Abstract
Clostridium clariflavum is a Cluster III Clostridium within the family Clostridiaceae isolated from thermophilic anaerobic sludge (Shiratori et al, 2009). This species is of interest because of its similarity to the model cellulolytic organism Clostridium thermocellum and for the ability of environmental isolates to break down cellulose and hemicellulose. Here we describe features of the 4,897,678 bp long genome and its annotation, consisting of 4,131 protein-coding and 98 RNA genes, for the type strain DSM 19732.
Keywords: Anaerobic, thermophilic, lignocellulose utilization, bioenergy, biotechnology, cellulolytic, cellulosome
Introduction
Cellulolytic clostridia are prominently represented among bacterial species. These organisms are able to solubilize lignocellulose, and their high rates of cellulose utilization make them candidates for consolidated bioprocessing applications [1]. In particular, anaerobic cellulolytic clostridia that grow at thermophilic temperatures are known to break down lignocellulose very efficiently. Clostridium clariflavum DSM 19732 is a cellulolytic thermophilic anaerobe isolated from anaerobic sludge [2], and closely related to the widely studied thermophile C. thermocellum. Environmental isolates of C. clariflavum have been found to dominate cellulolytic enrichment cultures from thermophilic compost and some have been found to utilize both hemicellulose and cellulose [3,4]. These organisms therefore represent a potentially important opportunity for the discovery of novel enzymes and mechanisms for efficient lignocellulose solubilization at thermophilic temperatures. Here we describe the complete annotated genomic sequence of the type strain Clostridium clariflavum DSM 19732.
Classification and Features
The phylogenetic relationship of the 16S rRNA gene of C. clariflavum DSM 19732 with other cellulolytic clostridia from Cluster III is shown in Figure 1. The sequences shown in here represent mostly cellulolytic and xylanotlytic clostridia sharing over 84.5% sequence identity. The branch comprised by C. clariflavum, C. straminisolvens and C. thermocellum is of particular interest since it includes cellulolytic organisms sharing at least 96.6% sequence homology able to grow at thermophilic temperatures. A few environmental samples have provided sequences with close homology (>99.0% sequence similarity) to the C. clariflavum 16S rRNA gene, and have been found in thermophilic methanogenic bioreactors [7], enrichment cultures from bioreactors (Accession number AB231801 and AM408567), and enrichments from thermophilic compost [3]. Two pure cultures have been isolated from compost enrichments with >99.7% sequence similarity to C. clariflavum and able to utilize xylan [4]. However, no evidence of this organism has been reported in metagenomic studies from similar environments.
Figure 1.
Phylogenetic tree of 16S rRNA gene highlighting the position of Clostridium clariflavum DSM 19732 relative to other clostridia in Cluster III. This tree was inferred from 1,401 aligned characters using the Minimum Evolution criterion [5] and rooted using C. cellulosi (from adjacent clostridial Cluster IV). Numbers above branches are support values from 1,000 bootstrap replicates [6] if larger than 60%.
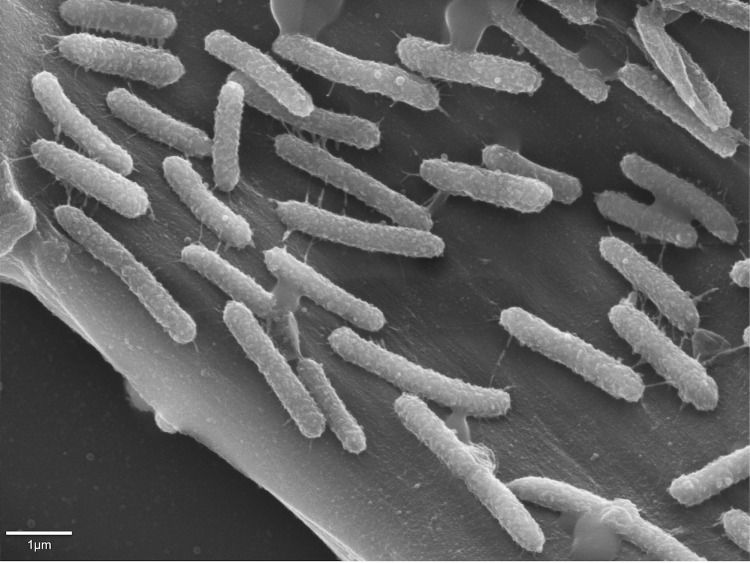
C. clariflavum DSM 19732 is anaerobic, chemoorganotrophic and grows in straight or slightly curved rods [Figure 2]. This organism can ferment cellulose and cellobiose as sole carbon sources, but cannot utilize glucose, xylose or arabinose [2]. Aesculin hydrolysis is positive, but no starch, casein or gelatin hydrolysis has been observed [2]. Nitrate is not reduced to nitrite, and catalase production was negative [2].
