Previous studies have shown that the members of the Enterobacter cloacae complex are difficult to differentiate with biochemical tests and in phylogenetic studies using multilocus sequence analysis, strains of the same species separate into numerous clusters. There are only a few complete E. cloacae genome sequences and very little knowledge about the mechanism of pathogenesis of E. cloacae on plants and humans. Enterobacter cloacae EcWSU1 causes Enterobacter bulb decay in stored onions (Allium cepa). The EcWSU1 genome consists of a 4,734,438 bp chromosome and a mega-plasmid of 63,653 bp. The chromosome has 4,632 protein coding regions, 83 tRNA sequences, and 8 rRNA operons.
Introduction
Enterobacter cloacae is ubiquitous in nature and is known to cause disease in numerous plants, such as onion, ginger, papaya, and macadamia [1-4]. In addition, E. cloacae is an emerging opportunistic human pathogen that is associated with nosocomial infections [5]. Phylogenetic analyses of the genus Enterobacter have resulted in the formation of the E. cloacae complex, which consists of several species. The E. cloacae complex includes the species E. cloacae, E. asburiae, E. hormaechei, E. kobei, E. ludwigii, and E. nimipressuralis, but the list is constantly growing as new species of Enterobacter are identified. Within medical isolates of the E. cloacae complex, there are two well supported clades and 13 clusters [6]. The younger clade has less genetic diversity and is composed primarily of E. hormaechei strains isolated from hospitals. The second clade has more genetic diversity and contains the other members of the complex, including E. cloacae. Interestingly, E. cloacae strains separate into six clusters indicating considerable diversity within the species. A neighbor-joining tree of the hsp60 gene from 206 E. cloacae strains showed that few E. cloacae strains (3%) actually cluster with the type strain, E. cloacae subsp. cloacae ATCC 13047 [7].
Enterobacter bulb decay develops after onions are harvested, cured, and stored. The decay usually occurs in a few scales of the onion bulb and the tissue develops a brown color giving the bulb a dirty ring appearance when cut in half [1,8]. If storage lots of onions have a high enough incidence of Enterobacter bulb decay (>2-5%), the whole lot cannot be sold and results in a significant loss to the grower. The mechanism of how E. cloacae causes bulb decay is unknown and as a result, the development of disease control methods for bulb decay are limited. In addition, many new strains are identified as E. cloacae due to traditional phenotype tests and 16S rRNA identity, but when other regions of the genome, or the genome as a whole, are compared, they appear to have more differences within a species than observed between species of other genera of bacteria [6, Humann and Schroeder, unpublished]. The genome sequence reported here will allow for comparisons on a genome-wide level with other E. cloacae strains and may help clarify the relationships between the E. cloacae complex members as well as allow for identification of putative pathogenesis genes.
Classification and features
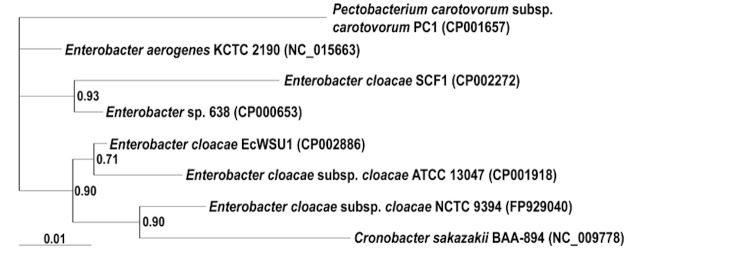
E. cloacae EcWSU1 was isolated from onion bulbs that were exhibiting symptoms of rot [8]. EcWSU1 is a Gram-negative, rod shaped bacterium of the family “Enterobacteriaceae” (Table 1). Species differentiation of the Enterobacter genus is difficult with biochemical and phylogenetic tests [6]. The genetic complexity of the E. cloacae complex is illustrated in a phylogenetic tree of the 16S rRNA region (Figure 1). EcWSU1 grouped with the type-strain E. cloacae subsp. cloacae ATCC 13047 with a 0.71 posterior probability in a Bayesian phylogenetic analysis. E. cloacae SCF1, isolated from soil in Puerto Rico, grouped closely with Enterobacter sp. 638 [26], an endophyte of poplar trees. Cronobacter sakazakii BAA-894, formerly Enterobacter sakazakii [27], clustered with E. cloacae subsp. cloacae NCTC 9394 (0.90 posterior probability), which was isolated from human feces. Interestingly, all the E. cloacae strains did not cluster together.
