Abstract
Although the Mycobacterium tuberculosis complex (MTBC) infects a third of all humans, little is known regarding the prevalence of mycobacterial infection in nonhuman primates (NHP). For more than a century, tuberculosis has been regarded as a serious infectious threat to NHP species. Advances in the detection of MTBC open new possibilities for investigating the effects of this poorly understood pathogen in diverse populations of NHP. Here we report results of a cross-sectional study using well-described molecular methods to detect a nucleic acid sequence (IS6110) unique to the MTBC. Sample collection was focused on the oral cavity, the presumed route of transmission of MTBC. Buccal swabs were collected from 263 macaques representing 11 species in four Asian countries and Gibraltar. Contexts of contact with humans included free ranging, pets, performing monkeys, zoos, and monkey temples. Following DNA isolation from buccal swabs, the PCR amplified IS6110 from 84 (31.9%) of the macaques. In general, prevalence of MTBC DNA was higher among NHP in countries where the World Health Organization reports higher prevalence of humans infected with MTBC. This is the first demonstration of MTBC DNA in the mouths of macaques. Further research is needed to establish the significance of this finding at both the individual and population levels. PCR of buccal samples holds promise as a method to elucidate the mycobacterial landscape among NHP, particularly macaques that thrive in areas of high human MTBC prevalence.
Keywords: Nonhuman primate, Mycobacterium tuberculosis complex, MTBC, Indonesia, Singapore, Thailand, Nepal, Gibraltar
INTRODUCTION
Mycobacteria infect both human and nonhuman primates
Tuberculosis is an infectious disease caused by subspecies of the Mycobacterium tuberculosis complex (M. tuberculosis, M. africanum, M. bovis, M. canettii, M. microti, and others, together known as the MTBC). The disease is globally distributed, infecting an estimated 2 billion humans and causing some 2 million deaths per year [WHO, 2009]. Mycobacterium tuberculosis, M. africanum, and M. canettii are considered the “human-associated” species while M. bovis, M. microti, and other MTBC species are each associated with their own type of nonhuman hosts such as ungulates and rodents, though all of the MTBC complex species may infect a wide range of mammalian hosts. However, given the ancient origin of MTBC in Africa 3 mya ago it is interesting that an NHP-associated MTBC species has not yet been identified [Brosch et al., 2002; Gutierrez et al., 2005]. This absence could reflect a lack of research on the full NHP pathogen spectrum, or a shared host role among humans and NHP for the “human-associated” MTBC. Either way it is clear that NHP do become infected with and develop disease from MTBC species.
The MTBC, including M. tuberculosis, M. bovis and M. africanum, has been implicated as causes of tuberculosis in several genera of captive and free-ranging NHP (Table 1). These reports have led to a presumption that NHP are highly susceptible to mycobacterial infection and fatal disease [CDC, 1993; Wolfe et al., 1998]. Empirical evidence indicates that rhesus macaques (Macaca mulatta) experimentally infected with MTBC develop a broad range of disease manifestations similar to that observed in humans, from latent infection to rapidly progressing infection with high mortality [Gormus et al., 2004]. Flynn and colleagues found that experimental infection of long-tailed macaques (M.fascicularis) with MTBC produces a similarly wide spectrum of outcomes, from latent infection (absence of disease and not contagious) to active disease (presence of disease, contagious) [Capuano, III et al., 2003; Flynn et al., 2003]. In addition, they describe a third category of subclinical “percolators” in whom pathological evidence of disease is absent, but cultures (of brochoalveolar or gastric lavage aspirates) are occasionally positive for MTBC [Lin et al., 2009].
Table 1.
Literature review of naturally occurring MTBC in NHP
| Year | Location | Context | N | NHP Genus | ID mechanism | ID |
|---|---|---|---|---|---|---|
| >1901–1923k | USA | Zoo | 3 | Simiadae | necropsy | MTBC |
| >1901–1923k | USA | Zoo | 171 | Cercopithecidae | necropsy | MTBC |
| >1901–1923k | USA | Zoo | 18 | Cebidae | necropsy | MTBC |
| >1901–1923k | USA | Zoo | Lemuridae | necropsy | MTBC | |
| 1928–1932i | England | Lab | 50 | Macaca | Histopathology, necropsy | Mycobacterium |
| 1931–1939l | France | Zoo | 1 | Gorilla | necropsy | MTBC |
| 1931–1939l | France | Zoo | 8 | Pan | necropsy | MTBC |
| 1931–1939l | France | Zoo | 2 | Hylobates | necropsy | MTBC |
| 1931–1939l | France | Zoo | 2 | Cercocebus | necropsy | MTBC |
| 1931–1939l | France | Zoo | 10 | Cercopithecus | necropsy | MTBC |
| 1931–1939l | France | Zoo | 130 | Macaca | necropsy | MTBC |
| 1931–1939l | France | Zoo | 175 | Papio | necropsy | MTBC |
| 1931–1939l | France | Zoo | 1 | Radiata | necropsy | MTBC |
| 1938a | USA | Lab | 79 | Macaca, Papio, Cercocebus | TST, necropsy | Mycobacterium |
| 1940 | France | Zoo | 2 | Papio | Necropsy | MTBC |
| 1940 | France | Zoo | 1 | Cercocebus | Necropsy | MTBC |
| 1942–1944g | USA | Lab | 290 | Macaca | Necropsy | MTBC |
| 1955l | USA | Lab | 6 | Pan | necropsy | MTBC |
| 1955l | USA | Lab | 69 | Macaca | necropsy | MTBC |
| 1957l | USA | Lab | 101 | Macaca | Necropsy | MTBC |
| 1968h | USA | Lab | 1 | Macaca | TST, necropsy | Mycobacterium |
| 1968h | USA | Lab | 1 | Macaca | Necropsy | Mycobacterium |