Figure 2.
Scanning electron micrograph of C. clariflavum DSM 19732.
Chemotaxonomy
The fatty acid profile of C. clariflavum was analyzed by Shiratori et al [2]. The most prominent cellular acids of C. clariflavum DSM 19732 were iso-C16:0 iso (23.7%), C16:0 (20.4%) and CDMA-16:0 (16.5%), which is consistent with the general observation of other Gram-positive, spore-forming, low-G+C thermophilic bacteria [Table 1]. Polar lipids have not been studied for this organism.
Table 1. Classification and general features of Clostridium clariflavum DSM 19732 according to the MIGS recommendations [8].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [9] | |
| Phylum Firmicutes | TAS [10-12] | ||
| Class Clostridia | TAS [13,14] | ||
| Order Clostridiales | TAS [15,16] | ||
| Family Clostridiaceae | TAS [16,17] | ||
| Genus Clostridium | TAS [16,18,19] | ||
| Species Clostridium clariflavum | TAS [2] | ||
| Type strain EBR45T | TAS [2] | ||
| Gram stain | positive | TAS [2] | |
| Cell shape | straight or slightly curved rods | TAS [2] | |
| Motility | non-motile | TAS [2] | |
| Sporulation | sporulating | TAS [2] | |
| Temperature range | thermophile | TAS [2] | |
| Optimum temperature | 55-60oC | TAS [2] | |
| Carbon source | Cellulose and cellobiose | TAS [2] | |
| Energy source | chemoorganotrophic | TAS [2] | |
| Terminal electron receptor | |||
| MIGS-6 | Habitat | Municipal waste | TAS [2,7] |
| MIGS-6.3 | Salinity | 0–0.7% (w/v) optimum 0.4% w/v |
TAS [2] |
| MIGS-22 | Oxygen | Moderately anaerobic (O2<0.4%) | TAS [2] |
| MIGS-15 | Biotic relationship | free living | NAS |
| MIGS-14 | Pathogenicity | non pathogenic | NAS |
| MIGS-4 | Geographic location | not reported | |
| MIGS-5 | Sample collection time | 2006 | TAS [7] |
| MIGS-4.1 | Latitude | not reported | |
| MIGS-4.2 | Longitude | not reported | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [20]
Genome sequencing and annotation
Genome project history
The genome was selected based on the ability of Clostridium clariflavum DSM 19732 to grow on cellulose at thermophilic temperatures like its close relative C. thermocellum and the ability of environmental strains identified as C. clariflavum to utilize hemicellulose. A summary of the project information is presented in Table 2. The complete genome sequence was finished in July 2011. The GenBank accession number for the project is CP003065. The genome project is listed in the Genome OnLine Database (GOLD) [21] as project Gi10738. Sequencing was carried out at the DOE Joint Genome Institute (JGI). Finishing was performed by JGI-Los Alamos National Laboratory (LANL). Annotation and annotation quality assurance were carried out by the JGI.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Complete, Level 6: Finished |
| MIGS-28 | Libraries used | 454 Standard and 9kb libraries, Illumina Standard library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454-GS-FLX-Titanium |
| MIGS-31.2 | Fold coverage | 38.7 × pyrosequence, 695.4 × Illumina sequence |
| MIGS-30 | Assemblers | Newbler, Velvet, Phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| Genbank ID | CP003065 | |
| Genbank Date of Release | December 12, 2011 | |
| GOLD ID | Gi10738 | |
| Project relevance | Bioenergy, lignocellulose utilization |
Growth conditions and DNA isolation
Clostridium clariflavum DSM 19732 was obtained from the DSMZ culture collection and grown on medium DSM 520 at 55oC. Genomic DNA was obtained by using a phenol-chloroform extraction protocol with CTAB, a JGI standard operating procedure [22].
