Abstract
Polarity in mammalian cells emerges from the assembly of signaling molecules into extensive biochemical interaction networks. Despite their complexity, bacterial pathogens have evolved parsimonious mechanisms to hijack these systems. Here, we develop a tractable experimental and theoretical model to uncover fundamental operating principles both in mammalian cell polarity and bacterial pathogenesis. Using synthetic derivatives of the enteropathogenic E. coli guanine-nucleotide exchange factor (GEF) Map, we discover that Cdc42 GTPase signal transduction is controlled by the interaction between Map and F-actin. Mathematical modeling reveals how actin dynamics coupled to a Map-dependent positive feedback loop spontaneously polarizes Cdc42 on the plasma membrane. By rewiring the pathogenic signaling circuit to operate through β-integrin stimulation, we further show how Cdc42 is polarized in response to an extracellular spatial cue. Thus, a molecular pathway of polarity is proposed, centered on the interaction between GEFs and F-actin, which is likely to function in diverse biological systems.
INTRODUCTION
The ability of cells to spatially segregate biochemical reactions is an essential feature of all polarity circuits including those found in directional cell migration, asymmetric cell division, and immune function (Drubin and Nelson, 1996; Wedlich-Soldner and Li, 2003). Because of their importance in both single-cell and multi-cellular organisms, the mechanisms underlying cell polarity have been the subject of vigorous investigation for many years. We now recognize that cell polarity is an emergent behavior of a complex biological system. This behavior arises from extensive protein-protein and protein-lipid interaction networks which, when assembled properly, determine the location and dynamics of signal transduction cascades within the cell. Due to the inherent complexity of these systems, the essential molecular connections underlying most polarity circuits are still poorly understood. Thus, identification of simple operating principles that generate cell polarity will greatly expand our understanding of a fundamental biological problem.
Many forms of eukaryotic cell polarity require signaling through Rho family GTPases – the master regulators of the actin cytoskeleton (Jaffe and Hall, 2005). Membrane-bound Rho-proteins shuttle between GDP- and GTP-bound states, but only the GTP-bound state propagates cellular information. The cycling between activity states is tightly regulated by Guanine-nucleotide Exchange Factors (GEFs) that facilitate GTP-binding and Rho activation, and GTPase Activating Proteins (GAPs) that assist GTP hydrolysis to promote Rho deactivation. While these conserved regulatory strategies unify Rho GTPase signaling mechanisms across species, they also impose the need for additional protein- and lipid-interactions to control signaling specificity, efficacy, and location within a given cell type. Indeed, microscopy-based studies show that the guanine-nucleotide exchange cycles on Rho, Rac, and Cdc42 are controlled with sub-micron precision along the plasma membrane (Machacek et al., 2009; Nalbant et al., 2004). Due to the complex GTPase activity patterns revealed by these studies, new experimental strategies will be needed to unravel the molecular mechanisms that assemble polarity circuits in space and time.
Because of their essential nature in cell biology, Rho-family GTPases are also common targets of microbial pathogens (Aktories, 2011). Indeed, we have recently identified a large family of bacterial GEFs that potently and specifically activate Rho GTPases (Huang et al., 2009). Upon cell-to-cell contact, bacterial GEFs are injected into the host cell cytoplasm via a Type 3 Secretion System (T3SS). Once inside the cell, these GEFs rapidly polarize GTPase signal transduction along the bacterial docking interface of host cells. However, unlike mammalian Dbl-family GEFs that are regulated through extensive protein- and lipid-contacts or post-translational modifications, bacterial GEFs exhibit a compact structural architecture that severely limits their regulatory interactions (see Figure S1 for a structural comparison between eukaryotic and prokaryotic GEFs). Therefore, bacterial infection systems offer an alternative strategy to probe the molecular mechanisms of cell polarity since these evolutionarily simplified GEFs spatially amplify GTPase signaling using minimal networks connections.
In this study, we use the intimate attachment between enteropathogenic E. coli (EPEC) and host cells to demonstrate how a network of host/pathogen interactions polarize GTPase signal transduction in space and time. For this purpose we developed an exogenous, minimal model of GTPase regulation based on our current knowledge of Cdc42 GTPase activation by Map, a bacterial GEF (Alto et al., 2006; Huang et al., 2009; Kenny et al., 2002). In addition to its compact GEF domain, Map possesses a C-terminal PSD-95/Disc Large/ZO-1 (PDZ)-binding motif that interacts with the PDZ domains of Ezrin binding proteins 50 (Ebp50) (Alto et al., 2006; Berger et al., 2009; Simpson et al., 2006). Importantly, these protein interactions act as a logical “AND” gate, whereby Map requires both Cdc42 and Ebp50 interactions to regulate F-actin structure and function (see Figure 1). These observations raise the question of whether there are more complex layers of Cdc42 regulation embedded within this bacterial signaling circuit. Do emergent behaviors arise from this specific network design? If so, to what extent will these insights provide a deeper understanding of cell polarity induced by both microbial and mammalian signal transduction systems?
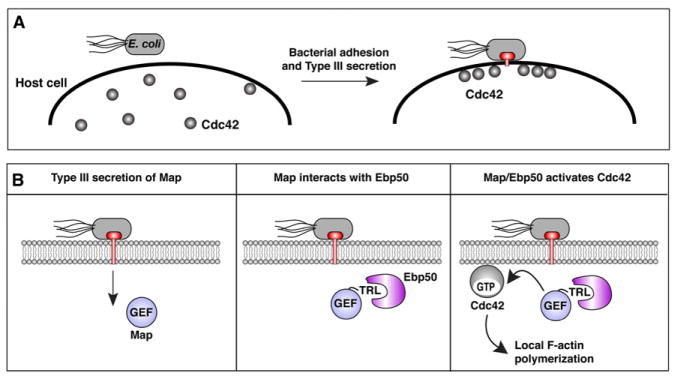
Figure 1. The molecular events that polarize Cdc42 to the bacterial docking interface of host cells.
(A and B) Diagram of EPEC induced Cdc42 polarity in host cells. EPEC adheres to the outer cell surface where it polarizes Cdc42 through Type 3 secretion dependent mechanism (A). Upon Type 3 secretion of Map, its C-terminal PDZ ligand motif (residues TRL) specifically binds the PDZ domains of Ebp50 and this complex subsequently activates Cdc42 on the membrane.
To answer these questions, we combined experimental analyses with mathematical modeling to capture the minimal essential features of the Cdc42 polarity circuit. Unexpectedly, we find that Ebp50 and its binding partner Ezrin function as a molecular scaffold to link Map to the actin cytoskeleton. This interaction network assembles a positive feedback loop that polarizes Cdc42 activity within membrane microdomains. We further show that actin polymerization locally amplifies and temporally sustains Cdc42 signaling in response to external stimulation, thus revealing the molecular and dynamic basis for GTPase polarization during E. coli infection. We now propose that bacteria hijack a fundamental circuit architecture that regulates GTPase signaling activities in a wide range of pathogenic and natural occurring cell polarity systems.
