Abstract
Purpose
TRIBUTE was a phase III trial evaluating the addition of erlotinib to carboplatin and paclitaxel as a first-line treatment for advanced non – small cell lung cancer that did not meet its primary end point of improving overall survival. Here, we assess the value of using epidermal growth factor receptor (EGFR) gene copy number in tumor biopsy samples, as determined by fluorescence in situ hybridization (FISH), as a predictor of treatment outcome.
Methods
EGFR FISH analysis was done using LSI EGFR SpectrumOrange/CEP7 Spectrum-Green probe.
Results
Of 275 samples, 245 (89.1%) were successfully analyzed by FISH. One hundred (40.8%) of patients were EGFR FISH(+). Median overall survival was not different between FISH(+) and FISH(−) patients in either the chemotherapy+erlotinib arm or the chemotherapy+placebo arm. In FISH(+) patients, median time to progression (TTP) was 6.3 months in the erlotinib arm versus 5.8 months in the placebo arm (hazard ratio, 0.59; 95% confidence interval, 0.35–0.99; P = 0.0430); in FISH(−) patients, median TTP was 4.6 months versus 6.0 months (hazard ratio,1.42; 95%confidence interval, 0.95–2.14; P = 0.0895; treatment interaction test, P = 0.007). After 6 months of treatment, a notable separation of the TTP curves in favor of erlotinib emerged. Objective response rates were11.6% versus 29.8% in FISH(+) patients (chemotherapy+erlotinib arm versus chemotherapy+placebo arm; P = 0.0495) and 21.8% versus 25.4%, respectively, for FISH(−) patients (P = 0.6954).
Conclusions
EGFR gene copy number by FISH did not predict survival benefit. However, among EGFR FISH(+) patients, TTP was longer in patients who received erlotinib and continued to receive it after completing first-line therapy.
Epidermal growth factor receptor (EGFR) is expressed in the majority of non–small cell lung cancers (NSCLC; refs. 1, 2). The efficacy of EGFR inhibitors in preclinical models, together with their favorable toxicity profiles, led to their clinical development for NSCLC (3). Erlotinib and gefitinib are small molecule inhibitors of the EGFR tyrosine kinase and have shown antitumor activity as single agents in phase II studies in patients with NSCLC (4–6). Depending on patient characteristics such as smoking history, ethnicity, gender, and histology, single-agent response rates vary from 5% to 27% (7, 8). A landmark study conducted by the National Cancer Institute of Canada (BR.21) showed a survival benefit in NSCLC patients treated with erlotinib as second- or third-line therapy compared with placebo [hazard ratio (HR), 0.70; P = 0.001; ref. 9]. However, in phase III studies of patients with untreated, advanced NSCLC, adding gefitinib or erlotinib to chemotherapy did not significantly improve outcome compared with chemotherapy alone (10–13). One possible explanation for the failure to observe added benefit in these trials is that patients were not selected for biological features such as EGFR protein expression, EGFR gene copy number, or EGFR activating mutations that could indicate clinical benefit from an EGFR inhibitor.
Although EGFR expression is not a prognostic indicator of survival (14–16), EGFR expression may predict response to treatment with EGFR inhibitors. In patients with advanced NSCLC who were previously treated with chemotherapy, retrospective studies with the EGFR tyrosine kinase inhibitors gefitinib (17–19) and erlotinib (15) showed that EGFR gene copy number, as detected by fluorescence in situ hybridization (FISH), may predict outcome after treatment: in analyses of two large placebo-controlled randomized studies—ISEL, with gefitinib, and BR.21, with erlotinib—the HRs were substantially reduced in the EGFR FISH(+) [HR, 0.61; 95% confidence interval (95% CI), 0.36–1.04; P = 0.067; and HR, 0.44; 95% CI, 0.23–0.82; P = 0.008, respectively].
TRIBUTE was a phase III randomized study conducted in the United States and sponsored by Genentech. The trial enrolled 1,079 chemotherapy-naBve patients with locally advanced or metastatic (stage IIIB or IV) NSCLC to compare the survival of patients who received daily erlotinib versus placebo administrated concurrently with carboplatin and paclitaxel (up to six 21-day cycles), followed by maintenance erlotinib or placebo. The primary efficacy end point was duration of survival, and the secondary efficacy end points were time to progression (TTP), objective response rate (ORR, defined by RECIST criteria), duration of response, and time to symptomatic progression. The erlotinib-containing arm did not show an advantage for any of these variables over the placebo arm (13).