Genome sequencing and assembly
The draft genome of Clostridium clariflavum DSM 19732 was generated at the DOE Joint genome Institute (JGI) using a combination of Illumina [23] and 454 technologies [24]. For this genome, we constructed and sequenced an Illumina GAii shotgun library which generated 44,772,666 reads totaling 3,402.7 Mb, a 454 Titanium standard library which generated 434,166 reads and 1 paired end 454 library with an average insert size of 9 kb which generated 392,711 reads totaling 223.9 Mb of 454 data. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website [25]. The initial draft assembly contained 239 contigs in 5 scaffolds. The 454 Titanium standard data and the 454 paired end data were assembled together with Newbler, version 2.3-PreRelease-6/30/2009. The Newbler consensus sequences were computationally shredded into 2 kb overlapping fake reads (shreds). Illumina sequencing data was assembled with VELVET, version 1.0.13 [26], and the consensus sequences were computationally shredded into 1.5 kb overlapping fake reads (shreds). We integrated the 454 Newbler consensus shreds, the Illumina VELVET consensus shreds and the read pairs in the 454 paired end library using parallel phrap, version SPS - 4.24 (High Performance Software, LLC). The software Consed [27-29] was used in the following finishing process. Illumina data was used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI (Alla Lapidus, unpublished). Possible mis-assemblies were corrected using gapResolution (Cliff Han, unpublished), Dupfinisher [30], or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR (J-F Cheng, unpublished) primer walks. A total of 1,046 additional reactions and 10 shatter libraries were necessary to close gaps and to raise the quality of the finished sequence. The total size of the genome is 4,897,678 bp and the final assembly is based on 176.7 Mb of 454 draft data which provides an average 38.7 × coverage of the genome and 3,174 Mb of Illumina draft data which provides an average 695.4 × coverage of the genome.
Genome annotation
Genes were identified using Prodigal [31] as part of the Oak Ridge National Laboratory genome annotation pipeline followed by a round of manual curation using the JGI GenePRIMP pipeline [32]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro, databases. Additional gene prediction analysis and functional annotation were performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [33].
Genome properties
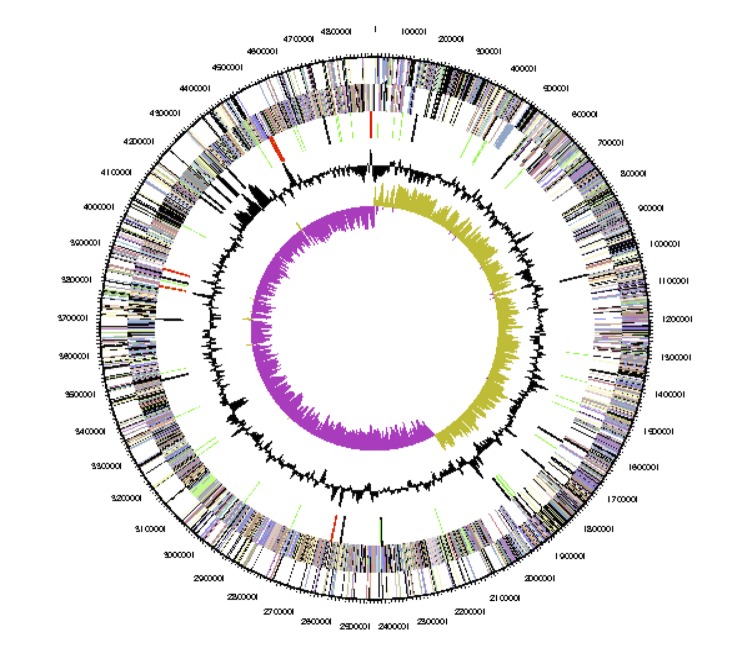
The genome of Clostridium clariflavum DSM 19732 is comprised of one circular chromosome of 4,897,678 bp in length with 35.6% GC content (Table 3 and Figure 3). The sequences of 16S rRNA gene (Accession number AB186359) and a family 48 glycosyl hydrolase (Accession number GQ487569) genes have been previously reported [2,3], and contain 3 mismatches each. The genome size of C. clariflavum is much larger than that of the cellulolytic thermophile and close relative Clostridium thermocellum ATCC 27405 (3.8Mb, 38.9%GC). The chromosome of C. clariflavum was predicted to contain 4,242 coding gene sequences with 6 rRNA operons and 60 tRNA genes (Table 3). The properties and the statistics of the genome are summarized in Tables 3 and 4.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,897,678 | 100.00% |
| DNA Coding region (bp) | 3,915,750 | 79.95% |
| DNA G+C content (bp) | 1,749,312 | 35.72% |
| Total genesb | 4,229 | 100.00% |
| RNA genes | 98 | 2.32% |
| rRNA genes | 6 | |
| Protein-coding genes | 4,131 | 97.68% |
| Pseudo genes | 239 | 5.65% |
| Genes with function prediction | 3,014 | 71.27% |
| Genes in paralog clusters | 585 | 13.83% |
| Genes assigned to COGs | 2,850 | 67.39% |
| Genes assigned Pfam domains | 3,029 | 71.62% |
| Genes with signal peptides | 1,003 | 23.72% |
| Genes with transmembrane helices | 1,047 | 24.76% |
a) The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
b) Also includes 239 pseudogenes.