RESULTS
Establishing an experimental model of Cdc42 polarity
Illustrated in Figure 1 is the progression of molecular events that polarize Cdc42 signaling during enteropathogenic E. coli (EPEC) infection. The key feature of this system is that EPEC rapidly mobilizes Cdc42 signaling events to the cell surface through a mechanism involving Type 3 secretion of Map, a bacterial GEF (Figure 1). Importantly, Map can only activate Cdc42 when bound to the PDZ domains of Ebp50 through a poorly understood coincidence detection mechanism (Alto et al., 2006; Simpson et al., 2006). Because E. coli pathogens secrete up to 40 bacterial effector proteins during infection (Tobe et al., 2006), it has been challenging to dissect the precise role of the Map signaling complex in polarizing Cdc42 at the bacterial docking interface of host cells.
To overcome this challenge, we performed live-cell imaging on cells ectopically expressing Map protein. To our surprise, Map induced clusters of actin-rich membrane protrusions that emerged stochastically from several discrete regions on the cell surface (Figure 2A). Each cluster was composed of numerous filopodia interconnected by a network of actin lamellipodia (Figure S2). Unexpectedly, F-actin was highly dynamic within the local membrane protrusion, yet these polymerization events did not spread laterally over a 30-minute imaging time-course (Movie S1). These data indicate that Map polarizes Cdc42 in the absence of external spatial cues. Indeed, eGFP-Cdc42 was enriched in the actin-rich filopodia clusters induced by Map, whereas MapE78A, a catalytic deficient mutant that does not bind or activate GTPases (Figure 2C), did not polarize Cdc42 in cells (Figure 2A). Using the Cdc42-binding CRIB domain of N-WASP as a probe for the endogenous Cdc42 GTP-activity state (Weiner et al., 2007), we further confirmed that Map locally amplifies and temporally sustains GTPase signal transduction on the plasma membrane (Figure 2A). These stable regions of actin dynamics at the membrane were termed “Cdc42 signaling zones”.
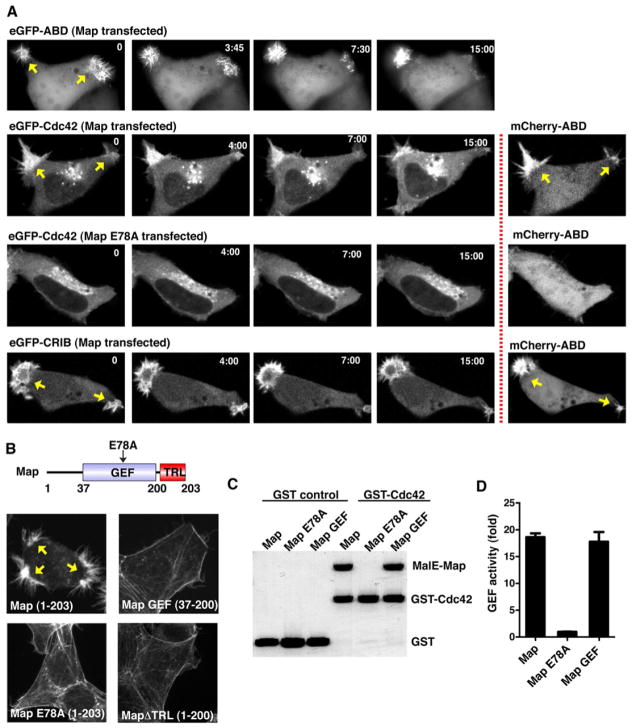
Figure 2. Map induces Cdc42 signaling zones in the absence of bacterial cues.
(A) Time-lapse fluorescence microscopy of cells co-expressing wild-type Map or the catalytically inactivated GEF mutant Map (MapE78A) with indicated fluorescent probes. F-actin dynamics (mCherry-ABD) were monitored simultaneously with eGFP-Cdc42 or eGFP-CRIBN-WASP. The arrows indicate Cdc42 signaling zones. Scale bar represents 10 μm.
(B) Fluorescence microscopy of F-actin (rhodamine-phalloidin stain) in HEK293A cells transfected with the indicated Map truncation mutants. The arrows indicate Cdc42 signaling zones. Scale bar represents 10 μm.
(C) Glutathione sepharose pulldown experiments with nucleotide free GST-Cdc42 in complex with the indicated Map proteins. N-terminal trunctions of Map proteins were tagged with MalE: Map (residues 37-203), MapE78A (residues 37-203), and GEF (residues 37-200) as indicated.
(D) Guanine nucleotide exchange reactions using GDP-loaded Cdc42 and incubating with Map constructs and GTPγS35. GEF activity is presented as the fold over un-stimulated Cdc42 nucleotide exchange rates.
We next tested if the induction of Cdc42 signaling zones by Map required its coincident interaction with both Cdc42 and Ebp50 in this model system. As predicted, neither the catalytically inactive mutant of Map (MapE78A, residues 1-203 with E78A mutation) nor a C-terminal PDZ-ligand mutant (MapΔTRL, residues 1-200) produced Cdc42 signaling zones (Figure 2B). The loss of signaling function for MapΔTRL was not due to the lack of GTPase recognition or enzymatic activity since recombinant MapΔTRL bound to the nucleotide-free Cdc42 and induced guanine-nucleotide exchange to a similar extent as wild-type Map in vitro (Figure 2C and 2D). These observations establish a robust and tractable experimental model to study the mechanism of spontaneous Cdc42 polarization in the absence of external spatial cues.
A synthetic engineering approach identifies F-actin as an essential signaling platform
Next, we took a synthetic biology approach to test the possibility that Ebp50 targets Map to an essential, yet unknown regulatory network of the host cell. Two pieces of information were critical to this approach. First, the Ebp50 scaffolding complex has been extensively mapped over the past two decades, providing a molecular guide to the essential network connections within the Map signal transduction circuit (Figure 3A) (Bretscher et al., 2000). Second, the isolated GEF domain of Map (residues 37-200) does not polarize Cdc42 activity when expressed in cells, yet is sufficient to activate Cdc42 in vitro (Figure 2C). These findings provided the motivation to restore Cdc42 signaling zones by functionally engineering the Map GEF domain with minimal network connections.