The present study examined whether increased EGFR gene copy number, as detected by FISH, had any effect on overall survival (OS), TTP, and progression-free survival for the patients in this large prospective phase III trial (13).
Patients and Methods
Patients samples
At time of enrollment in TRIBUTE, patients were given the option of providing an additional written informed consent to allow release of their archival tumor samples for research purposes. All samples used in this analysis came from patients who signed this secondary consent.
EGFR gene copy number analysis by FISH
The FISH analyses were done blindly without any knowledge of the patients’ clinical characteristics or treatment outcome. Before FISH analysis, slides were reviewed to assess the quality of the material and tumor content. Formalin-fixed paraffin-embedded tissue sections, which included 55 lung specimens (23 fine needle aspirates, 3 core biopsies, 9 surgical specimens, and 20 specimens of unknown type), 13 lymph node specimens (6 fine needle aspirates, 4 biopsies, and 3 of unknown type), 1 thigh mass biopsy, 1 humerus core biopsy, 1 pericardial surgical biopsy, 1 scalp biopsy, 1 femur biopsy, and 1 fine needle aspirate of the sacrum biopsy. All specimens were stained with H&E. Because the FISH EGFR assay was developed and validated on histologic sections containing at least 50 tumor cells with no overlapping nuclei, we used only specimens that met these criteria. The methodology of the EGFR FISH assay is described in details in previous publications (17–19). In brief, LSI EGFR SpectrumOrange/CEP 7 SpectrumGreenTM Probe1 probes (Abbott Molecularxz) were used. In scoring samples, tumors with 4 or more copies of the EGFR gene in >40% of the cells (high polysomy) or tumors with EGFR gene amplification [gene/chromosome ratio of >2 or ≥15 gene copies in >10% of the cells were considered FISH(+)], whereas all others (disomy, trisomy, and low polysomy) were considered FISH(−). Tissue was not a requirement for entry into TRIBUTE.
Statistical analyses
The analysis reported in this paper is a retrospective exploratory subgroup analysis. Demographic variables were summarized by EGFR gene copy status. ORR was summarized by gene copy status and treatment received. Comparisons across groups were made with Fischer’s exact test (for categorical variables). Time to event variables (i.e., duration of survival and TTP) were summarized by Kaplan-Meier curves. Median time to event was estimated from the Kaplan-Meier curves. Comparisons between groups for time-to-event variables were done via log-rank test. HRs were estimated by Cox regression. All hypothesis tests were two sided. Due to the exploratory nature of the analysis, no adjustment for multiple testing was used. Patients with missing values for a given clinical variable (including indeterminant EGFR gene copy status) were excluded from any analysis involving that variable.
Results
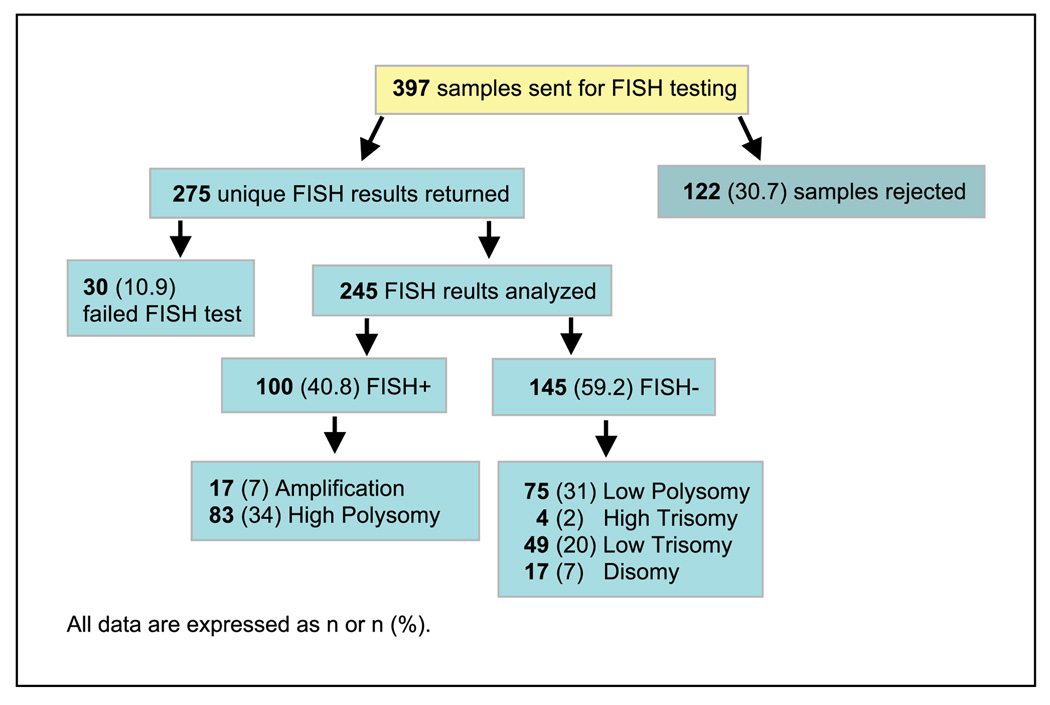
Of all patients enrolled in TRIBUTE, 710 provided the second, optional, written informed consent for tumor tissue research. Archival pathology specimens were released to Genentech, Inc. for 479 of these patients. The primary pathology reports and histopathologic diagnosis were reviewed by a pathologist. Tumor tissue samples from 397 patients, corresponding to 36.8% of the total study population, were sent to University of Colorado Cancer Center for further analysis. At the quality control step, samples from 122 patients were rejected either because there was insufficient tumor material, or because the samples were cytologic samples (e.g., samples in which the tissue was no longer intact). That left samples from 275 patients for EGFR FISH analysis (Fig. 1).