Figure 3.
Graphical circular map of the genome. From outside to the center; Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 169 | 5.44 | Translation |
| A | 1 | 0.03 | RNA processing and modification |
| K | 224 | 7.21 | Transcription |
| L | 372 | 11.98 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 53 | 1.71 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 81 | 2.61 | Defense mechanisms |
| T | 191 | 6.15 | Signal transduction mechanisms |
| M | 207 | 6.67 | Cell wall/membrane biogenesis |
| N | 89 | 2.87 | Cell motility |
| Z | 4 | 0.13 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 70 | 2.25 | Intracellular trafficking and secretion |
| O | 115 | 3.70 | Posttranslational modification, protein turnover, chaperones |
| C | 150 | 4.83 | Energy production and conversion |
| G | 166 | 5.35 | Carbohydrate transport and metabolism |
| E | 193 | 6.22 | Amino acid transport and metabolism |
| F | 70 | 2.25 | Nucleotide transport and metabolism |
| H | 136 | 4.38 | Coenzyme transport and metabolism |
| I | 54 | 1.74 | Lipid transport and metabolism |
| P | 132 | 4.25 | Inorganic ion transport and metabolism |
| Q | 31 | 1.00 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 332 | 10.69 | General function prediction only |
| S | 264 | 8.50 | Function unknown |
| - | 1,379 | 32.61 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Lignocellulose degrading capability
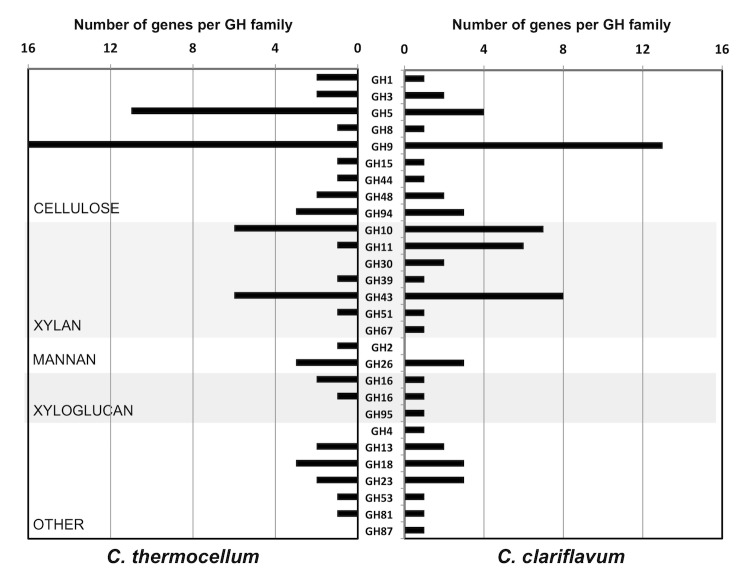
A total of 72 glycosyl hydrolases were identified, representing 27 known families. Of these, 40 enzymes contain Type-I dockerin domains indicating their association with scaffoldin proteins with complementary cohesin regions. When comparing the inventory of glycosyl hydrolases of C. clariflavum DSM 19732 with C. thermocellum ATCC 27405 (Figure 4), 50 enzymes (69.4%) have their closest match in C. thermocellum. Of the remaining 22, a total of 15 are xylanases of glycosyl hydrolase (GH) families GH10, GH11, GH39, GH43, GH51, GH67, and GH74. While cellulose–active GH families seem to be similarly distributed between both organisms, C. clariflavum has a higher proportion and diversity of xylanolytic enzymes than C. thermocellum.