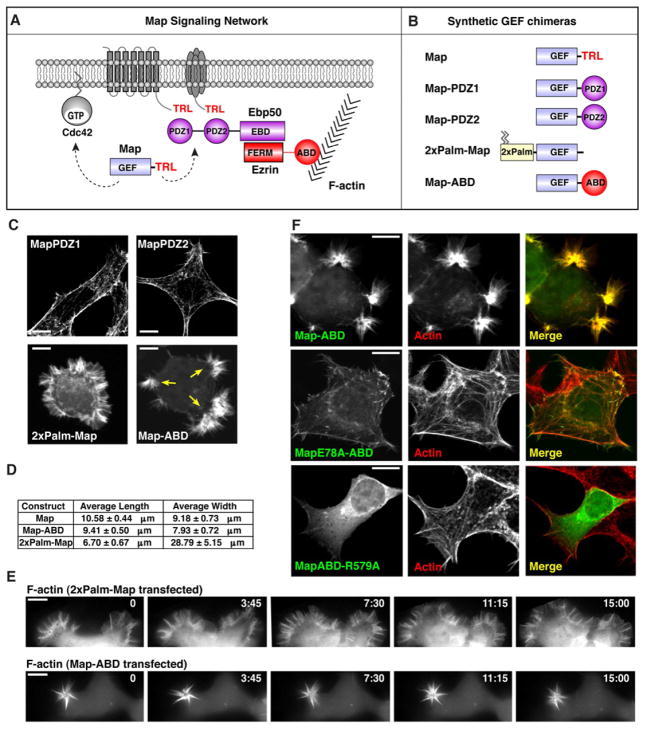
Figure 3. Modular recombination of Map GEF reveals differential Cdc42 signaling behaviors.
(A) Schematic of the Map interaction network showing the location of Cdc42, the Ebp50/Ezrin scaffold complex, and its membrane receptor/actin binding topology. T3SS: Type 3 Secretion System signal sequence; TRL: Threonine-Arginine-Lysine PDZ-ligand; PDZ: PSD-95, Discs large, ZO-1 domain; EBD: Ezrin Binding Domain; FERM: Protein 4.1, Ezrin, Radixin, Moesin; ABD: Actin binding domain.
(B) Cartoon of the synthetic GEF chimeras used for functional studies.
(C) Fluorescence microscopy of F-actin (rhodamine-phalloidin) in HEK293A cells transfected with the indicated synthetic GEF chimeras. Scale bar represents 10 μm.
(D) Geometric measurements of the F-actin phenotypes induced by Map compared to 2xPalmMap or MapABD as indicated.
(E) Time-lapse fluorescence microscopy of actin dynamics (eGFP-ABD) in HEK293A cells expressing wild-type 2xPalmMap or MapABD. Scale bar represents 10 μm.
(F) Fluorescence microscopy of cells transfected with eGFP-MapABD (top), GEF inactive mutant (middle) and the actin-binding mutant (bottom). Cells were stained with rhodamine-phalloidin (red) to observe co-localization of synthetic Map proteins with F-actin. Scale bar represents 10 μm.
Guided by the PDZ-domain interactions between Ebp50 and integral membrane proteins, we individually fused each PDZ-domain of Ebp50 to the GEF domain of Map (MapPDZ1 and MapPDZ2). These protein chimeras short-circuited the potential interaction between Map and plasma membrane channels or receptors (Figure 3B). Unexpectedly, neither MapPDZ1 nor MapPDZ2 induced Cdc42 signaling zones suggesting that Ebp50 does not simply target Map to a trans-membrane receptor complex (Figure 3C). To test whether direct plasma membrane association restored GEF signaling in vivo, the dual palmitoylated sequence of Neuromodulin was fused to the N-terminus of Map (2xPalmMap) (Figure S3). 2xPalmMap induced new actin ‘microspike’ structures that projected laterally over large segments of the plasma membrane and, occasionally, fully encompassed the cell surface (Figure 3C and 3D). Time-lapse microscopy revealed that 2xPalmMap induced a cell spreading phenotype characterized by lamellipodia membrane extensions interlaced with short F-actin microspikes (Figure 3E and Movie S2). Surprisingly however, this gain-of-function phenotype had no resemblance morphologically, quantitatively, or dynamically to the localized filopodia induced by wild-type Map (Figures 3D, 3E, and S3).
Concluding that Ebp50 does not localize Map to the plasma membrane, we next investigated a second key property of the scaffolding complex: F-actin binding (Figure 3A). The 30-residue actin-binding domain (ABD) of Ezrin (Turunen et al., 1994) was fused to the C-terminus of Map (MapABD), thereby short-circuiting the Ebp50/Ezrin connection to the actin cytoskeleton (Figure 3B). Ectopic expression of MapABD induced clusters of actin-rich filopodia that projected from several discrete regions of the cell surface (Figure 3C). This actin phenotype had nearly identical geometric boundaries as those observed in Map-expressing cells (Figure 3D). Furthermore, actin filopodia were stably maintained within local regions of the plasma membrane over time, a behavior that recapitulated the Cdc42 signaling zones established by wild-type Map (Figure 3E and Movie S2).
The unexpected finding that actin filaments function as a GTPase signaling platform is further supported by the following observations: first, eGFP-tagged MapABD perfectly co-localized with actin-rich filopodia in transfected cells (Figure 3F). Second, point mutations in either the GEF catalytic domain (Map E78A) or in the Ezrin actin-binding-domain (Ezrin ABD-R579A) (Saleh et al., 2009) inhibited MapABD from inducing F-actin polymerization (Figure 3F). We therefore conclude that the Ebp50/Ezrin scaffolding complex acts as a molecular bridge to indirectly link Map to the actin cytoskeleton.
Map signals from the tips of actin filaments
Many mammalian GEFs have been reported to associate with the actin cytoskeleton, yet the functional consequences of these interactions are poorly understood (Figure S4) (Banerjee et al., 2009; Bellanger et al., 2000; Hou et al., 2003). It was therefore of broad significance to explore the functional relationship between F-actin and GTPase signaling in the context of the synthetically engineered MapABD protein.
A structural model revealed that actin filaments must approach the cell surface to within ~60 Å to form a Map/Cdc42 activation complex on the membrane (Figure 4A). This spatial requirement places strict physical limitations on the Cdc42 activation pathway, as actin-bound Map must be associated with the tips of actin filaments to transduce a signal (Figure 4A, right). To verify this structural model, we examined the actin filament binding properties of MapABD in the absence of its GEF activity (this allows a direct assessment of Map binding to naturally occurring cytoskeleton structures). As predicted, MapABD (E78A) was highly enriched at the tips of actin-microspikes and was conspicuously less abundant on sub-cortical actin stress-fibers (Figure 4B). Moreover, previous studies have shown that the Ezrin-Moesin-Radixin (ERM) family members interact with the barbed-end of actin filaments, a localization that is mediated by the C-terminal ABD (Algrain et al., 1993). These data indicate that Map activates Cdc42 from the tips of actin filaments.