Fig. 1.
Flow chart of FISH subanalysis.
Of the 275 samples that were analyzed by EGFR FISH, 245 (89.1%) were successfully completed (referred to henceforth as the FISH subgroup). Of those, 100 (40.8%) patients had EGFR FISH(+) tumors—17 (7%) patients had EGFR gene amplification and 83 (34%) patients had high polysomy (Fig. 1). The remaining 145 patients were FISH(−)—31% were low polysomy, 2% high trisomy, 20% low trisomy, and 7% disomy (Fig. 1).
The demographic and baseline characteristics of the 245 patients in the FISH subgroup compared with all patients enrolled on the study are presented in Table 1. Overall the patients in the groups were similar. The only exception was that compared with the overall TRIBUTE patient population, more patients in the FISH subgroup had received their NSCLC diagnosis of >6 months before entering the study (17.1% versus 10.6%). For the EGFR FISH(+) and FISH(−) subgroups, there were some imbalances in patient characteristics between treatment arms (Table 2). These characteristics included sex, patients who never smoked, and tumor histology. In addition, only 8.8% of the FISH(+) patients in the erlotinib arm had a K-ras mutation compared with 26.3% of FISH(−) group.
Table 1.
Clinical characteristics for patients with tissue samples evaluable for EGFR gene copy number by FISH compared with the overall study population
| Patients with EGFR FISH results | All patients | |||
|---|---|---|---|---|
| Placebo (n = 124) | Erlotinib (n = 121) | Placebo (n = 540) | Erlotinib (n = 539) | |
| Age (y) | ||||
| Median | 65 | 64 | 63 | 63 |
| Range | 36–82 | 24–81 | 26–84 | 24–84 |
| Sex | ||||
| Male | 74 (59.7) | 65 (53.7) | 332 (61.6) | 322 (59.7) |
| Female | 50 (40.3) | 56 (46.3) | 207 (38.4) | 217 (40.3) |
| Ethnicity | ||||
| Caucasian | 117 (94.4) | 103 (81.1) | 482 (89.4) | 452 (83.9) |
| Asian or Pacific Islander | 3 (2.4) | 4 (3.3) | 13 (2.4) | 21 (3.9) |
| Other | 4 (3.2) | 14 (11.5) | 44 (8.1) | 66 (12.2) |
| Smoking history | ||||
| Never | 7 (5.6) | 12 (9.9) | 44 (8.2) | 72 (13.4) |
| Current | 19 (15.3) | 19 (15.7) | 106 (19.7) | 100 (18.6) |
| Previous | 98 (79.0) | 90 (74.4) | 389 (72.2) | 367 (68.1) |
| Baseline ECOG | ||||
| 0 | 43 (34.7) | 47 (38.8) | 195 (36.2) | 186 (34.5) |
| 1 | 81 (65.3) | 74 (61.2) | 342 (63.6) | 353 (65.5) |
| 2 | (0.0) | (0.0) | 1 (0.2) | (0.0) |
| Prior radiotherapy | ||||