Figure 4.
Comparison of glycosyl hydrolase inventory between C. clariflavum DSM 19732 and C. thermocellum ATCC 27405. The numbers of genes per GH family are shown, with GH families organized along the y-axis based on putative substrate specificity (cellulose, xylan, mannan, xyloglucan and other).
Within the glycosyl hydrolase inventory of C. clariflavum, a subset of bifunctional cellulases was observed (Table 5). Three of these are associated with Type I dockerins (cellulosomal) with varying arrangements of xylanases from families GH10 and GH11 (Clocl_1480, Clocl_2083, Clocl_2441). In addition, an untethered bifunctional set of cellulases (Clocl_3038) is a combination of a GH48 previously reported [3] most closely related to C. thermocellum CelY (Cthe_0071, 72% sequence similarity), in combination with a GH9 most closely related to C. thermocellum CelG (Cthe_0040, 69% sequence similarity) and two family 3 carbohydrate binding modules (CBM3) in a GH48-GH9-CBM3-CBM3 arrangement. A similar arrangement has been discovered in hyperthermophiles like Caldicellulosiruptor bescii [34] and Caldicellulosiruptor saccharolyticus [35], although these enzymes differ in that the CBMs are located in between both cellulases. The lack of a dockerin domain suggests that these multi-domain GHs are secreted, as is the case for Clostridium thermocellum CelY. This also suggests that synergy between secreted GH48 and GH9 enzymes in C. thermocellum [36] seems to be facilitated by this arrangement in C. clariflavum. It should also be noted that in our previous survey of GH48 enzymes from thermophilic cellulolytic clostridia, we reported that C. clariflavum only had a CelY-like GH48 [3]. However, the genome sequence of C. clariflavum revealed an additional cellulosomal GH48 enzyme (Clocl_4007) with a dockerin domain and high degree of similarity to C. thermocellum CelS, the most abundant enzymatic subunit of the C. thermocellum cellulosome [37] [38]. This makes C. clariflavum the only organism with two distinctly different GH48 enzymes, one of which is involved in a bifunctional association.
Table 5. Bifunctional glycosyl hydrolases in the C. clariflavum genome.
| ORF | Domain architecturea | Function(s) | Location |
|---|---|---|---|
| Clocl_1418 | GH11- CBM6-Doc-UK | Endo-1,4-beta-xylanase, unknown | Cellulosome |
| Clocl_2083 | GH11-GH10-Doc | Endo-1,4-beta-xylanases | Cellulosome |
| Cloco_2441 | GH11-CBM6-Doc-GH10 | Endo-1,4-beta-xylanases | Cellulosome |
| Clocl_3038 | GH48-GH9-CBM3-CBM3 | Cellulose 1,4-beta-cellobiosidase (GH48), Endo-1,4-β-D-glucanase (GH9) | Untethered |
aDomain architecture denotes presence of glycosyl hydrolases (GH) from families 9, 10, 11 and 48, Type I dockerin domains (Doc), carbohydrate binding modules (CBM) from families 3 and 6, and domains of unknown function (UK)
Lignocellulose sensing system
A novel system of carbohydrate sensory domains has recently been proposed for C. thermocellum [39]. We have identified a very similar set of genes present in C. clariflavum consisting of 8 sigma I-like factors associated with adjacent carbohydrate active domains (Table 6). Based on the specificity of the CBM modules associated with these gene pairs, there seem to be three potential cellulose-specific (CBM3) pairs: Clocl_1053-54, Clocl_2843-44 and Clocl_4008-09, the latter located directly upstream of the GH48 closely related to endoglucanase CelS. We have identified also one xylan-specific (CBM42) in Clocl_2098-99, one pectin-specific (PA12) in Clocl_2747-48, and three additional domains (Clocl_2044-45, Clocl_2797-98, Clocl_4136-37) which seem to have no catalytic function or CBM domains, but retain high sequence similarity to the proposed unspecific pairs in C. thermocellum.
