of PrPSc, can be a result of a series of stochastic mutations of atypical PrPres structure and selection of the most favorable conformers that fit well to a particular cellular environment. The transition from atypical PrPres to PrPSc is quite complicated and poorly understood. While achievable in the brain of animals over their life span, this transition may be very hard to simulate in vitro. While PrPC in hamster brain is predominantly diglycosylated, the distribution of PrPC glycoforms and their ratio vary in different brain regions and neuronal compartments as well as during neuronal differentiation (DeArmond et al., 1999; Russelakis-Carneiro et al., 2002; Monnet et al., 2003). dgPMCAb/PMCAb might not represent the environment at cellular sites where the transition from atypical PrPres to PrPSc occurs. Indeed, PrPSc was not generated in serial PMCAb seeded with 0.5 m fibrils or with atypical PrPres triggered by fibrils in dgPMCAb. These results strongly supported earlier conclusions that authentic PrPSc was absent in preparations of fibrils or in dgPMCAb products of fibrils. If the above mechanism is correct, PrPSc can be considered as a relatively rare byproduct of replication of atypical PrPres. Variations in the amounts of PrPSc within and between animal groups support the hypothesis of the stochastic nature of the second step. According to the proposed mechanism, after the first PrPSc particles were formed, PrPSc can replicate independently of atypical PrPres and eventually outcompete its ancestor (Fig. 3D). An alternative hypothesis is that atypical PrPres represents a low stability, so-called class III strain that can readily mutate into a high stability strain upon serial passage. (Bruce and Dickinson, 1987). However, considering that the properties of atypical PrPres are so strikingly different from those of low or high stability class PrPSc, this possibility is unlikely.
Formation of PrPSc in seeding events initiated by atypical PrPres is best described by a mechanism designated as “deformed templating” (Makarava et al., 2011). According to deformed templating, daughter fibrils or particles can acquire a folding pattern different from that of seeds (Makarava et al., 2009). The observations that atypical PrPres and PrPSc preferred different PrPC glycoforms as a substrate and that atypical PrPres was RNA independent while PrPSc was RNA dependent support the notion of significant structural differences between these forms. Substantial variation in amounts of PrPSc at the end of the first passage suggests that the deformed templating events are relatively rare and stochastic.
The atypical PrPres described here was very similar to the atypical PrPres found in patients with sporadic Creutzfeldt-Jakob disease (Zou et al., 2003), atypical bovine spongiform encephalopathy (H-BSE), which is believed to be sporadic in origin (Biacabe et al., 2007), or ovine scrapie (Baron et al., 2008). This current study suggests that atypical PrPres can replicate in animal brains and that its replication does not require PrPSc assistance; therefore, it represents one of the transmissible PrP states. In current and previous studies on synthetic prions (Makarava et al., 2011), atypical PrPres always preceded PrPSc. No PrPSc was found in atypical PrPres-negative animals. While accumulation of atypical PrPres alone was not pathogenic, its replication seems to represent a silent stage in the genesis of authentic PrPSc. Bearing in mind that much of the public health risk derives from long silent or asymptomatic stages (Peden et al., 2004; Comoy et al., 2008), detection of atypical PrPres should not be underestimated in developing prion detection strategies. This work introduces the first approach for selective amplification of atypical PrPres in vitro—dgPMCAb in RNA-depleted NBH. dgPMCAb should be a useful technique for establishing the relationship between atypical PrPres and PrPSc in natural TSEs.
The hypothesis that amyloid structures significantly different from that of PrPSc can trigger transmissible prion diseases has numerous clinical and epidemiological implications for understanding the origin of TSEs, including TSEs that are considered to be sporadic. The questions of great interest are whether all PrP amyloid structures are equally active in triggering PrPSc and, if not, what is the spectrum of non-PrPSc structures capable of inducing transmissible diseases in wild-type hosts? In contrast to 0.5 m fibrils, inoculations of 2 m fibrils or S fibrils did not lead to prion infection. Lack of any PrPres material including atypical PrPres in these two groups suggest these two structures were not effective in recruiting and/or converting PrPC. Bearing in mind that all three amyloid states were formed within the same amino acid sequence, the differences in their pathogenic activity should be attributed to their individual fibril-specific physical features.