| Yes | 29 (23.4) | 32 (26.4) | 119 (22.1) | 118 (21.9) |
| No | 95 (76.6) | 89 (73.6) | 419 (77.9) | 421 (78.1) |
| Cancer type | ||||
| Metastatic disease | 105 (84.7) | 111 (91.7) | 444 (82.4) | 456 (84.6) |
| Local advanced disease | 19 (15.3) | 10 (8.3) | 95 (17.6) | 83 (15.4) |
| Months since initial NSCLC diagnosis | ||||
| <6 | 97 (78.2) | 106 (87.6) | 476 (88.3) | 488 (90.5) |
| 6–12 | 8 (6.5) | 3 (2.5) | 19 (3.5) | 16 (3.0) |
| >12 | 19 (15.3) | 12 (9.9) | 44 (8.2) | 35 (6.5) |
| Histology | ||||
| Squamous cell | 21 (16.9) | 25 (20.7) | 87 (16.1) | 97 (18.0) |
| Adenocarcinoma | 78 (62.9) | 68 (56.2) | 331 (61.4) | 323 (59.9) |
| Large cell | 13 (10.5) | 12 (9.9) | 56 (10.4) | 43 (8.0) |
| Other | 12 (9.7) | 16 (13.2) | 65 (12.1) | 76 (14.1) |
| Weight loss* | ||||
| Yes | 39 (31.5) | 38 (31.4) | 169 (31.5) | 165 (30.6) |
| No | 85 (68.5) | 83 (68.6) | 368 (68.5) | 374 (69.4) |
| Cancer stage | ||||
| IIIB | 20 (16.1) | 10 (8.3) | 96 (17.8) | 84 (15.6) |
| IV | 104 (83.9) | 111 (91.7) | 443 (82.2) | 455 (84.4) |
| Disease measurability | ||||
| Measurable | 114 (91.9) | 113 (93.4) | 504 (93.3) | 506 (93.9) |
| Nonmeasurable | 10 (8.1) | 8 (6.6) | 36 (6.7) | 33 (6.1) |
NOTE: All data n (%), unless otherwise noted.
Abbreviations: EGOG, Eastern Cooperative Oncology Group.
Lost >5 lbs in last 6 mo.
Table 2.
Patient and tumor characteristics by FISH status
| EGFR FISH (+) | EGFR FISH (−) | |||
|---|---|---|---|---|
| Placebo (n = 57) | Erlotinib (n = 43) | Placebo (n = 67) | Erlotinib (n = 78) | |
| Age (y) | ||||
| Median | 65 | 67 | 65 | 62 |
| Range | 40–82 | 24–81 | 36–80 | 35–79 |
| Sex | ||||
| Male | 33 (57.9) | 19 (44.2) | 41 (61.2) | 46 (59.0) |
| Female | 24 (42.1) | 24 (55.8) | 26 (38.8) | 32 (41.0) |
| Ethnicity | ||||
| Caucasian | 53 (93) | 36 (83.7) | 64 (95.5) | 67 (85.9) |
| Asian or Pacific Islander | 3 (5.3) | 1 (2.3) | 0 (0.0) | 3 (3.8) |
| Other | 1 (1.8) | 6 (14.0) | 3 (4.5) | 8 (10.3) |
| Smoking history | ||||
| Never | 5 (8.8) | 4 (9.3) | 2 (3.0) | 8 (10.3) |
| Current | 3 (5.3) | 7 (16.3) | 16 (23.9) | 12 (15.4) |
| Previous | 49 (86.0) | 32 (74.4) | 49 (73.1) | 58 (74.4) |
| Baseline ECOG | ||||
| 0 | 22 (38.6) | 13 (30.2) | 21 (31.3) | 34 (43.6) |
| 1 | 35 (61.4) | 30 (69.8) | 46 (68.7) | 44 (56.4) |
| 2 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Prior radiotherapy | ||||
| Yes | 12 (21.1) | 15 (34.9) | 17 (25.4) | 17 (21.8) |