Table 6. Genes with putative carbohydrate sensing function.
| Loci | Domain structure1 | Putative Substrate | Matches in C. thermocellum2 | % similarity |
|---|---|---|---|---|
| Clocl_1053/ Clocl_1054 | SigI / RsgI-UK-CBM3 | Cellulose | Cthe_0268/ Cthe_0267 | 61/41 |
| Clocl_2843/ Clocl_2844 | SigI / RsgI-UK-CBM3 | Cellulose | Cthe_0058/ Cthe_0059 | 58/31 |
| Clocl_4008/ Clocl_4009 | SigI / RsgI-UK-CBM3 | Cellulose | Cthe_0058/ Cthe_0059 | 59/36 |
| Clocl_2098/ Clocl2099 | SigI / RsgI-UK-CBM42 | Xylan | Cthe_1272/ Cthe_1273 | 72/44 |
| Clocl_2747/ Clocl_2748 | SigI / RsgI-UK-PA14-PA14 | Pectin | Cthe_0315/ Cthe_0316 | 43/32 |
| Clocl_2044/ Clocl_2045 | SigI / RsgI-UK | Unknown | Cthe_2975/ Cthe_2974 |
44/32 |
| Clocl_4136/ Clocl_4137 | SigI / RsgI-UK | Unknown | Cthe_2522/ Chte_2521 | 63/42 |
| Clocl_2797/ Clocl_2798 | SigI / RsgI-UK | Unknown | Cthe_2975/ Cthe_2974 | 65/59 |
1Domain structure denotes pairs of sigma I- like protein (SigI) and its associated trans-membrane protein (RsgI), containing domains of unknown function (UK), carbohydrate binding domains from families 3 (CBM3) and 42 (CBM42), and a conserved domain proposed to have pectin-binding function (PA14).
2Indicates matches in pairs of loci in the C. thermocellum ATCC 27405 genome
Cellulosome assembly
Most cellulolytic clostridia are known to organize glycosyl hydrolases and other catalytic subunits outside of the cell by means of a multiprotein complex known as the cellulosome [40,41]. In terms of cellulosome architecture, several cellulosomal structural proteins containing type I and type II cohesin-dockerin interactions have been identified in the genome of C. clariflavum. Three scaffoldin domains were identified, with a CipA-like scaffoldin domain containing 8 type-I cohesins, a CBM3 module and a type II dockerin (Clocl_3306). Two additional CipA-like domains without CBM domains have been identified, both tethered by type II dockerins: an interesting scaffoldin-like assembly with 5 Type II cohesins (Clocl_3305), and a scaffoldin with 3 Type I cohesins (Clocl_3395). In terms of anchoring proteins, four different structures have been identified containing S-layer homology (SLH) domains: i) an SdbA-like domain with a Type II cohesin (Clocl_2745), ii) an OlpA-like protein with a type I cohesin (Clocl_3334), iii) a similar arrangement with four Type I cohesins (Clocl_3304), and iv) an arrangement consisting of a type I and two type II cohesins (Clocl_3303) similar to a novel anchoring system found in Acetivibrio cellulolyticus [42]. In addition, there seem to be a variety of untethered multi-cohesin complexes with 3 complexes containing multiple Type-I cohesins associated with CBM2 modules (Cloc_4158, Clocl_4211, Clocl_4212), and one untethered Type II cohesin complex (Clocl_1799). The diversity of cellulosomal structural proteins is very similar to what is found in Clostridium thermocellum and other cellulosomal microorganisms. However, CBM2 modules are not very common in cellulolytic clostridia, with C. phytofermentans and C. cellulovorans each having one such domain. C. clariflavum has four of these domains and they are associated with three separate multi-cohesin (Type I) domains with no anchoring mechanism. It may also be noted that the organization of the scaffoldin and anchoring proteins resembles the cellulosomal complexes found in the mesophile Acetivibrio cellulolyticus [42,43] more than it does the C. thermocellum cellulosome.
Pyruvate metabolism
The genome sequence of C. clariflavum revealed that this organism possesses a standard glycolytic pathway. However, the pyruvate node is slightly different from other Cluster III clostridia in that C. clariflavum possesses genes for both pyruvate kinase (Clocl_1090) and pyruvate dikinase (PPDK, Clocl_2755). This may be of relevance to pyruvate metabolism because genomes of cellulolytic clostridia from cluster III reveal that the pathway from phosphoenol pyruvate (PEP) to pyruvate in these organisms uses either PPDK (Clostridium thermocellum ATCC 27405 and DSM 1313) or pyruvate kinase (C. cellulolyticum, C. papyrosolvens). There are nevertheless cellulolytic clostridia outside of Cluster III that also possess both, as is the case of Clostridium cellulovorans.