Previous studies on synthetic prions that employed transgenic mice established a correlation between conformational stability of rPrP fibrils and the incubation time to disease (Colby et al., 2009, 2010). Fibrils with low conformational stability were found to cause the disease within a shorter incubation time when compared to the high stability fibrils (Colby et al., 2009). In addition, strain-specific conformational stability of PrPSc was proposed as one of the physical features that control prion amplification rate and incubation time to disease (Legname et al., 2005; Makarava et al., 2010; Ayers et al., 2011; Gonzalez-Montalban et al., 2011b). The current finding that 2 m or S fibrils with a high stability failed to trigger prion infection strongly support the previously established correlation. As evident from FTIR and x-ray diffraction analyses, the PrP folding pattern within S fibrils closely resembled that of PrPSc (Ostapchenko et al., 2010; Wille et al., 2009). Unexpectedly, S fibrils failed to trigger prion infection. S fibrils also failed to trigger atypical PrPres in vitro. These data suggest that conformational stability of rPrP fibrils appears to be more important for triggering pathogenic process than an apparent structural similarity between inoculated material and PrPSc. Conformational stability appears to be linked to the fibril's mechanical properties, such as its intrinsic fragility (Baskakov and Breydo, 2007; Sun et al., 2008). One might speculate that 2 m or S fibrils failed to recruit PrPC because of their low fragmentation rate.
The current studies illustrate that transmissible prion disease can emerge according to a previously unknown mechanism that is different from the spontaneous conversion of PrPC to PrPSc or the template-assisted conversion initiated by authentic PrPSc. The key features of the new mechanism are: (1) the pathogenic process is initiated by amyloid structures different from PrPSc; (2) it is accompanied by a long clinically silent stage; and (3) it is characterized by the accumulation of atypical transmissible PrP states that display limited neurotoxicity before PrPSc emerges. The current work also shows that prion infection can be induced in wild-type animals by rPrP fibrils produced in vitro in the absence of any cellular cofactors or PrPSc seeds.
Footnotes
This work was supported by NIH Grant NS045585 to I.V.B. and Baltimore Research and Education Foundation.
References
- Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog. 2011;7:e1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron T, Bencsik A, Vulin J, Biacabe AG, Morignat E, Verchere J, Betemps D. A C-terminal protease-resistant prion fragment distinguishes ovine “CH1641-like” scrapie from bovine classical and L-type BSE in ovine transgenic mice. PLoS Pathog. 2008;4:e1000137. doi: 10.1371/journal.ppat.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskakov IV, Breydo L. Converting the prion protein: What makes the protein infectious. Biochim Biophys Acta. 2007;1772:692–703. doi: 10.1016/j.bbadis.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Biacabe AG, Jacobs JG, Bencsik A, Langeveld JP, Baron TG. H-type bovine spongiform encephalopathy: complex molecular features and similarities with human prion diseases. Prion. 2007;1:61–68. doi: 10.4161/pri.1.1.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. In vitro conversion of full length mammalian prion protein produces amyloid form with physical property of PrPSc. J Mol Biol. 2005a;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- Bocharova OV, Breydo L, Salnikov VV, Gill AC, Baskakov IV. Synthetic prions generated in vitro are similar to a newly identified subpopulation of PrPSc from sporadic Creutzfeldt-Jakob disease PrPSc. Protein Sci. 2005b;14:1222–1232. doi: 10.1110/ps.041186605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME, Dickinson AG. Biological evidence that the scrapie agent has an independent genome. J Gen Virol. 1987;68:79–89. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Acad Natl Sci U S A. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB. Protease-sensitive synthetic prions. PLoS Pathog. 2010;6:e1000736. doi: 10.1371/journal.ppat.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoy EE, Casalone C, Lescoutra-Etchegaray N, Zanusso G, Freire S, Marcé D, Auvré F, Ruchoux MM, Ferrari S, Monaco S, Salès N, Caramelli M, Leboulch P, Brown P, Lasmézas CI, Deslys JP. Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. PLoS One. 2008;3:e3017. doi: 10.1371/journal.pone.0003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeArmond SJ, Qiu Y, Sánchez H, Spilman PR, Ninchak-Casey A, Alonso D, Daggett V. PrPC glycoform heterogeneity as a function of brain region: implications for selective targeting of neurons by prion strains. J Neuropathol Exp Neurol. 1999;58:1000–1009. doi: 10.1097/00005072-199909000-00010. [DOI] [PubMed] [Google Scholar]