| 1974m | USA | Lab | 5 | Macaca | Necropsy, histopathology | MTBC |
| 1972–1978n | USA | Captive | 7 | Macaca | Necropsy, histopathology, TST | MTBC |
| 1978n | USA | Lab | 43 | Macaca | Necropsy, AFB, culture | M. bovis (verified only in 3) |
| 1978n | USA | Captive | 1 | Macaca | TST, necropsy, culture | M. tuberculosis |
| 1979o | USA | Zoo | 1 | Presbytis | Necropsy, AFB, culture | MTBC |
| 1979o | USA | Zoo | 14 | Presbytis | TST following above case | Mycobacterium |
| 1981f | USA | Lab | 1 | Ateles | Histopathology, culture | MTBC |
| 1981f | USA | Lab | 1 | Macaca | Culture | MTBC |
| 1985c | Kenya | Free-range | 2+? | Papio | Amino acid uptake | MTBC |
| 1990–1993b | Mauritius | Captive | 76 | Macaca | TST, necropsy | Mycobacterium |
| 1990–1993b | Philippines | Captive | 2 | Macaca | TST, necropsy | Mycobacterium |
| 1990–1993b | Indonesia | Captive | 3 | Macaca | TST, necropsy | Mycobacterium |
| 1992v | South Africa | Zoo | 3 | Pan | Culture, PCR RFLP | M. tuberculosis |
| 1993v | South Africa | Zoo | 1 | Papio | Radiography, culture, PCR RFLP | M. tuberculosis |
| 1996p | South Africa | Free-range | 1 | Papio | PGRS RFLP | M. bovis |
| 1999v | South Africa | Zoo | 1 | Semnopithecus | PCR RFLP | M. tuberculosis |
| 2000v | South Africa | Zoo | 1 | Pan | PCR RFLP | M. tuberculosis |
| 2001j | Japan | Captive | 1 | Pan | Histopathology, necropsy, PCR | |
| 2001e | USA | Lab | 27 | Macaca | Spoligotyping | M. bovis |
| 2002s | Colombia | Zoo | 2 | Cebus | AFB, PCR, blots, RFLP | MTBC |
| 2002s | Colombia | Zoo | 1 | Saimiri | AFB, PCR, blots, RFLP | MTBC |
| 2003j | Japan | Zoo | 3 | Varecia | AFB | M. tuberculosis Beijing? |
| 2004j | Japan | Zoo | 17 | Macaca | Not reported | Not reported |
| 2004j | Japan | Zoo | 4 | Colobus | AFB | M. tuberculosis Beijing? |
| 2004j | Japan | Zoo | 8 | Cebus | AFB | M. tuberculosis Beijing |
| 2006t | Portugal | Zoo | 2 | Mandrillus | PCR RFLP, spoligotyping | M. africanum II |
| 2006u | USA | Lab | 2 | Papio | TST, radiography, histopathology, culture | MTBC |
| 2007p | Brazil | Zoo | 1 | Ateles | Necropsy, AFB, RFLP, spoligotyping | M. tuberculosis |
| 2008 | USA | Captive | 32 | Macaca | TST, necropsy, histopathology, PCR, culture | M. bovis |
| 2009d | USA | Lab | 1 | Macaca | PCR | MTBC |
| 2009r | India | Zoo | 62 | Macaca | Not reported | Mycobacterium |
TST= Tuberculin skin test; AFB= Acid fast bacilli; PCR=Polymerase chain reaction; RFLP=Restriction fragment link polymorphism
Sources
(CDC, 1933);
(Habel, 1947);
(Martin, 1968);
(Fox, 1923);
One of the major challenges in tuberculosis research, especially in NHP, is to accurately detect mycobacteria and diagnose infection and therefore the spectrum of the clinical presentations of tuberculosis in NHP is poorly defined [Gibson, 1998]. Historically, diagnosis of tuberculosis in live, captive NHP has relied on clinical examination, radiographs and intradermal tuberculin skin tests (TST) using mammalian old tuberculin all of which have significant drawbacks [Lerche et al., 2008]. Radiographs and TST are both logistically difficult to perform and unreliable for detecting infection [Lerche et al., 2008]. Existing serologic assays based on the detection of MTBC-specific antibodies and measurement of cytokine levels have been shown to be suboptimal, with both false positive and false negative results [Vervenne et al., 2004]. The “gold standard” for diagnosing active infection with MTBC in humans and NHP remains sputum culture[American Thoracic Society, 2000]. However, sputum cultures are notoriously insensitive and require BSL3 biocontainment precautions [American Thoracic Society, 2000]. Increasingly, molecular methods employing polymerase chain reaction (PCR) have been suggested as means to detect the presence of MTBC DNA in bronchoalveolar lavage, gastric aspirate and, for humans, sputum samples[American Thoracic Society, 2000]. These new methods are based on the detection of DNA sequences that are unique to mycobacteria. As early as 1995, PCR was used to detect M. tuberculosis in necropsied liver and lung tissues from a M. fasicularis monkey with a history of respiratory problems [Rock et al., 1995]. One of the most widely used molecular methods amplifies the IS6110 sequence specific to mycobacteria of the MTBC [Eisenach et al., 1991; Eisenach, 1994]. Data show IS6110-based PCR to be a sensitive and specific method for detecting MTBC [Eisenach, 1994].
Diagnosis and detection of MTBC infection among NHP in nonlaboratory settings poses additional difficulties. Logistical barriers in field settings typically preclude the use of TST, blood collection, radiographs and physical exams. Molecular detection of MTBC using easily collected and stored biological samples holds promise for advancing our ability to detect and characterize the presence of MTBC in NHP populations.
As MTBC is predominately a respiratory or gastrointestinal pathogen, we hypothesized that DNA from MTBC would be present in the oral cavities of macaques and could be detected by amplification of the IS6110 repetitive nucleic acid sequence characteristic of the MTBC. PCR analysis has been applied to tracheal wash samples in M. fascicularis with good results [Motzel et al., 1995]. Here we report the application of this method, which we call the oral swab PCR (OSP), to archived buccal swabs from multiple populations of macaques in Asia and Gibraltar. Our results show that OSP detects MTBC DNA in the mouths of several taxa of synanthropic macaques from different geographic locations.