Hemicellulose sugars metabolism
C. clariflavum possesses a variety of xylanolytic enzymes that allow it to break down xylan completely to xylose, unlike C. thermocellum, which is only able to break xylan down to xylooligomers. One of the key enzymes in xylose utilization, xylose isomerase, is found in mesophilic xylanolytic/cellulolytic clostridia such as C. cellulolyticum, C. phytofermentans, C. papyrosolvens and C. cellulovorans, as well as in hyperthermophiles like Caldicellulosiruptor bescii. However, the genome of C. clariflavum does not seem to possess a xylose isomerase. On the other hand, a putative xylulose kinase has been identified in C. clariflavum (Clocl_2440), which is a key difference from C. thermocellum, where this enzyme is absent. Xylulose kinase is usually adjacent to or in the same operon as xylose isomerase. A xylose epimerase (5.1.3.4) that leads to the production of L-ribulose-5P is immediately adjacent (Clocl_2439) to the putative xylulose kinase. In C. clariflavum, these genes are also surrounded by a variety of hemicellulose-active enzymes in an operon from Clocl_2435 to Clocl_2447, that includes 3 family 10 glycosyl hydrolases. Considering that none of these enzymes is present in C. thermocellum, there should be great interest in further exploring this operon in C. clariflavum and in environmental isolates. An alternative xylose epimerase (5.1.3.1) that produces D-ribulose-5P used in the pentose phosphate pathway is present elsewhere in the genome (Clocl_2564). It therefore seems that C. clariflavum DSM 19732 has much of the capabilities to grow on xylan and xylose, but seems to have lost that ability due to the absence of a xylose isomerase.
Conclusion
In summary, the genome of C. clariflavum strain DSM 19732 contains several features that differentiate this organism from other close relatives within the Cluster III cellulolytic clostridia, and C. thermocellum in particular, providing the first indications of the mechanisms by which C. clariflavum strains utilize lignocellulosic biomass. Seventy two new glycosyl hydrolyses were identified from C. clariflavum with prominently represented structural families including GH9, GH10, GH11 and GH43. Bifunctional arrangements of key GHs are observed involving both cellulosomal (e.g. xylanases GH10, GH11) and non-cellulosomal (e.g. GH9 and GH48) components, and are more prevalent than in C. thermocellum. Xylanases are also more numerous in C. clariflavum than in C. thermocellum. Unique among cellulolytic clostridia of cluster III, the C. clariflavum genome includes putative sequences for pyruvate kinase, which is not found in C. thermocellum, as well as pyruvate dikinase.
Acknowledgements
This work has been funded by BioEnergy Sciences Center (BESC), a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We would like to thank Tamar Kitzmiller and Anna Guseva for their valuable technical assistance, and Chuck Daghlian for guidance with electron microscopy.
References
- 1.Lynd LR, Weimer P, Van Zyl W, Pretorius I. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 2002; 66:506-577 10.1128/MMBR.66.3.506-577.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiratori H, Sasaya K, Ohiwa H, Ikeno H, Ayame S, Kataoka N, Miya A, Beppu T, Ueda K. Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int J Syst Evol Microbiol 2009; 59:1764-1770 10.1099/ijs.0.003483-0 [DOI] [PubMed] [Google Scholar]
- 3.Izquierdo JA, Sizova MV, Lynd LR. Diversity of bacteria and glycosyl hydrolase family 48 Genes in cellulolytic consortia enriched from thermophilic biocompost. Appl Environ Microbiol 2010; 76:3545-3553 10.1128/AEM.02689-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sizova MV, Izquierdo JA, Panikov NS, Lynd LR. Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 2011; 77:2282-2291 10.1128/AEM.01219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rzhetsky A, Nei M. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol Biol Evol 1993; 10:1073-1095 [DOI] [PubMed] [Google Scholar]
- 6.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol 2010; 17:337-354 10.1089/cmb.2009.0179 [DOI] [PubMed] [Google Scholar]