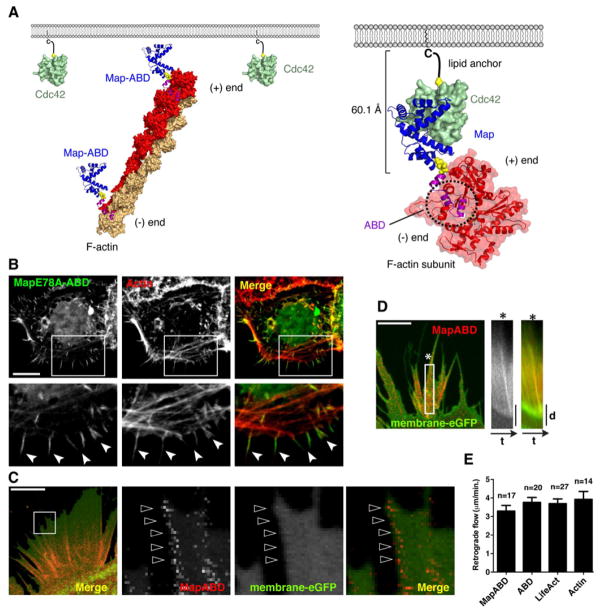
Figure 4. Map signals from the actin cytoskeleton.
(A) Structural organization of the Map signaling network. Cdc42 (green) is localized to the membrane through palmitoylation on the C-terminal CaaX box. MapABD (Map in blue and ABD in fuchsia) is tethered to a single F-actin subunit (red) extracted from an actin filament oriented with the barbed end toward the plasma membrane. Yellow spheres indicate linker regions whose structures are not solved. Known structures of Map/Cdc42 (PDB: 3GCG), Moesin ABD (PDB: 1EF1) and F-actin (PDB: 3MFP) were used in the model and the interaction between Moesin ABD and F-actin is a hypothetical orientation.
(B) Fluorescence microscopy of a cell transfected with eGFP-MapABD (E78A) and stained with rhodamine-phalloidin (red) to observe the actin cytoskeleton. The boxed region of each panel is magnified (2x) below. The synthetic protein preferentially binds to the tips of actin filaments (arrows) compared to the sub-cortical actin structures. Scale bar represents 10 μm.
(C) Fluorescence microscopy of cells co-expressing low levels of mCherry MapABD and membrane-targeted eGFP as a reference. Magnified (5x) view of the boxed region depicts mCherry MapABD speckles generated near the cell surface and align along actin cables. Scale bar represents 10 μm.
(D) Time-lapse microscopy of the cell reveals a wave of MapABD moving away from the cell by actin retrograde flow. The first kymograph depicts mCherry MapABD moving retrograde while the second kymograph is the merge with eGFP-membrane probe. The star is placed to orient the kymograph and the still framed image. See Movie S3. Scale bar represents 10 μm.
(E) Quantification of retrograde flow of mCherry tagged constructs indicated. Data was extracted from multiple time-lapse microscopy images using kymograph analysis in ImageJ. Data are presented as mean ±SEM.
The subcellular location of Map depends on actin polymer dynamics
Given the dynamic nature of actin-based membrane protrusions, it is likely that actin turnover (polymerization and depolymerization) regulates the location of Map relative to membrane-bound Cdc42. Filopodia-based membrane protrusions are constructed from a highly dynamic polymer network of both bundled and branched actin filaments (Svitkina et al., 2003). To directly visualize Map dynamics at these sites, low levels of mCherry-tagged MapABD were co-expressed with membrane-targeted eGFP as a positional reference. mCherry-MapABD formed fluorescent speckles that aligned along actin filaments (Figure 4C). Time-lapse microscopy revealed that MapABD speckles originated within membrane extensions and moved rapidly toward the cell interior (Figure 4D and Movies S3). This direction and rate of movement of MapABD was similar to retrograde flow of microinjected rhodamine-labeled actin and transiently expressed protein markers of F-actin dynamics. (Figure 4D and 4E) (Riedl et al., 2008; Theriot et al., 1992; Watanabe and Mitchison, 2002). Although we were unable to discriminate fluorescent speckles of wild-type Map (likely due to the low abundance of the Map/Ebp50/Ezrin trimeric-complex), its analogy with MapABD suggests that Map signal transduction is also controlled by actin-filament dynamics.
Mathematically modeling the Map signaling system reveals an actin-based positive feedback loop
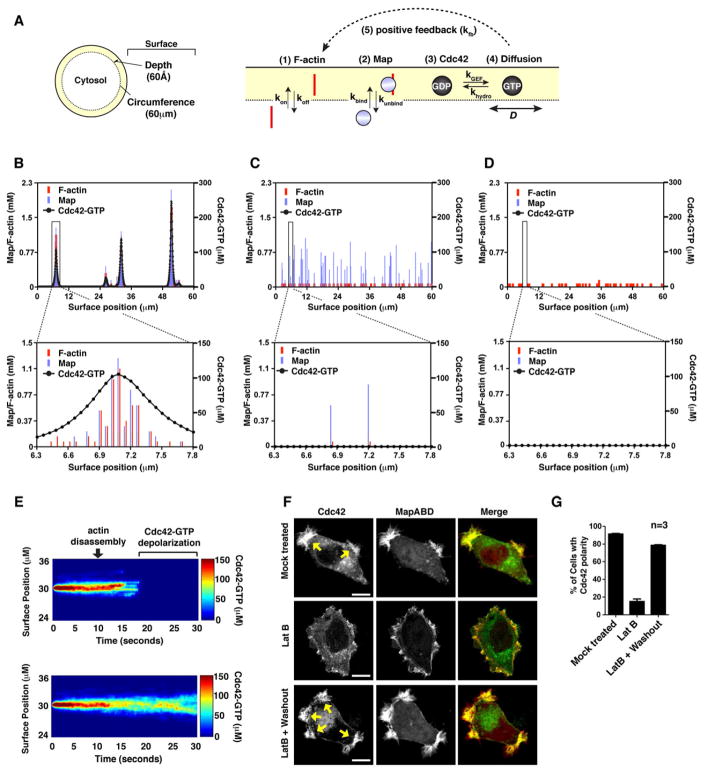
In summary, our data reveals three critical aspects of the bacterial signaling system: First, the binding interaction between Map and the actin cytoskeleton is necessary to polarize Cdc42 on the membrane; second, actin dynamics control the location of Map relative to Cdc42; and third, these molecular interactions induce spontaneous cell polarity in the absence of spatial cues. To determine if these findings can be integrated into a theoretical framework of cell polarity, we developed a mathematical model that describes the minimal set of interactions in a virtual cell (see extended experimental procedures for a detailed description of the model assumptions, parameters, and variables).
In theory, our model is based on the principle that spontaneous Cdc42 polarity results from the stochastic fluctuations of Map and F-actin between the cytosol and a membrane-proximal “surface compartment” (Figure 5A). We propose that the probability of Cdc42 activation is dependent on the coincidence of two events occurring independently: first, an actin filament must transition from the cytosol to the surface compartment (Figure 5A, point 1) and second, a Map molecule must bind near the tip of this actin filament (Figure 5A, point 2). Once recruited to the membrane, Map converts GDP-inactive Cdc42 to its GTP-active state (Figure 5A, point 3). Active Cdc42 diffuses laterally along the cell surface (Figure 5A, point 4), which recruits new actin filaments to adjacent membrane sites (for example by stimulating the N-WASP-Arp2/3 complex) (Miki et al., 1998). Together, this progression of molecular events initiates a positive feedback loop by increasing the actin tip density along the membrane, further recruiting new Map molecules to membrane-bound Cdc42 (Figure 5A, point 5).