Table 1. Classification and general features of Enterobacter cloacae EcWSU1 according to MIGS recommendations [9].
| MIGS ID | Property | Term | Evidence Code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [10] | |
| Phylum Proteobacteria | TAS [11] | ||
| Class Gammaproteobacteria | TAS [12,13] | ||
| Order “Enterobacteriales” | TAS [14] | ||
| Family Enterobacteriaceae | TAS [15-17] | ||
| Genus Enterobacter | TAS [15,18-21] | ||
| Species Enterobacter cloacae | TAS [15,18,21] | ||
| Strain EcWSU1 | TAS [8] | ||
| Gram stain | negative | TAS [22] | |
| Cell shape | rod | TAS [22] | |
| Motility | motile via peritrichous flagella | TAS [22] | |
| Sporulation | non-sporulating | TAS [22] | |
| Temperature range | mesophilic, 25-40°C | TAS [22] | |
| Optimum temperature | 30-37°C | TAS [22] | |
| Salinity | not reported | ||
| MIGS-22 | Oxygen requirement | facultative anaerobe | TAS [22] |
| Carbon source | carbohydrates | TAS [22] | |
| Energy source | chemoorganotroph | TAS [22] | |
| MIGS-6 | Habitat | soil, onion | TAS [8] |
| MIGS-15 | Biotic relationship | free-living | TAS [22] |
| MIGS-14 | Pathogenicity | pathogenic on onion | TAS [8] |
| Biosafety level | 2 | ||
| Isolation | Isolated from symptomatic onion | TAS [8] | |
| MIGS-4 | Geographic location | Colorado, USA | TAS [8] |
| MIGS-5 | Sample collection time | not reported |
Evidence codes – IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [23]. If the evidence code is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 1.
Phylogenetic tree of 16S rRNA sequences from strains of Enterobacter with genome sequences. Bayesian phylogenetic analyses of the 16S rRNA region yielded two distinct clusters, supported with a 0.90 posterior probability. Analyses were implemented in MRBAYES [24]. The Bayesian Information Criterion (BIC), DT-ModSel [25] was used to determine the nucleotide substitution model best suited for the dataset. The Markov chain Monte Carlo search included two runs with four chains each for 1,000,000 generations, ensuring that the average split frequencies between the runs was less than 1%. Pectobacterium served as the outgroup for the analysis. Numbers in parentheses behind the bacterial names correspond to the Genbank accession numbers for the genome sequences. The scale bar indicates the number of substitutions/site.
Genome project history
Genome sequencing and annotation
E. cloacae EcWSU1 was isolated from onions exhibiting symptoms of Enterobacter bulb decay [8]. EcWSU1 is the model strain for studying pathogenesis of E. cloacae on onion in the laboratory of Brenda Schroeder at Washington State University. A genome sequence of EcWSU1 was needed to facilitate the development of molecular biology experiments. Pyrosequencing of EcWSU1 was completed at the Laboratory for Biotechnology and Bioanalysis at Washington State University, and the PCR products to close the genome were sequenced at Elim Biopharmaceuticals (Hayward, CA, USA). The complete chromosome sequence as well as the mega-plasmid, pEcWSU1_A, has been deposited in Genbank under the accession numbers CP002886 and CP002887, respectively. Table 2 summarizes the EcWSU1 sequencing project.
Table 2. EcWSU1 Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-29 | Sequencing platform | 454 Life Sciences |
| MIGS-31.2 | Fold coverage | 20 × |
| MIGS-30 | Assembler | GS De novo Assembler V2.3 |
| MIGS-32 | Gene calling method | Bacterial Annotation System (BASys) [28] |
| tRNAscan-SE 1.21 [29] | ||
| Genbank ID | CP002886 (chromosome) | |
| CP002887 (pEcWSU1_A) | ||
| Genbank date of release | With SIGS publication | |
| Project relevance | Plant pathology |
Growth conditions and DNA isolation
E. cloacae EcWSU1 was cultured overnight in 5 ml of LB broth [30] in a 20 ml glass culture tube (16 mm O.D.) on a rotary shaker at 200 rpm at 28°C. Prior to genomic DNA isolation, the cells were washed twice with equal volumes of sterile, distilled water to remove excess exopolysaccharides. Genomic DNA was then isolated from the washed cells using a Wizard Genomic DNA Purification Kit (Promega, A1120) following the kit protocol for Gram-negative bacteria.