- 7.Shiratori H, Ikeno H, Ayame S, Kataoka N, Miya A, Hosono K, Beppu T, Ueda K. Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor. Appl Environ Microbiol 2006; 72:3702-3709 10.1128/AEM.72.5.3702-3709.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbons N, Murray R. Proposals concerning the higher taxa of bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 11.Garrity GM, Holt JG. The road map to the manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 12.Murray RGE. The higher taxa, or, a place for everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 13.Rainey FA. “Class II. Clostridia class nov.” In: P. De Vos, G.M. Garrity, D. Jones, N.R. Krieg, W. Ludwig, F.A. Rainey, K.H. Schleifer, and W.B. Whitman (eds): Bergey's Manual of Systematic Bacteriology, second edition, vol. 3 (The Firmicutes), Springer, Dordrecht, Heidelberg, London, New York (2009) p. 736. [Google Scholar]
- 14.Validation List 132. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 15.Prévot AR. In: Hauderoy P, Ehringer G, Guillot G, Magrou. J., Prévot AR, Rosset D, Urbain A (eds), Dictionnaire des Bactéries Pathogènes, Second Edition, Masson et Cie, Paris, 1953, p. 1-692. [Google Scholar]
- 16.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 17.Pribram E. Klassification der Schizomyceten. F. Deuticke, Leipzig, (1933) pp. 1-143. [Google Scholar]
- 18.Prazmowski A. "Untersuchung über die entwickelungsgeschichte und fermentwirking einiger bakterien-arten." Ph.D. Dissertation, University of Leipzig, Germany, 1880, p. 366-371. [Google Scholar]
- 19.Smith LDS, Hobbs G. Genus III. Clostridium Prazmowski 1880, 23. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 551-572. [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liolios K, Chen IMA, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.JGI standard operating procedure. http://my.jgi.doe.gov/general/protocols
- 23.Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433-438 10.1517/14622416.5.4.433 [DOI] [PubMed] [Google Scholar]
- 24.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DOE Joint Genome Institute http://www.jgi.doe.gov
- 26.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175-185 [DOI] [PubMed] [Google Scholar]
- 28.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:186-194 [PubMed] [Google Scholar]
- 29.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195-202 [DOI] [PubMed] [Google Scholar]
- 30.Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Arabnia HR, Valafar J, editors. Proceedings of the 2006 international Conference on Bioinformatics and Computational Biology: CSREA Press; 2006. p 141-146 [Google Scholar]
- 31.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 33.Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IMA, Dubchak I, Anderson I, Lykidis A, Mavromatis K, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res 2008; 36:D528-D533 10.1093/nar/gkm846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dam P, Kataeva I, Yang SJ, Zhou F, Yin Y, Chou W, Poole FL, Westpheling J, Hettich R, Giannone R, et al. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res 2011; 39:3240-3254 10.1093/nar/gkq1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 2011; 108:1559-1569 10.1002/bit.23093 [DOI] [PubMed] [Google Scholar]
- 36.Berger E, Zhang D, Zverlov VV, Schwarz WH. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol Lett 2007; 268:194-201 10.1111/j.1574-6968.2006.00583.x [DOI] [PubMed] [Google Scholar]
- 37.Raman B, Pan C, Hurst G, Rodriguez M, Jr, McKeown C, Lankford P, Samatova N, Mielenz J. Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS ONE 2009; 4:e5271 10.1371/journal.pone.0005271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gold ND, Martin VJJ. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J Bacteriol 2007; 189:6787-6795 10.1128/JB.00882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahel-Raifer H, Jindou S, Bahari L, Nataf Y, Shoham Y, Bayer EA, Borovok I, Lamed R. The unique set of putative membrane-associated anti-sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol Lett 2010; 308:84-93 10.1111/j.1574-6968.2010.01997.x [DOI] [PubMed] [Google Scholar]
- 40.Shoham Y, Lamed R, Bayer E. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol 1999; 7:275-281 10.1016/S0966-842X(99)01533-4 [DOI] [PubMed] [Google Scholar]
- 41.Doi RH, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol 2004; 2:541-551 10.1038/nrmicro925 [DOI] [PubMed] [Google Scholar]
- 42.Xu Q, Barak Y, Kenig R, Shoham Y, Bayer E, Lamed R. A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J Bacteriol 2004; 186:5782-5789 10.1128/JB.186.17.5782-5789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, Gao W, Ding S, Kenig R, Shoham Y, Bayer E, Lamed R. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J Bacteriol 2003; 185:4548-4557 10.1128/JB.185.15.4548-4557.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]