| No | 45 (78.9) | 28 (65.1) | 50 (74.6) | 61 (78.2) |
| Cancer type | ||||
| Metastatic disease | 48 (84.2) | 41 (95.3) | 57 (85.1) | 70 (89.7) |
| Local advanced disease | 9 (15.8) | 2 (4.7) | 10 (14.9) | 8 (10.3) |
| Months since initial NSCLC diagnosis | ||||
| <6 | 47 (82.5) | 39 (90.7) | 50 (74.6) | 67 (85.9) |
| 6–12 | 4 (7.0) | 1 (2.3) | 4 (6.0) | 2 (2.6) |
| >12 | 6 (10.5) | 3 (7.0) | 13 (19.4) | 9 (11.5) |
| Histology | ||||
| Squamous cell | 7 (12.3) | 9 (20.9) | 14 (20.9) | 16 (20.5) |
| Adenocarcinoma | 40 (70.2) | 25 (58.1) | 38 (56.7) | 43 (55.1) |
| Large cell | 3 (5.3) | 3 (7.0) | 10 (14.9) | 9 (11.5) |
| Other | 7 (12.3) | 6 (14.0) | 5 (7.5) | 10 (12.8) |
| Weight loss* | ||||
| Yes | 21 (36.8) | 13 (30.2) | 18 (26.9) | 25 (32.1) |
| No | 36 (63.2) | 30 (69.8) | 49 (73.1) | 53 (67.9) |
| Disease measurability | ||||
| Measurable | 54 (94.7) | 30 (90.7) | 60 (89.6) | 74 (94.9) |
| Nonmeasurable | 3 (5.3) | 4 (9.3) | 7 (10.4) | 4 (5.1) |
| Cancer stage | ||||
| IIIB | 10 (17.5) | 2 (4.7) | 10 (14.9) | 8 (10.3) |
| IV | 47 (82.5) | 41 (95.3) | 57 (85.1) | 70 (89.7) |
| EGFR mutation | ||||
| N | 43 | 32 | 49 | 55 |
| Yes | 13 (30.2) | 11 (34.4) | 3 (6.1) | 3 (5.5) |
| No | 30 (69.8) | 21 (65.6) | 46 (93.9) | 52 (94.5) |
| K-ras mutation | ||||
| n | 46 | 34 | 55 | 57 |
| Yes | 12 (26.1) | 3 (8.8) | 12 (21.8) | 15 (26.3) |
| No | 34 (73.9) | 31 (91.2) | 43 (78.2) | 42 (73.7) |
Lost >5 lbs in last 6 mo.
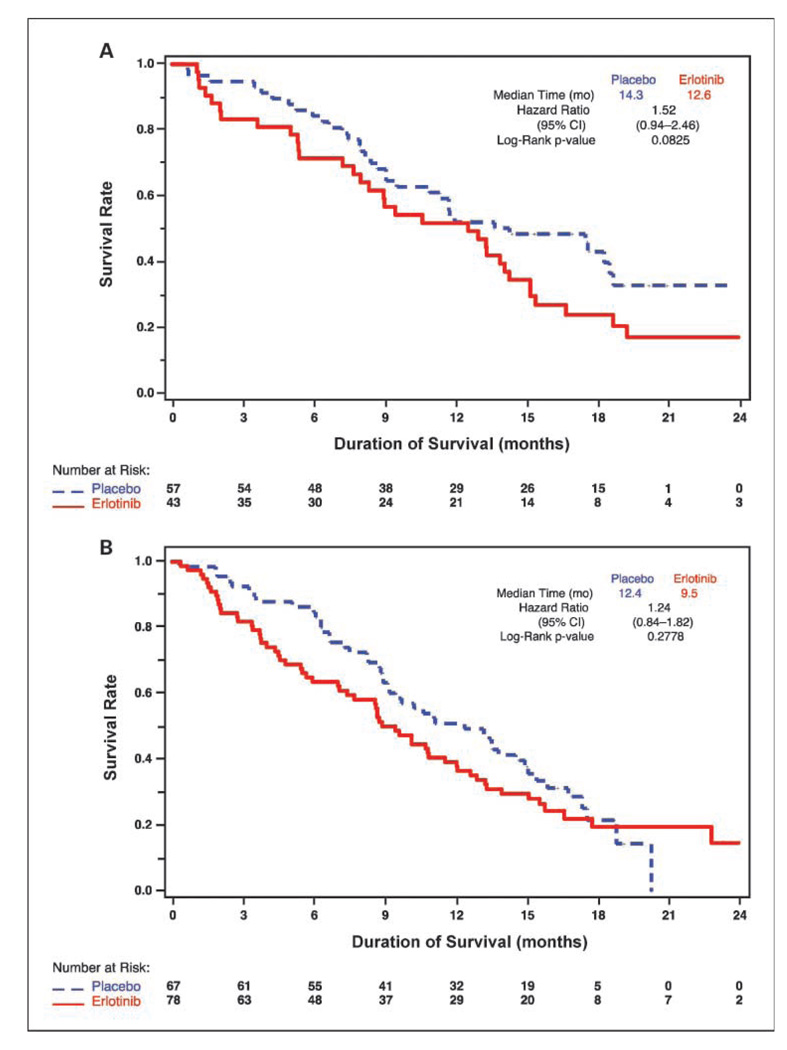
In the FISH subgroup, the median OS was better for placebo-treated patients than for patients treated with erlotinib (13.2 months versus 9.6 months; HR, 1.38; 95% CI, 1.03–1.86; P = 0.0330). For EGFR FISH(+) patients, median survival for erlotinib-treated patients was 12.6 months versus 14.3 months for placebo-treated patients (HR, 1.52; 95% CI, 0.94–2.46; log-rank P = 0.0825, Fig. 2A). In the EGFR FISH(−) group, the patients in the erlotinib arm had a median survival of 9.5 months versus 12.4 months for patients in the placebo arm (HR, 1.24; 95% CI, 0.84–1.82; log-rank P = 0.2778; Fig. 2B). By FISH interaction test for survival, this difference was not statistically significant (P = 0.49). Survival curves for erlotinib+ chemotherapy and placebo+chemotherapy patients seemed similar in both the FISH(+) and FISH(−) subsets, reflecting the survival curve for the entire FISH subgroup.