Figure 5. Mathematical model of the Map signaling circuit.
(A) Schematic of the virtual cell (60μm circumferance) partitioned into cytoplasm and a surface compartment (6nm depth). The model parameters are shown as: (1) kon, koff the rate of actin filament association and dissociation from the surface compartment; (2) kbind, kunbind the rate of Map association and dissociation from F-actin; (3) kGEF, khydro the rate of Guanine-nucleotide exchange and GTP hydrolysis; and (4) D the rate of Cdc42-GTP diffusion on the membrane; (5) kfb the rate of positive feedback induced by Cdc42-GTP recruiting new actin filaments to the surface compartment.
(B) Single cell simulation showing the concentrations of F-actin (red bars), Map (blue bars), and Cdc42-GTP (dotted line) per 60nm increments of the cell surface compartment (X-axis). Cdc42-GTP concentrations are plotted as a line graph to clearly resolve the signaling zones from Map and F-actin concentrations. The numerical value bars for Map were manually offset from F-actin by 18nm for visual purposes. The boxed region corresponds to the graph below.
(C–D) Single cell simulation in which the parameter kGEF is set to 0 (C), or kbind is set to 0 (D). Data is plotted as in (B).
(E) Kymographs of Cdc42-GTP concentration (color bar) along the cell surface (y-axis) over time (x-axis). Upper panel: computational simulation in which F-actin is disassembled by setting kfb=0 and kon=0 at time 10 seconds (arrow). Lower panel: computational simulation with no actin perturbation (control). Color bar is indicated at right.
(F) Fluorescence microscopy showing eGFP-Cdc42 polarity in cells expressing mCherry-MapABD. Cells were either treated with DMSO (mock treated, upper panel) or treated with 50 nM Lat B for 30 minutes (LatB, middle panel). After 30 minutes, the LatB was washed out and cells were allowed to recover for 10 hours (LatB + Washout, lower panel). Scale bar represents 10μm.
(G) Quantification of the number of Cdc42 signaling zones in the population of cells shown in Figure 5F.
We first considered the scenario where Map directly interacts with F-actin (Figure 5A). Literature values were used to estimate the rates of actin filament dynamics near the membrane (kon and koff), the affinity of interaction between Map and F-actin (kbind and koff), and the regulatory cycle of Cdc42 (kGEF, kGAP, D) (Table S1). Furthermore, experimental data was used to calibrate the positive feedback term (kfb) (Figure S5A). Computational simulations resulted in the spontaneous polarization of Cdc42-GTP and the accumulation of new actin filaments within discrete regions of the plasma membrane (Figure 5B). Cdc42-GTP signaling zones occupied 9.28 ± 0.74% of the total surface area in silico, a value that closely matched the measured width of Cdc42 signaling zones in Map expressing cells (12.1 ± 0.83%) (Figure S5B). In addition, the model gave rise to temporally stable Cdc42 guanine-nucleotide exchange cycles on the plasma membrane as is observed in vivo (Figure S5C and Figure 2A). These data indicate that the stochastic assembly of a Cdc42/Map/F-actin complex is required to establish polarity within discreet membrane zones. Consistent with this interpretation, Map was unable to polarize Cdc42-GTP in the absence of its GEF activity (Figure 5C) or when decoupled from the actin cytoskeleton (Figure 5D). Thus, our stochastic model of polarity agrees with the structural, mutational, and cellular analysis presented in Figures 2, 3, and 4.
A scan of model parameter values revealed a direct relationship between the rate of actin filament tip accumulation along the plasma membrane (parameter kon) and the strength of the actin-based positive feedback loop (parameter kfb) in determining the number and width of Cdc42 signaling zones (Figure S5D). We also found that Cdc42 is rapidly depolarized when actin cytoskeleton dynamics are computationally disrupted at a discrete point in time (Figure 5E). To test this model prediction experimentally, Cdc42 localization was monitored in the presence of low concentrations of Latrunculin B (LatB, 50nM), an actin-monomer binding drug that potently inhibits actin filament nucleation. Addition of LatB caused the rapid depolarization of Cdc42 in cells expressing MapABD (Figure 5F and 5G). Cdc42 polarity was re-established upon drug removal, providing direct evidence that actin polymerization locally amplifies and temporally sustains Cdc42 polarity in response to an actin-bound GEF (Figure 5F and 5G).
Reconstitution of Cdc42 polarity in response to external spatial cues
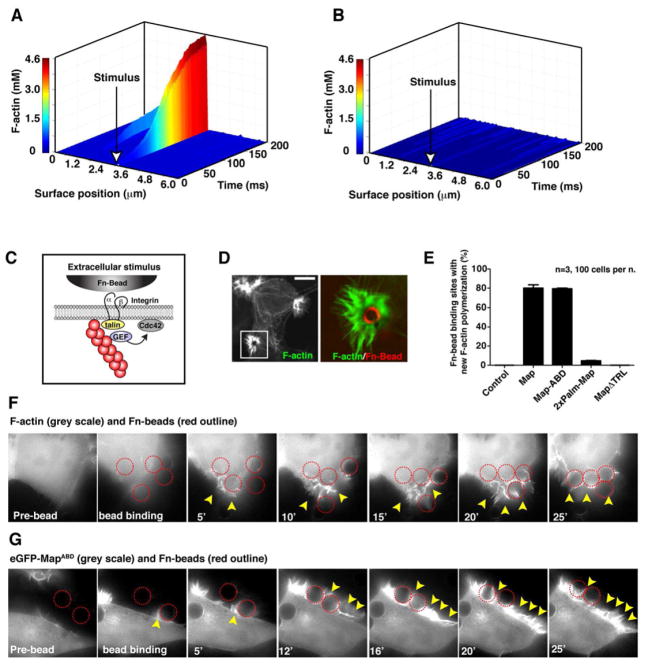
It is important to note that Cdc42 is not polarized randomly during E. coli infection, but is precisely recruited to the bacterial docking interface of host cells. How then can our model of stochastic cell polarity described above be reconciled with the deterministic behavior observed during bacterial infection? Our mathematical model provided an essential platform to uncover the molecular nature of these events. Because local Cdc42 activation is initiated by the spontaneous interaction between F-actin and the membrane, it is logical to assume that an external signal that stabilizes F-actin on the membrane would polarize Cdc42 activity at this site. Indeed, nucleating a small number of actin filaments at the membrane prior to running computational simulations resulted Cdc42 activation and a local peak of F-actin accumulation (Figure 6A). Both the actin-based positive feedback loop (Figure 6B) and Map binding to F-actin (data not shown) was essential to polarize Cdc42. These data suggest that Cdc42 polarization can be triggered by local outside-in stimulation of actin polymerization.
Figure 6. Reconstitution of cue-dependent polarity in the Map signaling system.