METHODS
Sampling procedures
Since 2000, as part of a comprehensive project to characterize the cross species transmission of infectious agents at the human-NHP interface, we have sampled pet, performing, temple, urban, and free-ranging NHP in several countries. Animal handling, sample collection, processing and storage have all been reported elsewhere [Jones-Engel et al., 2007]. Macaque species, geographic location and context of human contact are presented in Table 2. Briefly, after administering sedation to each macaque, we gently brushed the buccal mucosa using sterile cytology swabs (Omniswab # #WB10-0004, Whatman)in order to collect saliva and buccal cells. Swabs were stored in a sterile cell lysis buffer consisting of 50 mM Tris pH 8.0, 50 mM EDTA, 50 mM sucrose, 100 mM NaCl and 1% SDS at room temperature. These research protocols have been reviewed and approved by the University of Washington Institutional Animal Care and Research Committee (4233-01). This research adhered to the American Society of Primatologists (ASP) Principles for the Ethical Treatment of Non Human Primates. This research adhered to the legal requirements of the countries in which the research was conducted.
TABLE 2.
Sample characteristics by location, date, species, and context, and frequency of IS6110 positive swabs from synanthropic macaques.
| Location | Date | Species | Context | Total N tested | IS6110 + | % Pos |
|---|---|---|---|---|---|---|
| Bali | 2000 | |||||
| Macaca fascicularis | Temple | 37 | 23 | 62.2 | ||
| Sulawesi | 2000 | |||||
| M. tonkeana | Pet | 47 | 34 | 72.3 | ||
| M. maura | Pet | 7 | 6 | 85.7 | ||
| M. nigrescens | Pet | 2 | 1 | 50.0 | ||
| M. nigra | Pet | 10 | 7 | 70.0 | ||
| M. ochreata | Pet | 1 | 1 | 100 | ||
| M. fascicularis | Pet | 4 | 2 | 50.0 | ||
| M. nemestrina | Pet | 1 | 1 | 100 | ||
| Macaque hybrids | Pet | 4 | 2 | 50.0 | ||
| Total Sulawesi | 76 | 54 | 71.1 | |||
| Java | 2003 | |||||
| M. fascicularis | Performing | 22 | 0 | 0 | ||
| Thailand | 2002 | |||||
| M. fascicularis | Zoo | 10 | 5 | 50 | ||
| M. arctoides | Zoo | 4 | 0 | 0 | ||
| M. assamensis | Zoo | 2 | 0 | 0 | ||
| Total Thailand | 16 | 5 | 31.3 | |||
| Nepal | 2003 | |||||
| M. mulatta | Temple | 39 | 1 | 2.6 | ||
| Singapore | 2003 | |||||
| M. fascicularis | Free ranging | 37 | 1 | 2.7 | ||
| Gibraltar | 2005 | |||||
| M. sylvanus | Free ranging | 36 | 0 | 0 | ||
| 263 | 84 | |||||
| 31.9% |
Locations
Sulawesi
Sulawesi is the third largest island in the Indonesian archipelago and has a diverse ethnic and socioeconomic makeup. At least seven morphologically distinct taxa, 40% of all known macaque species, are endemic to this island (Fooden, 1976). Primate pet ownership on the island cuts across all geographic, religious and socioeconomic boundaries [Jones-Engel et al., 2005a]. During a three month period in 2000 a total of 76 pet macaques (Macaca sp.) were sampled. These macaques were being kept as pets throughout the Northern, Central and Southern provinces of Sulawesi. Because pets were acquired through trapping, purchased in animal markets or received as a gift the exact provenance of the animals could not be determined. Taxonomic identification was based on morphology and pelage characteristics. Nearly all of the pets were belted and kept within their owner’s compounds exposed to the elements and accessible to the other villagers.
Bali
Temple monkeys are common throughout much of South and SE Asia. Macaques in these settings benefit from the religious tolerance that is extended to them and in turn in many of the temples they have become a significant source of economic revenue as a tourist attraction. Population levels can grow significantly in these temples because of the ready access to human food and provisioning [Fuentes, 2006; Fuentes and Gamerl, 2005]. Physical contact between the animals and local, domestic and international visitors to these sites is common. In 2000, a group of long-tailed macaques (M. fascicularis) at a temple in Bali were sampled. These animals had been recently captured in order to be evaluated for respiratory symptoms, and were sampled opportunistically.
Java
Performing monkeys and their owners are encountered throughout Asia. In Java, we located a large group of performing monkeys and their owners who operated from a permanent village base [Schillaci et al., 2006]. The owners reported that nearly all of the animals we sampled had been purchased from a local market when they were less than six months old. The provenance of the animals prior to their purchase from the market could not be confirmed. Performing monkeys in this village were the sole source of income for the residents, as such, the care, feeding and housing of these animals was a priority for the community. In 2002 samples were obtained from 22 long-tailed macaques (M. fascicularis) kept by this community.
Thailand
In 2002, an outbreak of respiratory disease was noted in urban, temple macaques in a city in central Thailand. Over a period of months, numerous long-tailed macaques exhibiting respiratory symptoms were trapped by local authorities and removed to a nearby zoo. Samples were obtained from 10 of these former urban monkeys as well as four M. arctoides and two M. assamensis monkeys at the zoo.
Singapore
The small and cosmopolitan island nation of Singapore is home to approximately 1500 free-ranging, long-tailed macaques. These highly synanthropic animals are concentrated in the central part of the island in the Bukit Timah and Central Catchment Nature Reserves but can also be found in small pockets of green space throughout the urban area [Sha et al., 2009]. In early 2003 we trapped and obtained samples from 37 of these animals.
Nepal
The Swoyambhu temple site in Kathmandu, Nepal is a World Heritage site and pilgrim site for hundreds of thousands of visitors annually. The temple, located on a hill in the densely populated Kathmandu Valley provides refuge for approximately 350 rhesus macaques (M. mulatta) who subsist on handouts from humans and who also forage in the surrounding urban area. Samples were obtained from 39 macaques that we screened in 2003[Jones-Engel et al., 2006a].
