Abstract
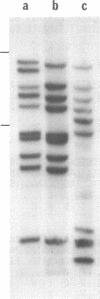
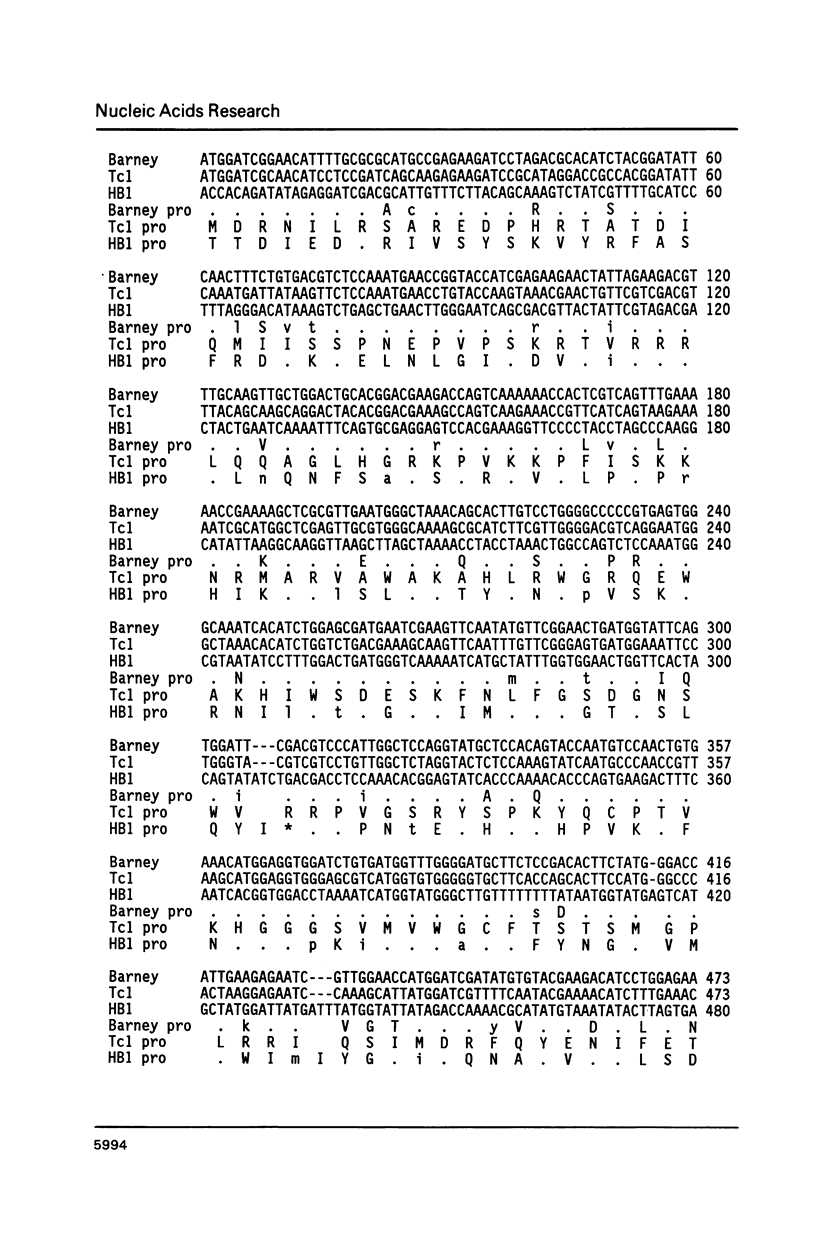
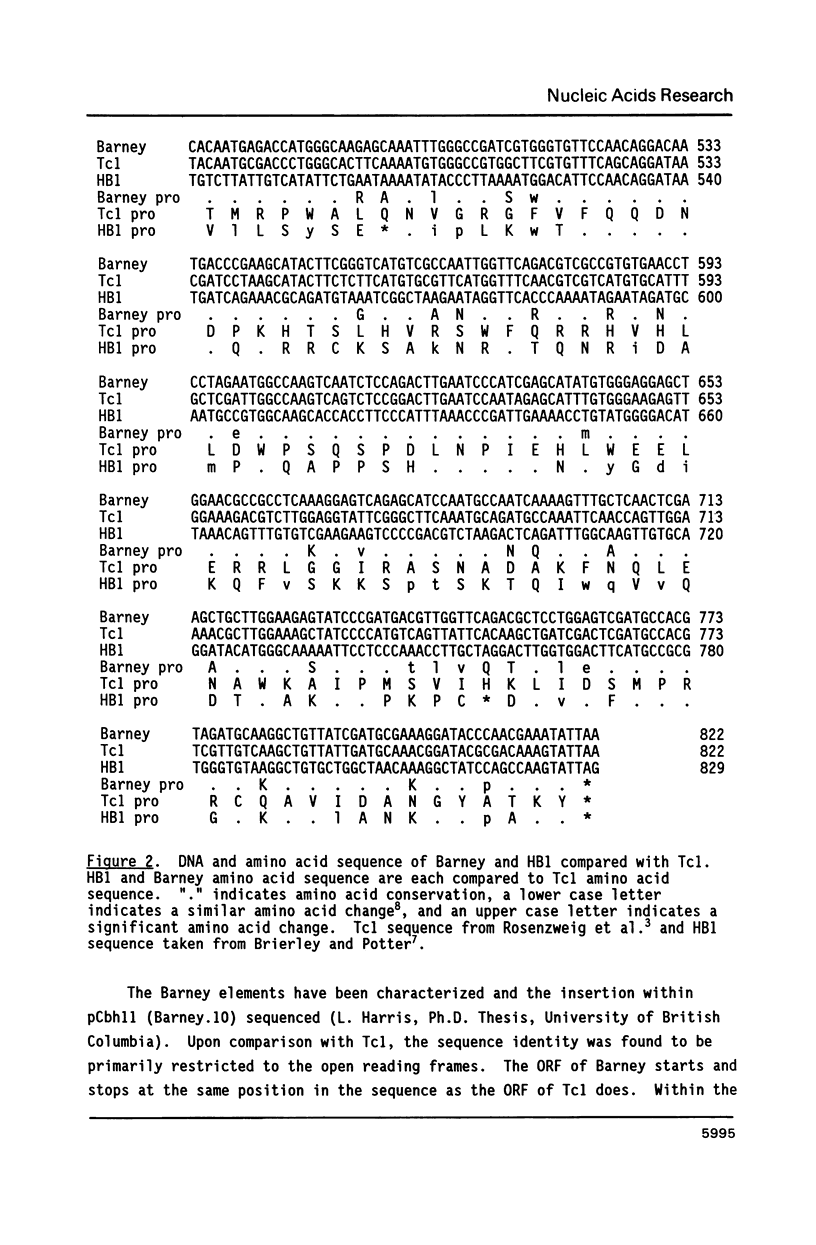
The Tc1-like transposable elements, originally described in Caenorhabditis elegans, have a much wider phylogenetic distribution than previously thought. In this paper, we demonstrate that Tc1 shares sequence identity in its open reading frame and terminal repeats with a new transposable element Barney (also known as TCb1-Transposon Caenorhabditis briggsae 1). Barney was detected and isolated by Tc1 hybridization from the closely related nematode species, Caenorhabditis briggsae. The conserved open reading frames of Tc1 and Barney share identity with a structurally similar family of elements named HB found in Drosophila melanogaster, after the introduction of 3 small centrally located deletions in HB1. These reading frames would code for proteins with 30% amino acid identity (42% when conservative changes are included). Tc1, Barney and HB1 contain highly conserved blocks of amino acids which are likely to be in the functional domains of the putative transposase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker R. F., Thompson D. V., Talbot D. R., Swanson J., Bennetzen J. L. Nucleotide sequence of the maize transposable element Mul. Nucleic Acids Res. 1984 Aug 10;12(15):5955–5967. doi: 10.1093/nar/12.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley H. L., Potter S. S. Distinct characteristics of loop sequences of two Drosophila foldback transposable elements. Nucleic Acids Res. 1985 Jan 25;13(2):485–500. doi: 10.1093/nar/13.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V., Rivin C., Walbot V. Stable non-mutator stocks of maize have sequences homologous to the Mu1 transposable element. Genetics. 1986 Nov;114(3):1007–1021. doi: 10.1093/genetics/114.