Fig. 2.
OS according to EGFR gene copy number as determined by FISH. A, FISH(+) patient subgroup. B, FISH(−) patient subgroup.
Similar TTP was observed for the FISH subgroup compared with the overall population. However, the FISH (+) patients had a significantly longer TTP when treated with erlotinib compared with placebo; median TTP for EGFR FISH(+) patients who received chemotherapy and erlotinib was 6.3 months versus 5.8 months for those who received chemotherapy and placebo (HR, 0.59; 95% CI, 0.35–0.99; P = 0.0430; Fig. 3A). In addition, after 6 months of treatment, a notable separation of the TTP curves in favor of erlotinib emerged (Fig. 3A). In the EGFR FISH(−) group, the median TTP for chemotherapy+erlotinib treatment was 4.6 months versus 6.0 months for chemotherapy+placebo (HR, 1.42; 95% CI, 0.95–2.14; P = 0.0895; Fig. 3B). The result of treatment by FISH interaction test for TTP, indicating the difference in HRs for erlotinib versus placebo according to FISH status, was strongly statistically significant (P =0.007).
Fig. 3.
TTP according to EGFR gene copy number as determined by FISH. A, FISH(+) patient subgroup. B, FISH(−) patient subgroup.
Although TTP was the specified secondary end point in TRIBUTE, progression-free survival might be a better end point. To determine progression-free survival, we included as events all deaths that occurred within 30 days of last treatment and reanalyzed the data. We found that the progression-free survival trended in the same direction as TTP but was not significantly different for the FISH(+) subset who received chemotherapy+erlotinib compared with those who received chemotherapy+-placebo (HR, 0.70; P = 0.14).
With regard to ORR, a higher ORR was observed in the placebo arm of the FISH subgroup compared with that of the overall population (27.4% versus 19.3%). In the FISH(+) group, 5 of 43 (11.6%) patients responded to chemotherapy and erlotinib, whereas 17 of 53 (29.8%) patients responded to chemotherapy+placebo (P = 0.0495). Among FISH(−) patients, 17 of 78 (21.8%) patients responded to chemotherapy+erlotinib compared with 17 of 67 (25.4%) patients who responded to chemotherapy and placebo (P = 0.6954; Table 3).
Table 3.
ORRs to chemotherapy and erlotinib or chemotherapy and placebo according to EGFR gene copy number by FISH
| EGFR FISH (+) | EGFR FISH (−) | |||||
|---|---|---|---|---|---|---|
| Chemotherapy + erlotinib (n = 43) |
Chemotherapy + placebo (n = 57) |
Chemotherapy + erlotinib (n = 78) |
Chemotherapy + placebo (n = 67) |
|||
| ORR | 5 (11.6) | 17 (29.8) | 17 (21.8) | 17 (25.4) | ||
| 95% CI | 4.7–25.0 | 18.4–42.4 | 13.9–32.4 | 15.8–36.8 | ||
| P | 0.0495 | 0.6954 | ||||
All data n (%), unless otherwise noted.
Discussion
In the overall TRIBUTE study population, addition of erlotinib to chemotherapy as first-line treatment for advanced NSCLC did not result in improved OS (the primary end point) or TTP—median survival was 10.6 months for chemotherapy+erlotinib versus 10.5 months for chemotherapy+placebo (HR, 0.995; 95% CI, 0.86–1.16; P = 0.95) and TTP was similar (13). Previously, we and others have shown that increased EGFR gene copy number, as detected by FISH, is associated with better response, and prolonged TTP and OS, after second or third line treatment with EGFR tyrosine kinase inhibitors in advanced NSCLC (15, 17–19). Thus, we considered whether improvements might be seen in patients who were selected on the basis of EGFR copy number.