Gibraltar
Macaca sylvanus is the only non-Asian macaque in our study sample. The species is found in forested pockets in Northern Africa and were introduced into Gibraltar and have naturalized. The Gibraltar M.sylvanus are habituated to humans and receive >90% of their daily nutrition through provisioning. The population of 225 animals is closely monitored and managed by local authorities [Cortes and Shaw, 2006]. Each year hundreds of thousands of tourist visit the Upper Rock Nature Reserve and interact with these free-ranging macaques. In 2005 we collected buccal samples from 38 animals.
DNA extraction and amplification
The OSP protocol involves an initial DNA extraction from all swabs with a standard phenol-chloroform procedure in a dedicated pre-PCR laboratory[Sambrook J and Russell DW, 2001]. To avoid cross-contamination of samples, as well as contamination from positive controls, several steps were taken: work flow is unidirectional from pre-PCR to post-PCR laboratories, and the pre-PCR laboratory is only entered before work in the post-PCR laboratory on any given day. Dedicated instruments, consumables, and reagents were used for this project, and reagents were tested for purity before commencement of any work. All reagents were aliquotted under aseptic conditions into smaller containers so as to minimize contamination opportunities. Extraction and PCR-setup hoods were UV irradiated for at least 20 minutes between each extraction and PCR preparation. Hoods were also cleaned with 10% commercial bleach solution and RNase between work sessions. Alongside each DNA extraction, we ran at least one extraction blank. Buccal swabs from TST negative humans were also run as negative controls. All PCR amplification procedures were performed in a separate post-PCR laboratory.
Amplification of MTBC-specific loci
Following DNA extraction, aliquots of the samples were quantified using mass spectrometry and sample DNA was then diluted into a working solution of 5ng/DNA per microliter. A target 123 base-pair fragment of the MTBC-specific repetitive element, IS6110, was amplified following Eisenach et al. (1994) PCR conditions were as follows: 2.5 U Platinum Taq polymerase (Invitrogen) with the supplied buffer at 1X, 0.4 μM dNTPs, 2.1 μM MgCl2−, 0.2 μM each primer, 0.02 mg RSA, and 5 μL sample DNA in a 50 μL volume with an annealing temperature of 69 [Eisenach, 1994]. Three PCR blanks were included in each PCR run. This number of blanks is necessary to detect potential low level contamination in reagents or sporadic chance contamination in the laboratory. Microtubes with separate caps were used so that, following set-up in the pre-PCR laboratory, a positive control could be added to a single tube immediately prior to thermal cycling. No tubes containing swab sample DNA or blanks were opened in the post PCR-laboratory until after amplification. To visualize the product, 7 μL of PCR product was electrophoresed on 3% agarose and compared to the positive control and a 1kb DNA ladder. In order to ascertain that the amplicon was IS6110, we sequenced the PCR product from six animals, and the sequence was identical to the target IS6110 sequence of NCBI Reference Sequence NC_000962.2.
RESULTS
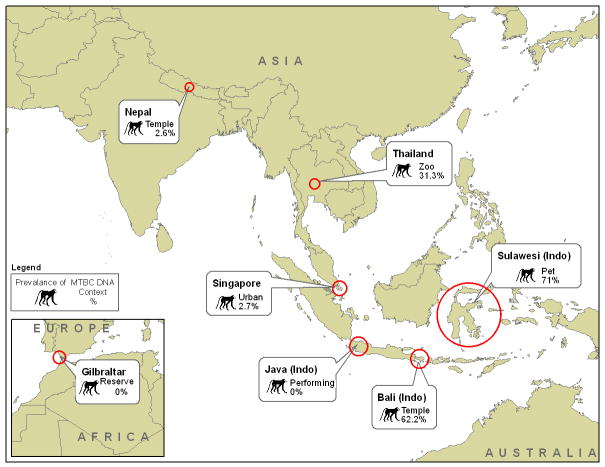
Buccal swabs were collected from a total of 263 macaques and tested for the presence of IS6110 (Table 2). Eleven macaque species and four hybrid macaques from five countries are represented. Contexts of contact with humans included temple (N=76), pet and performing macaques (N=98), free-ranging monkeys (N=73) and zoos macaques (N=16). IS6110 was isolated in buccal swabs of eighty-four animals (31.9%) (Figure 1). We have previously reported data describing demographic characteristics, human-NHP contact and the presence/absence of a variety of infectious agents in each of the populations listed above [Engel et al., 2008; Jones-Engel et al., 2005a; Jones-Engel et al., 2005; Jones-Engel et al., 2006b; Jones-Engel et al., 2006a; Jones-Engel et al., 2007; Schillaci et al., 2005].
Figure 1.
Study country locations, context in which the nonhuman primates were sampled and the overall prevalence of MTBC DNA detection from buccal samples in each location.
Indonesia
There was no significant difference between males (75.6%, N=37) and females (66.7%, N=39) in the detection of MTBC DNA in buccal swabs among the pet macaques in Sulawesi (p =0.3856). Similarly, there was no significant difference among species (p=0.7564) or across age classes (p= 0.4921) among the Sulawesi pets. The pets were sampled from three main geographic regions: on the northern peninsula where 13 of 20 macaques were MTBC positive, in central Sulawesi where 32 of 46 were positive and in southern Sulawesi where 9 of 10 were positive for MTBC DNA. More than a third of the owners indicated that their pet showed signs of illness including coughing, diarrhea and/or weight loss.
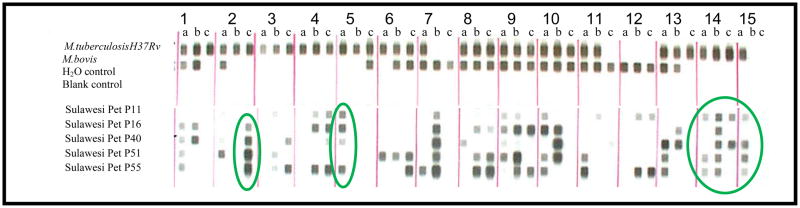
We selected a total of five pet macaque MTBC DNA positive buccal samples representing the three geographic regions in Sulawesi, and submitted these to the Microbial Diseases Laboratory of the California Department of Health Services for spoligotyping. Spoligotyping is a strain differentiation method based on polymorphisms of a repeat sequence of the mycobacterial chromosome, which can be used for fine grain discrimination of MTBC strains [Kamerbeek et al., 1997]. Each mycobacterial strain varies in the number of repeats and sequences between them, resulting in a distinct banding pattern. The quantity of DNA extracted from the original buccal swabs was insufficient to provide a complete spoligotype (n.b. spoligotyping is traditionally done using DNA extracted from infected tissue culture), however the presence of bands in several regions of the spoligotypes indicated that the source of MTBC DNA was not M.bovis in these samples (Figure 2).