(A–B) Computational simulation of a single cell in which an individual membrane compartment was seeded with F-actin attachments prior to running the simulation with (left) or without (right) positive feedback (kfb) in the system. The simulation results are plotted as a 3-D graph showing the surface position (x-axis) and the concentration of F-actin filaments (y-axis) over time (z-axis). The site of seeded F-actin attachments is shown with a white arrow.
(C) Cartoon depicting the β-integrin signaling connection between Fn-bead binding to the outer cell surface and actin filament attachments to this membrane site.
(D). Fluorescent micrograph of actin-rich filopodia clusters induced by Fn-bead binding to a MapABD expressing cell. F-actin is visualized (left) and the boxed region is magnified to illustrate the filopodia protrusions (green) around the Fn-bead (pseudo-colored red).
(E) Quantification of the number of Fn-beads that induced the F-actin phenotype shown in the presence of cells expressing the indicated synthetic Map construct. Data are presented as mean ±SEM.
(F) Time-lapse microscopy of HEK293A cells engaging Fn-beads (outlined in red). These cells are co-expressing Map with eGFP-ABD as a visual marker for actin polymerization dynamics in response to Fn-bead binding. Arrowheads indicate new sites of F-actin polymerization.
(G). Time-lapse microscopy eGFP-MapABD showing GEF recruitment to the sites of Fn-bead engagement (outlined in red) and its subsequent localization within newly formed membrane protrusions (arrowheads).
To experimentally test this computational prediction, fibronectin-coated beads (Fn-beads) were used to initiate F-actin nucleation at discrete locations on the plasma membrane (Figure 6C). As shown previously, Fn-beads induce clustering of β-integrins and subsequent actin filament attachment to these membrane sites (Figure S6A) (Miyamoto et al., 1995). Remarkably, engagement of Fn-beads to cells ectopically expressing Map induced bursts of actin polymerization that were tightly localized to the sites of surface stimulation (Figure 6D). New actin filopodia were generated at 79 ± 0.7% of Fn-bead binding sites in Map expressing cells (Figure 6E) and these sites were enriched in Cdc42 activity (Figure S6B and S6C). Decoupling Map GEF activity from the Ebp50/Ezrin complex using the MapΔTRL mutant (residues 1-200) failed to induce actin polymerization and Cdc42 accumulation, suggesting that Map/actin attachment is an essential feature of the polarity circuit (Figure 6E and S6A). Consistent with this notion, over 80 ± 3.4% of MapABD expressing cells induced local sites of actin polymerization whereas membrane-targeted 2xPalmMap was non-responsive to Fn-Bead stimulation (Figure 6E and S6A). Finally, time-lapse microscopy was used to observe the timing and propagation of actin polymerization in response to outside-in stimulation (Figure 6F). Most importantly, eGFP-tagged MapABD was recruited to Fn-bead binding site just prior to inducing bursts of F-actin polymerization (Figure 6G, S6, and Movie S4). Taken together, these data confirm that a series of stochastic interactions between F-actin, Map, and membrane-bound Cdc42 can generate signal polarity in response to an external spatial cue. They also suggest a concerted mechanism for the excitation of GTPase signal transduction initiated through bacterial infection.
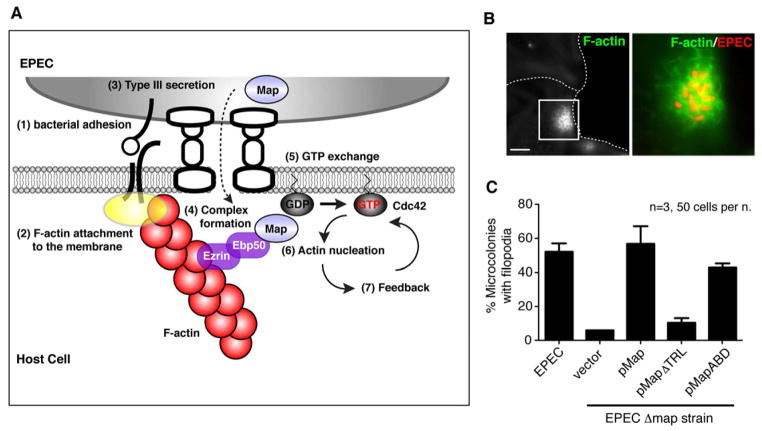
The actin-based positive feedback loop is essential for Cdc42 polarity during EPEC infection
Given that the Map signaling system is responsive to outside-in signaling cues, it is intriguing to propose that E. coli induces an intracellular “landmark” by first creating a small, local perturbation in actin polymerization (Figure 7A, points 1–2). Concomitantly, Type 3 secreted Map protein would monitor the internal cellular state by directly interacting with the Ebp50/Ezrin complex (Figure 7A, points 3). This host/pathogen interaction specifically recognizes the actin landmark established by bacterial adhesion (Figure 7A, points 4). Together, these initiating events trigger an actin-based positive feedback loop, leading to initial Cdc42 polarization and subsequent burst of actin polymerization at the site of bacterial infection (Figure 7A, point 5–7). In agreement with this molecular scheme, Type 3 secretion of Map induced spatially localized actin filopodia at the EPEC infection site of host cells (Figure 7B and 7C) (Alto et al., 2006; Kenny et al., 2002). This spatial regulation requires the Ebp50/Ezrin complex since Type 3 secretion of a MapΔTRL mutant displayed reduced levels of cellular F-actin dynamics (Figure 7C) (Alto et al., 2006; Simpson et al., 2006). Most importantly, complementation of the EPECΔmap strain with a plasmid encoded MapABD chimera rescued local actin filopodia dynamics, indicating that direct attachment of Map to F-actin polarizes Cdc42 to a discrete subcellular location (Figure 7C). Thus, the actin-based positive feedback circuit is required to locally amplify and temporally sustain Cdc42 activity at the bacterial docking interface of host cells.
Figure 7. Validation of the Map signaling circuit during bacterial infection.
(A) Model of the EPEC induced Cdc42 polarity circuit. EPEC establishes an extracellular landmark by initiating a small outside-in signaling event that generates actin attachments to the membrane (points 1–2). This site is recognized by Type 3 secreted Map protein through the Ebp50-Ezrin-actin complex (points 3–4). Once this signal is initiated, the bacterial GEF controls GTPase activity patterns on the cell surface by engineering an actin-based feedback loop that precisely tunes the location and dynamics of the host cellular response (points 5–7).
(B–C) Representative example of EPEC infected HeLa cells showing F-actin cytoskeleton (B). Scale bar represents 10 μm. Quantification of localized filopodia in HeLa cells infected with EPEC or EPECΔmap strain (C). EPECΔmap carrying the indicated plasmids for complementation are shown. At least 50 EPEC infection sites were scored for the formation of filopodia in three independent experiments. Data are presented as mean ±SEM.