In the current analysis, patients in the FISH subgroup (which included the 245 patients for whom a FISH result was obtained) exhibited differential survival from the overall TRIBUTE study population—placebo-treated patients had longer survival than erlotinib-treated patients, whereas in the overall population, survival was similar. The reason for this is not clear. Overall, baseline demographic characteristics for the two groups were quite similar (Table 1). The only difference was that for more patients in the FISH subgroup >6 months had elapsed because diagnosis (17.1% versus 10.6%), suggesting that the patients with tissue available for FISH analysis may have had more indolent disease.
The current analysis did not show a difference in OS for patients in either the FISH(+) and FISH(−) subgroups between patients receiving chemotherapy+erlotinib versus those receiving chemotherapy+placebo. However, patients in the FISH(+) group who received erlotinib exhibited longer TTP compared with those who received placebo (HR, 0.59; 95% CI, 0.35–0.99; P = 0.0430). These results remained significant even when we tested robustness by adjusting for the imbalances in characteristics between treatment arms among the FISH(+) group (data not shown).
In a biomarker study based on tumor biopsy samples from participants of the TRIBUTE trial, Eberhard et al. (20) analyzed EGFR and K-ras mutations by sequencing. The study reported that irrespective of treatment with erlotinib or placebo, patients with tumors harboring EGFR mutations exhibited prolonged survival, suggesting that EGFR-mutant NSCLC may be more indolent. The addition of erlotinib to chemotherapy in patients harboring K-ras mutations was associated with significantly decreased TTP and survival compared with those tumors contained wild-type K-ras. Similarly, biomarker analysis of the INTACT (phase III) gefitinib trials, Bell et al. (21) reported that in both the gefitinib and placebo groups, EGFR mutations were associated with more favorable survival. A similar trend was observed for patients who had >4-fold EGFR gene amplification. It should be noted, however, that EGFR gene copy number quantification by qPCR and by FISH may yield different results, as was shown in a study comparing these methods in 82 patients treated with gefitinib (22).
In the FISH(+) group, a lower response rate was observed for patients treated with chemotherapy+erlotinib versus those treated with chemotherapy+placebo (11.6% versus 29.8%; P = 0.0495). The lower response rate in the erlotinib+chemotherapy arm could indicate an antagonistic effect of combining chemotherapy with erlotinib. It has been proposed, based on preclinical studies, that there is an antagonistic effect between EGFR tyrosine kinase inhibitors and chemotherapy when they are given concomitantly (23). The biological hypothesis is that the EGFR tyrosine kinase inhibitor therapy results in a cell cycle arrest in G1, which hinders the ability of the chemotherapy to elicit an effect at the G2-M phase of the cell cycle. This may be why TRIBUTE and similar studies that combined a tyrosine kinase inhibitor with chemotherapy (i.e., TALENT, INTACT I, and INTACT II) did not show an overall benefit (11, 12, 24).
This hypothesis is potentially consistent with the finding here that the TTP curves for patients in the FISH(+) group, which are superimposed for the first 6 months of treatment, separate after ~6 months, the point at which patients continued on erlotinib alone (i.e., without concurrent chemotherapy). The split in the TTP curves at the 6-month time point suggested a possible advantage for maintenance erlotinib after the completion of chemotherapy in EGFR FISH (+) patients and led to the investigation of erlotinib as a maintenance therapy after first-line chemotherapy in advanced NSCLC. Trials addressing this question are ongoing.
In conclusion, EGFR gene copy number by FISH did not predict survival benefit. However, among EGFR FISH(+) patients, there was prolonged TTP and a trend toward longer progression-free survival in patients who received erlotinib and then continued to receive it after completing first-line therapy. This finding supports further exploration of maintenance erlotinib as part of a treatment regimen for advanced NSCLC.
Acknowledgments
We thank Dawn Colburn for her comments on the manuscript.
The clinical investigation of erlotinib (Tarceva) is a tripartite collaboration of Genentech, Inc., OSI Pharmaceuticals, Inc., and F. Hoffmann-La Roche, Inc. Tarceva is a registered trademark of OSI Pharmaceuticals, Inc. Genentech, Inc. provided writing assistance for this manuscript.