Figure 2.
Spoligotype of M.tuberculosis H37Rv and M.bovis reference strains, two negative controls, and samples collected from macaques in Sulawesi. Spoligotyping is a strain differentiation method based on polymorphisms of a repeat sequence of the mycobacterial chromosome. Each mycobacterial strain varies in the number of repeats and the sequences between them, resulting in a distinct banding pattern. All five monkey sample spoligotypes indicate MTBC DNA, though the quantity of DNA available from the buccal swabs was too limited to provide complete spoligotype strain verification. However, comparison of the two reference strains with macaque samples in column 7 (lane b) and column 12 (lanes b and c) indicate that the samples are not M. tuberculosis H37Rv laboratory strain. Column 2 (lane c), circled in green, column 5 (lane a) and columns 14 and 15 (lanes a,b,c and a respectively), all circled in green indicate that the MTBC DNA in the Sulawesi pet samples is not M.bovis.
The animals sampled at the temple in Bali were being treated for a presumptive outbreak of tuberculosis in the monkey population that was concurrent with a tuberculosis outbreak among humans in the neighboring area (personal communication, K. Suaryana). The 37 animals that we swabbed represented approximately 20% of the population at that site. Local authorities were treating these 37 animals with a course of antibiotics and had initially tested them with TST. Individual animal results of the TST were not available nor were the treatment outcomes. On average these animals were 2.2kg lighter than M.fascicularis at neighboring temples, in general poor body condition and respiratory symptoms (coughing reported by their caregivers) were present. In this population MTBC DNA was detected in 17 of 29 male, and 6 of 8 female buccal swabs. Among these temple monkeys there was no significant difference across age classes in positive MTBC DNA results (p= 0.8469).
Among the 22 M. fascicularis performing monkeys we sampled in Java, including 14 juveniles, 5 subadults and three adults, none showed evidence of MTBC DNA in their buccal swabs.
Thailand
All of the M. fascicularis macaques that we sampled at the zoo in Thailand had previously ranged throughout the nearby urban area and temple but had been trapped and brought to the zoo during the last three months because they had either been injured or were showing signs of respiratory illness. Three out of the five adult females and 2 of 5 adult male M.fascicularis were MTBC positive. The M. assamensis and M. arctoides macaques that were also adults and housed in a different section of the zoo were not positive for MTBC DNA in their buccal swabs.
Nepal
Of the 39 M.mulatta sampled at Swoyambhu only 1 adult male tested positive for MTBC DNA on the buccal swab. The remaining 16 males and 22 females, representing three age classes, juveniles (N=13), sub-adults (N=3) and adults (N=18), were negative using our OSP.
Singapore
We sampled 37 animals from five separate troops that occupy home ranges in the Bukit Timah and Central Catchment Nature Reserves. The 12 adult and five sub-adult animals were negative on the OSP. Only one of the 20 juveniles we tested was positive for MTBC DNA on her buccal swab.
Gibraltar
During our sampling of these animals TSTs were placed and read 48–72 hours post injection. None of the animals were reactive on TST. We did not detect MTBC DNA in any of the 38 M.sylvanus that we sampled. These animals represented both sexes, all age classes and the five troops that ranged throughout the area.
DISCUSSION
Our results show the presence of MTBC DNA in buccal samples from macaques in several countries and a variety of contexts. We used an assay that detects MTBC by amplifying the IS6110 insertion sequence unique to MTBC (McHugh et al., 1997). Over the past two decades assays based on the PCR amplification of IS6110 are routinely used to detect mycobacterial infection in human populations. In the context of pulmonary disease, these assays are sensitive and specific tests for MTBC [American Thoracic Society, 2000].
Our assay detects DNA from the MTBC, but cannot differentiate between MTBC species. The MTBC comprises six related species: M. tuberculosis, M. bovis, M. bovis BCG, M. africanum, M. microti, M. pinipedi and M. canetti. Of these mycobacterial species, M. africum, M. microti, M. pinipedi and M. canetti have been reported almost exclusively in other animal populations (e.g. M. pinipedi in seals and sealions, M. microti in rodents) and other geographic areas (M. africanum is restricted to Africa) making their presence in the macaques sampled for this study unlikely.
Based on the known epidemiology of MTBC species, M. tuberculosis and M. bovis are the most likely mycobacterial species to be present in these specimens. The principal reservoir for M. tuberculosis is the human population, with over 2 billion humans worldwide estimated to be infected. Data (not presented here) on contact between humans and the monkeys in the current study show that, frequent and sometimes intimate contact occurs. Moreover, M. tuberculosis is endemic or hyperendemic in human populations in Indonesia, Nepal and Thailand, providing ample opportunity for human to NHP transmission of this agent (Table 3). As a result, we would expect to find, as we did, lower prevalence of MTBC DNA in macaques in countries, such as Gibraltar and Singapore, which have a lower human prevalence of M. tuberculosis.
TABLE 3.
TB prevalence in human population by country for sampling year (as reported by the WHO) compared to frequency of IS6110 positive NHP swabs
| HUMAN PREVALENCE per 100K population | NHP SAMPLE PREVALENCE |
|---|---|
| Indonesia 397 | Indonesia 57% (Sulawesi 71%, Bali 62.2%, Java 0) |
| Nepal 237 | Thailand 31.3 % |
| Thailand 194 | Nepal 2.6% |
| Singapore 57 | Singapore 2.7% |
| Gibraltar (Spain) 24 | Gibraltar 0 |
In areas of high human tuberculosis prevalence and widespread use of cattle, transmission of M. bovis to NHP may occur [Sapolsky and Else, 1987]. Cattle are considered the principle reservoir of M. bovis with likely spillover into wildlife populations. There has been little published research on the interaction of macaques and domestic cattle, though range overlap is not uncommon, providing an opportunity for cross-species transmission to macaques (personal observation). The five samples that underwent spoligotyping had banding patterns incompatible with known M. bovis, making the presence of M. bovis, at least in these samples, very unlikely. Finally, it is possible that some of the MTBC DNA detected in our study belong to an as yet undescribed MTBC species.