Genome sequencing and assembly
The genomic DNA extraction showed a high absorbance at 230 nm during quantification, indicating the presence of polysaccharides. As a result, prior to preparing the DNA for pyrosequencing, the polysaccharides were selectively precipitated in 20% ethanol and removed from the sample by centrifugation. The DNA was then precipitated with two volumes of ethanol, pelleted via centrifugation, dried and suspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8). The sequencing library was constructed using 500 ng of the genomic DNA with the GS FLX Titanium Rapid Library Preparation Kit (Roche, 05608228001) and RL MID adapters (Roche, 05619211001) in place of the standard RL adapters. Minor modifications to the protocol included more extensive washing at the sequencing bead enrichment and harvest steps. The resulting shotgun library was diluted 1:5 and 10 µl was quantified using a 384-well fluorescent plate assay in a Perkin-Elmer Victor X Multi-label Plate Reader. Quality and size of the library was assessed using an Agilent High Sensitivity DNA chip assay (Agilent, 5067-4626) read on an Agilent 2100 Bioanalyzer. Pyrosequencing was performed on a Genome Sequencer GS FLX Titanium instrument (454 Life Sciences, Branford, CT, USA) with the sample occupying one quarter of one picotiter plate. A total of 242,000 reads were obtained accounting for 97.5 Mb of sequence. Reads were assembled using GS De Novo Assembler V2.3 with default parameters and 99.7% of the bases aligned into 35 contigs with 27 of those greater than 5 kb. For the contigs, 281 bp remained at 1× coverage with a bimodal peak depth predominantly centered at 20× that trailed into a second smaller peak of coverage at 141-180× resulting from a higher plasmid copy number relative to genomic DNA in the sample.
The genome sequence of E. cloacae subsp. cloacae ATCC 13047 (CP001918) initially was used as a reference sequence for assembly of the pyrosequencing reads. However, the genomic sequence of EcWSU1 did not have sufficient identity to the DNA sequence of ATCC 13047 for this to be effective (only 19.56% of the reads mapped to ATCC 13047). As a result, the EcWSU1 genome was closed by developing primers that amplified out from each end of the contigs. A putative contig order was generated by using blastn to align the 35 contigs against the incomplete genome (18 contigs) of E. cloacae P101 [31-33], an endophyte of switchgrass that had higher DNA similarity to EcWSU1 than EcWSU1 had with ATCC 13047. The putative contig order of EcWSU1 was then confirmed with PCR amplifications across the contig junctions using GoTaq Polymerase (Promega, M3001) according to the manufacturer’s protocol and 50 ng of EcWSU1 genomic DNA. An annealing temperature of 52°C, with an extension of 1 m was sufficient for most of the contig junctions since there usually were 0-50 bases missing between the contigs. DMSO was added at either a 5% or 10% final concentration in the PCR reaction, in combination with an extension time of 8.5 m, to produce larger fragments that amplified across the 16S-23S rRNA cassettes or to amplify contig junctions that would not amplify with the normal PCR reaction used above. Sequencing was completed for both strands using the same primers used for amplification of the fragments. Fragments that spanned the 16S-23S rRNA regions were also sequenced with internal primers that were specific for contigs that corresponded to the 16S and 23S rRNA regions of EcWSU1. The contigs and sequences from the PCR products were aligned with Bioedit (Ibis Biosciences, Carlsbad, CA) and a complete chromosome sequence was generated with 34 of the 35 contigs. The remaining contig of 63.7 kb was shown to be circular and was designated as pEcWSU1_A.
Genome annotation
Genome annotation was completed using the Bacterial Annotation System (BASys) [28]. tRNA sequences were determined using tRNAscan-SE [29] and rRNA sequences were identified by searching the genome sequence with rRNA sequences from E. cloacae subsp. cloacae ATCC 13047 using a private nucleotide BLAST server [34]. Minor editing to the annotation to remove ORFs that were completely contained in other ORFs was done, and the features file was generated using in-house Java programs. The submission file for Genbank was prepared using Sequin from the NCBI website.