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., Klass M. R., Hirsh D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L. High-frequency excision of transposable element Tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984 Mar;36(3):599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L., Ruan K. S., Katzenberg D. Evidence for a transposon in Caenorhabditis elegans. Cell. 1983 Jan;32(1):55–65. doi: 10.1016/0092-8674(83)90496-8. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Jacobson J. W., Medhora M. M., Hartl D. L. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L. W., Rosenzweig B., Hirsh D. Analysis of a transposable element in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3585–3589. doi: 10.1073/pnas.80.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D. G., Waterston R. H. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics. 1984 Dec;108(4):859–877. doi: 10.1093/genetics/108.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Rose A. M., Snutch T. P. Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans. Nature. 1984 Oct 4;311(5985):485–486. doi: 10.1038/311485a0. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Sequence of the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Jun 25;11(12):4201–4209. doi: 10.1093/nar/11.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan K., Emmons S. W. Extrachromosomal copies of transposon Tc1 in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4018–4022. doi: 10.1073/pnas.81.13.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck R. D., Macgaffey J. E., Beckendorf S. K. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986 Dec 20;5(13):3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Freeling M. An extrachromosomal form of the Mu transposons of maize. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4924–4928. doi: 10.1073/pnas.84.14.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L. P., Walbot V. Isolation and characterization of a 1.7-kb transposable element from a mutator line of maize. Genetics. 1987 Oct;117(2):297–307. doi: 10.1093/genetics/117.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]