This is the first reported detection of MTBC DNA in NHP using oral swabs as a means of collecting specimens. The rationale for testing specimens from the oral cavity is based on MTBC’s transmission route. It might be expected that a macaque with active MTBC infection would periodically cough up mycobacteria or that MTBC from the GI tract could reflux up into the oral cavity. Alternatively, MTBC DNA could be detected on a buccal swab as a result of an individual placing MTBC-contaminated fomites, feces or other secretions from an external source, into their mouth. At least one mycobacterial species, M. bovis, is known to remain viable in optimal soil conditions for up to 10 days after being deposited [Young et al., 2005]. Thus, although the presence of MTBC DNA in a NHP’s mouth suggests infection, the OSP cannot be regarded as a diagnostic test for tuberculosis. Further experimental research is needed to determine whether or not amplification of IS6110 from buccal swab specimens can be regarded as a sensitive and/or specific test of mycobacterial infection.
We are currently developing protocols that optimize DNA recovery to extend our analysis of this unique sample set in order to ecotype the MTBC present in these animals. These data will allow for characterization of the pathogen landscape in NHP populations and by extension information that may be critical to protecting their health.
Additionally, modifications of the buccal swab method hold promise for extending its application to NHP in other contexts. We have experimented with sample collection from free-ranging NHP using flavored “chew swabs” that do not require direct contact with the animal. These chew swabs are tossed to the animals, who place them in their mouths and chew on them before discarding them. The collected swabs, placed into lysis buffer, are processed in the same manner as buccal swabs. We have detected IS6110 in specimens obtained from free-ranging NHP populations using this method (data not shown). Chew swabs may become an ideal tool for primatologists investigating infectious agent transmission in populations of NHP that are challenging to sedate and sample.
Notwithstanding uncertainty regarding the taxonomy and origin of the MTBC we detected in this study, development of a test for MTBC that utilizes oral swab specimens could potentially revolutionize our ability to follow and detect infection in NHP populations. For example, detection of mycobacterial infections and maintenance of tuberculosis-free colonies is a high priority in NHP laboratories, as infection both compromises the integrity of the animals as biomedical models and constitutes a threat to the animals and the staff [Lerche et al., 2008; Morton et al., 2008]. The modalities currently employed, TST, immune-based assays, and clinical evaluations have, on several occasions, failed to detect infected animals[Stockinger DE et al., 2011]. An oral-swab-based PCR assay, which relies solely on the detection of the microbes’ DNA, is potentially a preferable alternative to TST, in terms of accuracy as well as ease of application and interpretation and, unlike immune-based assays, is not hampered by our incomplete understanding of the NHP immune response to exposure and infection with MTBC. Moreover, the ability to more easily detect the presence of mycobacterial TB DNA in a population, regardless of the infection status of the individuals from which samples were obtained, could enhance our capacity to study transmission patterns within and between host populations.
Acknowledgments
The authors are grateful to Drs. D. Cohn, J. Heidrich, N. Lerche, K. Suaryana, A. Fuentes, M. Pizarro, J. Cortes, J. Froehlich, J. Buikstra, A. Stone, J. Supriatna, E. Desmond, J. Cangelosi, S. Rojanasathien S. Thongsawat, R. Chaiwarith, T. Sutthipat as well as U. Paputangan, N. Babo, R. Gharti, D.R. Shakyain, J. Yee, L. Engel and H. Engel for supporting and participating in this research. We thank the communities, temple committees and government agencies in the areas where we have been sampling monkeys for years including: Lembaga Ilmu Pengetahuan Indonesia, Ms S. Chan and the staff of the Central Nature Reserve, National Parks Board, Singapore, the Gibraltar Veterinary Clinic and the Government of Gibraltar, the Federation of Swoyambhu Management and Conservation Committee, the National Research Council of Thailand, as well as of the Faculty of Veterinary Medicine, Chiang Mai University. This research was supported by funding from NIH-NCRR grant P51 RR000166 and RR 02S014, NIH-NIAID grants R01 AI078229 and R03 AI064865, N66001-02-C-8072 DARPA, Unconventional Concepts Inc, Chicago Zoological Society, and the University of New Mexico Research Allocations Committee.