DISCUSSION
By asking the simple question: how does an extracellular bacterial pathogen regulate intracellular host actin dynamics, we have uncovered a fundamentally new molecular circuit involved in mammalian cell polarity and bacterial infection. These findings have far-reaching implications on the regulatory mechanisms that control both pathogenic and natural eukaryotic cell behavior.
Bacterial pathogens assemble signaling circuits from host cell machinery
Our data establish the molecular circuitry that transmits spatial information from extracellular EPEC to the intracellular signaling environment of the host cell. EPEC has evolved Map to interact with the actin-cytoskeleton through the Ebp50/Ezrin scaffolding complex. In the context of bacterial infection, this interaction network functions as a molecular “homing device,” allowing EPEC to first mark its position on the extracellular surface via initial actin polymerization and then use the Type 3 secreted effector Map to home in on this intracellular positional landmark (Figure 7a). Once the bacterial position is recognized, Map assembles an actin-based positive feedback loop that spatially amplifies Cdc42 signaling on the membrane. This conclusion is strongly supported by the Fn-bead binding studies (Figure 6) that recapitulate EPEC infection in an intact, bacterial-free, cellular system. We have previously shown that Map belongs to an extended family of structurally and functionally related bacterial GEF proteins that are required for Shigella, Salmonella, and Burkholderia invasion (Alto et al., 2006; Buchwald et al., 2002; Huang et al., 2009; Upadhyay et al., 2008). Like E. coli Map, these GEFs polarize GTPase signaling at the sites of bacterial infection. It is therefore likely that most bacterial GEFs possess targeting sequences that directly or indirectly interact with F-actin or assemble new host polarity circuits that are currently unknown. In a larger context, the ability of bacteria to engineer signaling circuits from the host cellular machinery provides a mechanism for pathogens to gain “systems level” control over complex host cellular behaviors.
Actin dynamics shape the timing and location of GTPase activity on the membrane
The experimental and theoretical analysis presented here indicates that actin filament dynamics controls the location and magnitude of Cdc42 activity on the plasma membrane. In the circuit described here, actin filament association with the membrane initiates symmetry breaking of Cdc42 by positioning Map in a location competent for GTPase activation. Once this signaling system has been initiated, actin filament nucleation and branching controls the magnitude of Cdc42 activity by recruiting Map molecules to the tips of actin filaments. Consistent with this model, the actin-depolymerizing agent Latrunculin B rapidly depolarized Cdc42 in cells, indicating that the assembly of actin filaments amplifies GTPase activity on the plasma membrane. These findings are further supported by the observation that Map is recruited to the site of β-integrin stimulation just prior to the excitation of actin polymerization at these sites (see Figure 6). Taken together, these data reveal a previously unrecognized network design that converts actin filament nucleation into GTPase signal amplifier that responds locally and robustly to extracellular spatial cues.
It is notable that Map activates membrane-bound Cdc42 while associated with the tips of actin filaments yet paradoxically, moves away from the plasma membrane at a rate similar to actin retrograde flow (Figure 4D). It is currently unknown how actin subunit treadmilling may influence the interaction between Map and Cdc42 but it is logical to assume that it dampens the signaling system by displacing Map from the membrane. For example, a membrane/N-WASP/F-actin complex (Co et al., 2007) would stabilize actin-bound Map molecules near the cell surface to activate Cdc42. Release of this complex and subsequent actin retrograde flow would cause the displacement of Map away from Cdc42, thus equilibrating the system. We also suspect that additional actin binding proteins such as capping proteins or membrane tethering factors (Pollard and Cooper, 2009) will substantially influence GTPase activity in response to actin-bound GEF. It is therefore likely that the relationship between GTPase activation and F-actin dynamics may be more complex than we have so far described. Nevertheless, our study provides a theoretical and experimental platform to further dissect the various processes and molecular mechanisms that connect actin cytoskeleton dynamics to the polarization GTPase signal transduction cascades in space and time.
Using bacterial GEFs as a model of eukaryotic GTPase regulation
Beyond the relatively simple bacterial infection system investigated here, it is intriguing to speculate on how the infection paradigm relates to signaling in higher eukaryotic systems (e.g. cell migration, cell division, and immune function). In those systems, GTPase polarity is precisely controlled through extensive protein-protein and protein-lipid interaction networks. However, they all share a common need for the intrinsically asymmetric distribution of actin polymers and the organization of the cytoskeleton into higher-order structures. It is attractive to hypothesize that the actin-based signaling circuit hijacked by EPEC will also be found in natural Rho GTPase signaling pathways. Both a literature survey and bioinformatic analyses indicates that mammalian Dbl-family GEFs have domains capable of associating directly or indirectly with the actin cytoskeleton (Figure S4). In addition, F-actin has been implicated in the positive feedback regulation of GTPase signaling at the leading edge of chemotactic cells (Xu et al., 2003). Other studies have identified Rac1 specific GEFs that co-localizes with F-actin in the establishment of cell polarity (Park et al., 2004). Despite the close relationship between actin architecture and GTPase activity, the role of actin filament dynamics in the feedback regulation of GTPase signal transduction is still poorly understood. Because bacterial pathogens are unlikely to invent completely new operating principles, we propose that E. coli has usurped a conserved circuit topology used to establish direct communication link between the force generating structures of F-actin and the signal transduction systems that control cell polarity.
The central role of actin dynamics in cell polarity circuits
Most models of cell polarity emphasize the upstream signaling pathways that control down stream F-actin architectures. Conversely, we now propose a fundamentally different view of cell polarity that emphasizes actin filaments as the organizational center of spatially and quantitatively regulated signal transductions pathways. In fact, our findings add significantly to a small, but growing body of literature indicating that F-actin dynamics are the central hub in physiologically relevant signaling processes. For example, Weiner et al. recently reported that waves of actin polymerization control the location and activity of the Scar/WAVE signaling complex at the leading edge of migrating neutrophils (Weiner et al., 2007). Likewise, it has been proposed that myosin light chain kinase (MLCK) is transported retrograde with actin filaments, spatially regulating the assembly of focal contacts during directional cell migration (Giannone et al., 2004). It therefore appears that actin-based circuits are not limited to GTPase polarity as described in our study, but is found in a diverse array of signaling systems. Together, these data extend the known functions of the actin cytoskeleton such as force generation, vesicle trafficking, adhesion, and membrane protein dynamics to include the spatial and temporal regulation of signaling transduction. Thus, elucidating the molecular relationships between actin cytoskeleton dynamics and enzyme regulation promises to be a rewarding area of research in many complex biological systems.