Grant support: Specialized Program of Research Excellence P01-CA58187 grant and by Genentech. R. Dziadziuszko was supported by a fellowship grant from the International Association for the Study of Lung Cancer.
Disclosure of Potential Conflicts of Interest
F. Hirsch has received research grants from AstraZeneca, Genentech, OSI, Merck, Syndax, Genmab, and Sanofi-Aventis, and is on the advisory boards of AstraZeneca, Roche, Lily, Genentech, Merck Serono, Pfizer, Boehringer/Ingelheim, BMS/Imclone, Ventana Diagnostics. F. Hirsch also holds a patent on the EGFR gene copy number (FISH) and proteins (IHC) for prediction of outcome from EGFR inhibitors (licensed to Abbot Diagnostics) (US 2008/0090233 A1).
Footnotes
Note: This article represents an original clinical study. Data from this study were reported at the 2007 American Society for Clinical Oncology annual meeting.
References
- 1.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura H, Kawasaki N, Taguchi M, et al. Survival impact of epidermal growth factor receptor over-expression in patients with non-small cell lung cancer: a meta-analysis. Thorax. 2006;61:140–145. doi: 10.1136/thx.2005.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL1 Trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [corrected] [DOI] [PubMed] [Google Scholar]
- 5.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Soler R. Phase II clinical trial data with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib (OSI-774) in non-small-cell lung cancer. Clin Lung Cancer. 2004;6(Suppl 1):S20–S23. doi: 10.3816/clc.2004.s.010. [DOI] [PubMed] [Google Scholar]
- 7.Bunn PA, Jr, Dziadziuszko R, Varella-Garcia M, et al. Biological markers for non-small cell lung cancer patient selection for epidermal growth factor receptor tyrosine kinase inhibitor therapy. Clin Cancer Res. 2006;12:3652–3656. doi: 10.1158/1078-0432.CCR-06-0261. [DOI] [PubMed] [Google Scholar]
- 8.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd FA, Rodrigues PJ, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 10.Gatzemeier U, Pluzanska A, Szczesna A, et al. Phase III study of erlotinib in combination with cisplatin and gemcitabine in advanced non-small-cell lung cancer: the Tarceva Lung Cancer Investigation Trial. J Clin Oncol. 2007;25:1545–1552. doi: 10.1200/JCO.2005.05.1474. [DOI] [PubMed] [Google Scholar]
- 11.Giaccone G, Herbst RS, Manegold C, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial-INTACT1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Herbst RS, Giaccone G, Schiller JH, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial-INTACT 2. J Clin Oncol. 2004;22:785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 13.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 14.Clark GM, Zborowski DM, Santabarbara P, et al. Smoking history and epidermal growth factor receptor expression as predictors of survival benefit from erlotinib for patients with non-small-cell lung cancer in the National Cancer Institute of Canada Clinical Trials Group study BR.21. Clin Lung Cancer. 2006;7:389–394. doi: 10.3816/clc.2006.n.022. [DOI] [PubMed] [Google Scholar]
- 15.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 16.Dziadziuszko R, Holm B, Skov B, et al. Epidermal growth factor receptor gene copy number and protein level are not associated with outcome of non-small-cell lung cancer patients treated with chemotherapy. Ann Oncol. 2007;18:447–452. doi: 10.1093/annonc/mdl407. [DOI] [PubMed] [Google Scholar]
- 17.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 20.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5999. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 21.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 22.Dziadziuszko R, Witta SE, Cappuzzo F, et al. Epidermal growth factor receptor messenger RNA expression, gene dosage, and gefitinib sensitivity in non-small cell lung cancer. Clin Cancer Res. 2006;12:3078–3084. doi: 10.1158/1078-0432.CCR-06-0106. [DOI] [PubMed] [Google Scholar]
- 23.Davies AM, Ho C, Lara PN, Jr, et al. Pharmacodynamic separation of epidermal growth factor receptor tyrosine kinase inhibitors and chemotherapy in non-small cell lung cancer. Clin Lung Cancer. 2006;7:385–388. doi: 10.3816/CLC.2006.n.021. [DOI] [PubMed] [Google Scholar]
- 24.Fuster LM, Sandler AB. Select clinical trials of erlotinib (OSI-774) in non-small-cell lung cancer with emphasis on phase III outcomes. Clin Lung Cancer. 2004;6(Suppl 1):S24–S29. doi: 10.3816/clc.2004.s.011. [DOI] [PubMed] [Google Scholar]