Genome properties
The genome of E. cloacae EcWSU1 consists of one circular chromosome of 4,734,438 bp and a mega-plasmid, pEcWSU1_A, of 63,653 bp. The average G+C content for the genome is 54.5% (Table 3). There are 83 tRNA genes and 8 rRNA operons each consisting of a 16S, 23S, and 5S rRNA gene. There are 4,632 predicted protein-coding regions and 13 pseudogenes in the genome. A total of 4,122 genes (87.0%) have been assigned a predicted function while the rest have been designated as hypothetical proteins (Table 3). The numbers of genes assigned to each COG functional category are listed in Table 4. About one sixth (15.3%) of the annotated genes were not assigned to a COG or have an unknown function.
Table 3. EcWSU1 Genome Statistics.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,798,091 | 100% |
| DNA coding region (bp) | 4,326,148 | 90.16% |
| DNA G+C content (bp) | 2,616,970 | 54.54% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 1 | |
| Total genesb | 4,740 | 100% |
| tRNA genes | 83 | 1.75% |
| rRNA operons | 8 | |
| Protein-coding regions | 4,632 | 97.72% |
| Pseudo genes | 13 | 0.27% |
| Genes with function prediction | 4,122 | 86.96% |
| Genes in paralog clusters | 322 | 7.00% |
| Genes assigned to COGs | 3,830 | 80.80% |
| Genes assigned Pfam domains | 3,972 | 83.80% |
| Genes with signal peptides | 898 | 18.95% |
| Genes with transmembrane helices | 1143 | 24.11% |
| CRISPR repeats | 0 |
a The total is based on either the total number of base pairs or the total number of genes in the genome and includes the chromosome and the plasmid pEcWSU1_A
b Includes the tRNA genes and pseudogenes
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | % age | Description |
|---|---|---|---|
| J | 190 | 4.1 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 401 | 8.7 | Transcription |
| L | 147 | 3.2 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 33 | 0.7 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 53 | 1.1 | Defense mechanisms |
| T | 221 | 4.8 | Signal transduction mechanisms |
| M | 248 | 5.4 | Cell wall/membrane/envelope biogenesis |
| N | 109 | 2.4 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 113 | 2.4 | Intracellular trafficking, secretion, and vesicular transport |
| O | 145 | 3.1 | Posttranslational modification, protein turnover, chaperones |
| C | 235 | 5.1 | Energy production and conversion |
| G | 455 | 9.8 | Carbohydrate transport and metabolism |
| E | 385 | 8.3 | Amino acid transport and metabolism |
| F | 88 | 1.9 | Nucleotide transport and metabolism |
| H | 169 | 3.6 | Coenzyme transport and metabolism |
| I | 124 | 2.7 | Lipid transport and metabolism |
| P | 232 | 5.0 | Inorganic ion transport and metabolism |
| Q | 91 | 2.0 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 483 | 1.0 | General function prediction only |
| S | 366 | 7.9 | Function unknown |
| - | 343 | 7.4 | Not in COGs |
a The total is based on the total number of protein coding genes in the entire annotated genome
Acknowledgements
The authors thank Drs. Linda Thomashow and Debra Inglis for critical evaluation of the manuscript. This project was supported by the Department of Plant Pathology (PPNS No. 0577) in the Washington State University College of Agricultural, Human and Natural Resource Sciences, and the Washington State University Agricultural Research Center for CRIS Project No. WNPO0652.