Literature Cited
- Alfonso R, Romero RE, Diaz A, Calderon MN, Urdaneta G, Arce J, Patarroyo ME, Patarroyo MA. Isolation and identification of mycobacteria in New World primates maintained in captivity. Vet Micro. 2004;98:285–295. doi: 10.1016/j.vetmic.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Amado A, Albuquerque T, Goncalves A, Duarte E, Botelho A, Fernandes T, Bernardino R, Lapao N. Tuberculosis in mandrills at the Lisbon zoo. Vet Rec. 2006;159:643. doi: 10.1136/vr.159.19.643. [DOI] [PubMed] [Google Scholar]
- American Thoracic Society. Diagnostic standards and classification of tuberculosis in adults and children. Am J Resp Crit Care Med. 2000;161:1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van SD, Cole ST. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Nat Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano SV, III, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Tuberculosis in imported nonhuman primates -- United States, June 1990–May 1993. MMWR. 1993;42:572–576. [PubMed] [Google Scholar]
- Cortes J, Shaw E. The Gibraltar macaques: management and future. In: Hodges JK, Cortes J, editors. The Barbary Macaque: Biology, Management and Conservation. Nottingham: Nottingham University Press; 2006. pp. 185–198. [Google Scholar]
- Eisenach KD. Use of an insertion sequence for laboratory diagnosis and epidemiologic studies of tuberculosis. Ann Emerg Med. 1994;24:450–453. doi: 10.1016/s0196-0644(94)70182-2. [DOI] [PubMed] [Google Scholar]
- Eisenach KD, Sifford MD, Cave MD, Bates JH, Crawford JT. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Amer Rev Respir Dis. 1991;144:1160–1163. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- Engel GA, Pizarro M, Shaw E, Cortes J, Fuentes A, Barry P, Lerche N, Grant R, Cohn D, Jones-Engel L. Unique pattern of enzootic primate viruses in Gibraltar macaques. Emerg Infect Dis. 2008;14:1112–1115. doi: 10.3201/eid1407.071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother R. Spontaneous diseases observed in 600 monkeys. J Path Bact. 1932;35:867–873. [Google Scholar]
- Flynn JL, Capuano SV, Croix D, Pawar S, Myers A, Zinovik A, Klein E. Non-human primates: a model for tuberculosis research. Tuber (Edinb ) 2003;83:116–118. doi: 10.1016/s1472-9792(02)00059-8. [DOI] [PubMed] [Google Scholar]
- Fooden J. Provisional classifications and key to living species of macaques (primates: Macaca) Folia Primatol (Basel) 1976;25:225–236. doi: 10.1159/000155715. [DOI] [PubMed] [Google Scholar]
- Fox H. Disease in Captive Wild Mammals. Philadelphia: J.B. Lippincott; 1923. [Google Scholar]
- Fuentes A. Human culture and monkey behavior: Assessing the contexts of potential pathogen transmission betweenmacaques and humans. Am J Primatol. 2006;68:880–896. doi: 10.1002/ajp.20295. [DOI] [PubMed] [Google Scholar]
- Fuentes A, Gamerl S. Disproportionate participation by age/sex classes in aggressive interactions between long-tailed macaques (Macaca fascicularis) and human tourists at padangtegal monkey forest, Bali, Indonesia. Am J Primatol. 2005;66:197–204. doi: 10.1002/ajp.20138. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Bouley DM, Larson MJ, Lifland B, Moorhead R, Simkins MD, Borie DC, Tolwani R, Otto G. Outbreak of Mycobacterium bovis in a conditioned colony of rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. Comp Med. 2004;54:578–584. [PubMed] [Google Scholar]
- Gormus BJ, Blanchard JL, Alvarez XH, Didier PJ. Evidence for a rhesus monkey model of asymptomatic tuberculosis. J Med Primatol. 2004;33:134–145. doi: 10.1111/j.1600-0684.2004.00062.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Path. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habel K. Tuberculosis in a laboratory monkey colony: its spread and its control. Am Rev Tuber LV. 1947:77–92. doi: 10.1164/art.1947.55.1.77. [DOI] [PubMed] [Google Scholar]
- Jones-Engel L, Engel GA, Schillaci MA. An ethnoprimatological assessment of disease transmission among humans and wild and pet macaques on the Indonesian island of Sulawesi. In: Patterson J, Wallis J, editors. Commensalism and Conflict: The primate-human interface. 2005a. pp. 196–221. [Google Scholar]
- Jones-Engel L, Engel GA, Heidrich J, Chalise M, Poudel N, Viscidi R, Barry PA, Allan JS, Grant R, Kyes R. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerg Inf Dis. 2006a;12:900–906. doi: 10.3201/eid1206.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Engel L, Engel GA, Schillaci MA, Lee B, Heidrich J, Chalise M, Kyes RC. Considering human-primate transmission of measles virus through the prism of risk analysis. Am J Primatol. 2006b;68:868–879. doi: 10.1002/ajp.20294. [DOI] [PubMed] [Google Scholar]
- Jones-Engel L, Engel GA, Schillaci MA, Rompis A, Putra A, Suaryana KG, Fuentes A, Beer B, Hicks S, White R, Wilson B, Allan JS. Primate-to-human retroviral transmission in Asia. Emerg Inf Dis. 2005b;11:1028–1035. doi: 10.3201/eid1107.040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Engel L, Steinkraus KA, Murray SM, Engel GA, Grant R, Aggimarangsee N, Lee BP, May C, Schillaci MA, Somgird C, Sutthipat T, Vojtech L, Zhao J, Linial ML. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging Asian monkeys. J Virol. 2007;81:7330–7337. doi: 10.1128/JVI.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van AM, van SD, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van EJ. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microb. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard M, Schroeder C, Trask J, Paul J. A cutaneous test for tuberculosis in primates. Sci, New Series. 1939;89:442–443. doi: 10.1126/science.89.2315.442. [DOI] [PubMed] [Google Scholar]
- Lerche NW, Yee JL, Capuano SV, Flynn JL. New approaches to tuberculosis surveillance in nonhuman primates. ILAR. 2008;49:170–178. doi: 10.1093/ilar.49.2.170. [DOI] [PubMed] [Google Scholar]