- Deleault AM, Deleault NR, Harris BT, Rees JR, Supattapone S. The effects of prion protein proteolysis and disaggregation on the strain properties of hamster scrapie. J Gen Virol. 2008;89:2642–2650. doi: 10.1099/vir.0.2008/002303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson RA, Supattapone S. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J Biol Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Acad Natl Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry. 2010;49:3928–3934. doi: 10.1021/bi100370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Diamond MI. Prion-like mechanisms on neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Makarava N, Ostapchenko VG, Savtchenko R, Alexeeva I, Rohwer RG, Baskakov IV. Highly Efficient Protein Misfolding Cyclic Amplification. PLoS Pathogen. 2011a;7:e1001277. doi: 10.1371/journal.ppat.1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Makarava N, Savtchenko R, Baskakov IV. Relationship between conformational stability and amplification efficiency of prions. Biochemistry. 2011b;50:7933–7940. doi: 10.1021/bi200950v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, Jasanoff A, Lindquist S. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63:438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Surewicz WK, Petersen RB, Zou W, Chen SG, Gambetti P, Parchi P, Capellari S, Goldfarb L, Montagna P, Lugaresi E, Piccardo P, Ghetti B. Inherited prion diseases. In: Prusiner SB, editor. Prion biology and diseases. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2004. pp. 673–775. [Google Scholar]
- Legname G, Nguyen H-OB, Baskakov IV, Cohen FE, DeArmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci U S A. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Baskakov IV. The same primary structure of the prion protein yields two distinct self-propagating states. J Biol Chem. 2008;283:15988–15996. doi: 10.1074/jbc.M800562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Ostapchenko VG, Savtchenko R, Baskakov IV. Conformational switching within individual amyloid fibrils. J Biol Chem. 2009;284:14386–14395. doi: 10.1074/jbc.M900533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild type animals. Acta Neuropathol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Kovacs GG, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Genesis of mammalian prions: from non-infectious amyloid fibrils to a transmissible prion disease. PLoS Pathog. 2011;7:e1002419. doi: 10.1371/journal.ppat.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Savtchenko R, Alexeeva I, Rohwer RG, Baskakov IV. Fast and ultrasensitive method for quantitating prion infectivity titer. Nat Commun. 2012;3:741. doi: 10.1038/ncomms1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. Could they all be prion diseases? Science. 2009;326:1337–1339. doi: 10.1126/science.326.5958.1337. [DOI] [PubMed] [Google Scholar]
- Monnet C, Marthiens V, Enslen H, Frobert Y, Sobel A, Mège RM. Heterogeneity and regulation of cellular prion protein glycoforms in neuronal cell lines. Eur J Neurosci. 2003;18:542–548. doi: 10.1046/j.1460-9568.2003.02777.x. [DOI] [PubMed] [Google Scholar]
- Ostapchenko VG, Sawaya MR, Makarava N, Savtchenko R, Nilsson KP, Eisenberg D, Baskakov IV. Two amyloid states of the prion protein display significantly different folding patterns. J Mol Biol. 2010;400:908–921. doi: 10.1016/j.jmb.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AH, Head MW, Ritchie DL, Bell JE, Ironside JW. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet. 2004;364:527–529. doi: 10.1016/S0140-6736(04)16811-6. [DOI] [PubMed] [Google Scholar]
- Piro JR, Wang F, Walsh DJ, Rees JR, Ma J, Supattapone S. Seeding specificity and ultrastructural characteristics of infectious recombinant pPrions. Biochemistry. 2011;50:7111–7116. doi: 10.1021/bi200786p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott MR. Genetics of prions. Annu Rev Genet. 1997;31:139–175. doi: 10.1146/annurev.genet.31.1.139. [DOI] [PubMed] [Google Scholar]
- Russelakis-Carneiro M, Saborio GP, Anderes L, Soto C. Changes in the glycosylation pattern of prion protein in murine scrapie. J Biol Chem. 2002;277:36872–36877. doi: 10.1074/jbc.M202229200. [DOI] [PubMed] [Google Scholar]
- Saá P, Castilla J, Soto C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J Biol Chem. 2006;281:35245–35252. doi: 10.1074/jbc.M603964200. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KP, Hornemann S, Heikenwalder M, Manco G, Schwartz P, Ott D, Rulicke T, Liberski T, Julius C, Falsiq J, Stitz L, Wuthrich K, Aguzzi A. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Acad Natl Sci U S A. 2009;106:304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov S, Beekes M, Naumann D. Structural differences between TSEs strains investigated by FT-IR spectroscopy. Biochim Biophys Acta. 2006;1760:1138–1149. doi: 10.1016/j.bbagen.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Sun Y, Makarava N, Lee CI, Laksanalamai P, Robb FT, Baskakov IV. Conformational stability of PrP amyloid firbils controls their smallest possible fragment size. J Mol Biol. 2008;376:1155–1167. doi: 10.1016/j.jmb.2007.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. Generating a Prion Bacterially Expressed Recombinant Prion Protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, Stubbs G. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Acad Natl Sci U S A. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou WQ, Capellari S, Parchi P, Sy MS, Gambetti P, Chen SG. Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J Biol Chem. 2003;278:40429–40436. doi: 10.1074/jbc.M308550200. [DOI] [PubMed] [Google Scholar]