References
- 1.Bishop AL, Davis RM. Internal decay of onions caused by Enterobacter cloacae. Plant Dis 1990; 74:692-694 10.1094/PD-74-0692 [DOI] [Google Scholar]
- 2.Nishijima KA, Couey HM, Alvarez AM. Internal yellowing, a bacterial disease of papaya fruits caused by Enterobacter cloacae. Plant Dis 1987; 71:1029-1034 10.1094/PD-71-1029 [DOI] [Google Scholar]
- 3.Nishijima KA, Alvarez AM, Hepperly PR, Shintaku MH, Keith LM, Sato DM, Bushe BC, Armstrong JW, Zee FT. Association of Enterobacter cloacae with rhizome rot of edible ginger in Hawaii. Plant Dis 2004; 88:1318-1327 10.1094/PDIS.2004.88.12.1318 [DOI] [PubMed] [Google Scholar]
- 4.Nishijima KA, Wall MM, Siderhurst MS. Demonstrating pathogenicity of Enterobacter cloacae on macadamia and identifying associated volatiles of gray kernel of macadamia in Hawaii. Plant Dis 2007; 91:1221-1228 10.1094/PDIS-91-10-1221 [DOI] [PubMed] [Google Scholar]
- 5.Sanders WE, Jr, Sanders CC. Enterobacter spp.: Pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 1997; 10:220-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paauw A, Caspers MPM, Schuren FHJ, Leverstein-van Hall MA, Delétoile A, Montijn RC, Fluit AC. Genomic diversity within the Enterobacter cloacae complex. PLoS ONE 2008; 3:e3018 10.1371/journal.pone.0003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann H, Roggenkamp A. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 2003; 69:5306-5318 10.1128/AEM.69.9.5306-5318.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder BK, Waters TD, du Toit LJ. Evaluation of onion cultivars for resistance to Enterobacter cloacae in storage. Plant Dis 2010; 94:236-243 10.1094/PDIS-94-2-0236 [DOI] [PubMed] [Google Scholar]
- 9.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 12.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 13.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 14.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 15.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 16.Rahn O. New principles for the classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 1937; 96:273-286 [Google Scholar]
- 17.Judicial Commission Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species OPINION NO. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73-74 10.1099/0096266X-8-1-73 [DOI] [Google Scholar]
- 18.Hormaeche E, Edwards PR. A proposed genus Enterobacter. Int Bull Bacteriol Nomencl Taxon 1960; 10:71-74 10.1099/0096266X-10-2-71 [DOI] [Google Scholar]
- 19.Sakazaki R. Genus VII. Enterobacter Hormaeche and Edwards 1960, 72; Nom. cons. Opin. 28, Jud. Comm. 1963, 38. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 324-325. [Google Scholar]
- 20.Judicial Commission Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species OPINION NO. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73-74 10.1099/0096266X-8-1-73 [DOI] [Google Scholar]
- 21.Editorial Board OPINION 28 Rejection of the Bacterial Generic Name Cloaca Castellani and Chalmers and Acceptance of Enterobacter Hormaeche and Edwards as a Bacterial Generic Name with Type Species Enterobacter cloacae (Jordan) Hormaeche and Edwards. Int Bull Bacteriol Nomencl Taxon 1963; 13:38 10.1099/0096266X-13-1-38 [DOI] [Google Scholar]
- 22.Holt JG, Kreig NR, Sneath PHA, Staley JT, Williams ST. 1994. Bergey's manual of determinative bacteriology, Ninth Edition, Williams & Wilkins, Baltimore. [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001; 17:754-755 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 25.Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol 2003; 52:674-683 10.1080/10635150390235494 [DOI] [PubMed] [Google Scholar]
- 26.Taghavi S, van der Lelie D, Hoffman A, Zhang YB, Walla MD, Vangronsveid J, Newman L, Monch S. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PloS Gen 2010; 6:e1000943 10.1371/journal.pgen.1000943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. The taxonomy of : proposal of a new genus gen. nov. and descriptions of comb. nov. subsp. sakazakii, comb. nov., subsp. malonaticus subsp. nov., sp. nov., sp. nov., sp. nov. and genomospecies I. BMC Evol Biol 2007; 7:64 10.1186/1471-2148-7-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 2005; 33:W455-W459 10.1093/nar/gki593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 2005; 33:W686-W689 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: A laboratory manual, 2nd edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Riggs PJ, Moritz RL, Chelius MK, Dong Y, Iniguez AL, Kaeppler SM, Casler MD, Triplett EW. 2002. Isolation and characterization of diazotrophic endophytes from grasses and their effects on plant growth, p. 263-267. In T. Finan, M. O'Brian, D. Layzell, K. Vessey, and W. Newton (ed.), Nitrogen Fixation: Global Perspectives. CAB International, New York, NY. [Google Scholar]
- 32.Drew JC, Triplett EW. Whole genome sequencing in the undergraduate classroom: Outcomes and lessons from a pilot course. J Microbiol & Biol Ed 2008; 9:3-11 [PMC free article] [PubMed] [Google Scholar]
- 33.Riggs PJ, Chelius MK, Iniguez AL, Kaeppler SM, Triplett EW. Enhanced maize productivity by inoculation with diazotrophic bacteria. Aust J Plant Physiol 2001; 28:829-836 [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]