- Lin PL, Rodgers M, Smith L, Bigbee M, Myers A, Bigbee C, Chiosea I, Capuano SV, Fuhrman C, Klein E, Flynn JL. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infec Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machotka S, Chapple F, Stookey J. Cerebral tuberculosis in a rhesus monkey. Journal of the Am Vet Med Assoc. 1975;167:648–650. [PubMed] [Google Scholar]
- Martin J. Tuberculosis if the spine (Pott’s disease) in a rhesus monkey (Macaca mulatta) Journal of the Am Vet Med Assoc. 1968;153:914–917. [PubMed] [Google Scholar]
- Martino M, Hubbard GB, Schlabritz-Loutsevitch N. Tuberculosis (Mycobacterium tuberculosis) in a pregnant baboon (Papio cynocephalus) J Med Primatol. 2007;36:108–112. doi: 10.1111/j.1600-0684.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- McHugh TD, Newport LE, Gillespie SH. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J Clin Microbio. 1997;35:1769–1771. doi: 10.1128/jcm.35.7.1769-1771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AL, Coetzee ML, Keet DF, Mare L, Warren R, Cooper D, Bengis RG, Kremer K, van HP. Molecular epidemiology of Mycobacterium bovis isolates from free-ranging wildlife in South African game reserves. Vet Microb. 2009;133:335–343. doi: 10.1016/j.vetmic.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Michel A, Venter L, Espie I, Coetzee M. Mycobacterium tuberculosis infections in eight species at the National Zoological Gardens of South Africa, 1991–2001. J Zoo Wild Med. 2003;34:364–370. doi: 10.1638/02-063. [DOI] [PubMed] [Google Scholar]
- Morton WR, Agy MB, Capuano SV, Grant RF. Specific pathogen-free macaques: definition, history, and current production. ILAR. 2008;49:137–144. doi: 10.1093/ilar.49.2.137. [DOI] [PubMed] [Google Scholar]
- Motzel S, Lell J, Pouch W, Ngai D, Middleton M, Nolan T, Klein H. Polymerase chain reaction as a method to aid in diagnosis of Mycobacterium tuberculosis in a colony of cynomolgus monkeys [abstract] Lab Anim Sci. 1995;45:453–454. [Google Scholar]
- Panarella M, Bimes R. A naturally occurring outbreak of tuberculosis in a group of imported Cynomolgus monkeys (Macaca fascicularis) J Am Assoc Lab Anim Sci. 2010;49:221–225. [PMC free article] [PubMed] [Google Scholar]
- Rocha V, Ikuta C, Gomes MS, Quaglia F, Matsushima ER, Neto J. Isolation of Mycobacterium tuberculosis from captive Ateles paniscus. Vec Born Zoo Dis. 2011;11:593–594. doi: 10.1089/vbz.2010.0070. [DOI] [PubMed] [Google Scholar]
- Rock F, Landi M, Meunier L, Morris T, Rolf L, Warnick C, McCreedy B, Hughes H. Diagnosis of a case of Mycobacterium tuberculosis in a cynomolgus (Macaca fascicularis) monkey colony by polymerase chain reaction and enzyme-linked immunosorbent assay. Lab Anim Sci. 1995;45:315–319. [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Cold Spring Harbor: CSHL Press; 2001. [Google Scholar]
- Sapolsky RM, Else JG. Bovine Tuberculosis in A Wild Baboon Population - Epidemiologic Aspects. J Med Primatol. 1987;16:229–235. [PubMed] [Google Scholar]
- Schillaci MA, Jones-Engel L, Engel GA, Kyes RC. Exposure to human respiratory viruses among urban performing monkeys in Indonesia. Am J Trop Med Hyg. 2006;75:716–719. [PubMed] [Google Scholar]
- Schillaci MA, Jones-Engel L, Engel GA, Paramastri Y, Iskandar E, Wilson B, Allan JS, Kyes RC, Watanabe R, Grant R. Prevalence of enzootic simian viruses among urban performance monkeys in Indonesia. Trop Med Int Heal. 2005;10:1305–1314. doi: 10.1111/j.1365-3156.2005.01524.x. [DOI] [PubMed] [Google Scholar]
- Sha JC, Gumert MD, Lee B, Fuentes A, Chan S, Jones-Engel L. Status of the long-tailed macaque (Macaca fascicularis) in Singapore and implications for management. Biodiv Conser. 2009;18:2909–2926. [Google Scholar]
- Singh P, Singla L, Gupta M, Sharma S, Sharma D. Epidemiology and chemotherapy of parasitic infections in wild omnivores in the Mahendra Choudhury zoological Park, Chhat Bir, Punjab. J Threaten Taxa. 2009;1:62–64. [Google Scholar]
- Stockinger DE, Roellich KM, vogel KW, Eiffert KL, Torrence AE, Prentice JL, Stephens KG, Wallis CK, Hotchkiss CE, Murnane RD. Primary hepatic Mycobacterium tuberculosis complex infection with terminal dissemination in a pig-tailed macaque (Macaca nemestrina) J Am Assoc Lab Animal Sci. 2011;50:258–262. [PMC free article] [PubMed] [Google Scholar]
- Tarara R, Suleman M, Sapolsky R, Wabomba M, Else J. Tuberculosis in wild olive baboons, Papio cynocephalus anubis (Lesson), in Kenya. J Wild Dis. 1985;21:137–140. doi: 10.7589/0090-3558-21.2.137. [DOI] [PubMed] [Google Scholar]
- Une Y, Mori T. Tuberculosis as a zoonosis from a veterinary perspective. Comp Immunol. 2007;30:425. doi: 10.1016/j.cimid.2007.05.002. [DOI] [PubMed] [Google Scholar]
- van Bogaert L, Innes J. Neurologic diseases of apes and monkeys. In: Innes J, Saunders LZ, editors. Comparative Neuropathology. New York: Academic Press; 1962. [Google Scholar]
- Vervenne RAW, Jones SL, van Soolingen D, van der Laan T, Peter A, Heidt PJ, Thomas AW, Langermans JAM. TB diagnosis in non-human primates: comparison of two interferon-gamma assays and the skin test for identification of Mycobacterium tuberculosis infection. Vet Immunol Immunopath. 2004;100:61–71. doi: 10.1016/j.vetimm.2004.03.003. [DOI] [PubMed] [Google Scholar]
- West C, Vainisi S, Vygantas C, Beluhan F. Intraocular granulomas associated with tuberculosis in primates. J Am Vet Med Assoc. 1981;179:1240–1244. [PubMed] [Google Scholar]
- WHO. Global tuberculosis control-- epidemiology, strategy, financing. 2009. pp. 1–28. [Google Scholar]
- Wolfe ND, Escalante AA, Karesh WB, Kilbourn A, Spielman A, Lal AA. Wild primate populations in emerging infectious disease research: the missing link? Emerg Infect Dis. 1998;4:149–158. doi: 10.3201/eid0402.980202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JS, Gormley E, Wellington EM. Molecular detection of Mycobacterium bovis and Mycobacterium bovis BCG (Pasteur) in soil. Appl Environ Microbio. 2005;71:1946–1952. doi: 10.1128/AEM.71.4.1946-1952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumpe D, Silberman MS, Michael RP. Unusual Outbreak of Tuberculosis Due to Mycobacterium-Bovis in A Closed Colony of Rhesus-Monkeys (Macaca-Mulatta) Lab Anim Sci. 1980;30:237–240. [PubMed] [Google Scholar]