MATERIALS AND METHODS
Plasmids and Bacterial-Eukaryotic Chimeras
For C-terminal GEF chimeras, Map residues 1-200 was cloned into pEGFP-C1 without a stop codon to allow in frame fusion to the downstream gene fragments including Ebp50 PDZ1 (amino acids 10-110; accession number O14745), Ebp50 PDZ2 (amino acids 129-229), and the actin binding domain (ABD) of Ezrin (amino acids 541-586; accession number NM_001111077). 2xPalmMap was generated by PCR cloning the dual palmitoylation sequence of Neuromodulin (amino acids 1-20; accession number NP_002036) upstream of eGFP-MapΔTRL in a modified pcDNA 3.1 vector. mCherry tagged proteins were generated by subcloning constructs into mCherry tagged pcDNA 3.1. Cdc42 and the CRIB Domain of N-WASP (amino acids 180-267) were cloned into a modified pCDNA3.1 EGFP vector. For protein expression constructs, N-terminal truncations of the Map protein were required to generate soluble protein. Therefore, Map (37-203) and MapGEF (37-200) were cloned into a 6xHis-Maltose Binding Protein (MalE) fusion vector with a pET28 backbone. Site-directed mutagenesis was carried out using the QuickChange Site-Directed Mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing.
Cell Culture and Microscopy
HEK293A and HeLa cells were maintained in DMEM containing 10% (v/v) FBS, 2mM glutamine, and 100 μg/ml penicillin/streptomycin (Thermo Scientific) at 37°C in a 5% CO2 incubator. Cells were seeded onto coverslips in a 6 well dish and after overnight incubation were transfected using FuGene6 (Roche) and incubated for 16–18 hours. Cells were then fixed and prepared for immunocytochemistry. Fixed cell imaging was performed on a LSM 510 PASCAL scanning confocal microscope (Zeiss, Thornwood, NY). Live cell imaging was performed on an Applied Precision (Seattle, WA) Deltavision RT deconvolution microscope. For fluorescent speckle microscopy, low expressing cells were imaged every 5s on a LSM 510 META scanning confocal microscope (Zeiss, Thornwood, NY). Data was analyzed and quantified using the kymograph plugin for Image J. This plugin captures a narrow region from individual frames of a time-series and stacks them into a single image. Stationary objects appear as a line parallel to the time axis. Object movement is observed as a diagonal streak with the slope being proportional to the velocity. The velocity of retrograde flow was calculated from the distance (μm) over time (seconds) of speckle movement over successive frames.
Mathematical modeling
A detailed description of the mathematical model can be found in the Supplementary Information. MatLab code is available upon request.
Fibronectin Bead assays
5 μm polystyrene divinyl-benzene beads (Duke Scientific Coproration, Palo Alto, CA) were diluted in PBS to 2*1010 beads/mL and incubated with fibronectin (20μg/mL) at 4°C overnight with gentle rocking. Beads were washed once with 5mLs of PBS and resuspended in 1mL of PBS by gentle sonication. 10 μL of the bead slurry were incubated with cells for 20 minutes and subsequently washed with PBS, fixed, and prepared for immunocytochemistry. For live cell imaging, beads were added to cells and immediately monitored using time-lapse microscopy on an Applied Precision (Seattle, WA) Deltavision RT deconvolution microscope.
Protein Purification and GEF Assays
6xHis-MBP-tagged Map or mutant Map protein purification, GST-Cdc42 glutathione pulldown assays, and guanine-nucleotide exchange assays were performed as previously described (Huang et al., 2009).
EPEC infection
EPECΔmap strain (Kenny et al., 2002) was complemented with the plasmid pBBRMCS1 encoding wild-type map gene, the map gene missing the PDZ ligand (MapΔTRL, amino acids 1-200), or a chimeric fusion between Map amino acids 1-200 fused to human ezrin residues 541-586 (MapABD). HeLa cells were infected for 20 minutes with pre-activated EPEC as described previously (Kenny et al., 2002). Infected cells were fixed and processed for immunofluorescence as described above.
Supplementary Material
Acknowledgments
We would like to thank our colleagues, specifically, K. Orth, M. Rosen, and members of the Alto lab for helpful discussions in preparation of this manuscript. This work was supported NIH grants AI083359 (N.M.A), AI007520 (R.C.O), GM T32008297/5 T32 08014 (M.K) GM081549 (L.F.W.), GM071794 (S.J.A.), GM088428 (G.M.S) and the Welch Foundation I-1704 (N.M.A), I-1644 (L.F.W.), I-1619 (S.J.A.), I-1675 (G.M.S), the Perot Foundation (M.K.) and the James S. McDonnell Foundation 220020141 (G.M.S). We are also grateful for the assistance of the UT Southwestern Live Cell Imaging Facility, a shared resource of the Harold C. Simmons Cancer Center, supported in part by an NCI Cancer Center Support Grant, 1P30 CA142543-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nature reviews Microbiology. 2011;9:487–498. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. The Journal of cell biology. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Banerjee J, Fischer CC, Wedegaertner PB. The amino acid motif L/IIxxFE defines a novel actin-binding sequence in PDZ-RhoGEF. Biochemistry. 2009;48:8032–8043. doi: 10.1021/bi9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nature cell biology. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- Berger CN, Crepin VF, Jepson MA, Arbeloa A, Frankel G. The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cellular microbiology. 2009;11:309–322. doi: 10.1111/j.1462-5822.2008.01254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annual review of cell and developmental biology. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. The EMBO journal. 2002;21:3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- Hou P, Estrada L, Kinley AW, Parsons JT, Vojtek AB, Gorski JL. Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Human molecular genetics. 2003;12:1981–1993. doi: 10.1093/hmg/ddg209. [DOI] [PubMed] [Google Scholar]
- Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nature structural & molecular biology. 2009;16:853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annual review of cell and developmental biology. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kenny B, Ellis S, Leard AD, Warawa J, Mellor H, Jepson MA. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Molecular microbiology. 2002;44:1095–1107. doi: 10.1046/j.1365-2958.2002.02952.x. [DOI] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Akiyama SK, Yamada KM. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science (New York, NY. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science (New York, NY. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- Park KC, Rivero F, Meili R, Lee S, Apone F, Firtel RA. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. The EMBO journal. 2004;23:4177–4189. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science (New York, NY. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh HS, Merkel U, Geissler KJ, Sperka T, Sechi A, Breithaupt C, Morrison H. Properties of an ezrin mutant defective in F-actin binding. Journal of molecular biology. 2009;385:1015–1031. doi: 10.1016/j.jmb.2008.11.051. [DOI] [PubMed] [Google Scholar]
- Simpson N, Shaw R, Crepin VF, Mundy R, FitzGerald AJ, Cummings N, Straatman-Iwanowska A, Connerton I, Knutton S, Frankel G. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Molecular microbiology. 2006;60:349–363. doi: 10.1111/j.1365-2958.2006.05109.x. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. The Journal of cell biology. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Matthews S, Marches O, Frankel G, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen O, Wahlstrom T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. The Journal of cell biology. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A, Wu HL, Williams C, Field T, Galyov EE, van den Elsen JM, Bagby S. The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42. The Biochemical journal. 2008;411:485–493. doi: 10.1042/BJ20071546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science (New York, NY. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Li R. Spontaneous cell polarization: undermining determinism. Nature cell biology. 2003;5:267–270. doi: 10.1038/ncb0403-267. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS biology. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.