SUMMARY
We analyzed the usage and consequences of alternative cleavage and polyadenylation (APA) in Drosophila melanogaster by using >1 billion reads of stranded mRNA-seq across a variety of dissected tissues. Beyond demonstrating that a majority of fly transcripts are subject to APA, we observed broad trends for 3′ untranslated region (UTR) shortening in the testis and lengthening in the central nervous system (CNS); the latter included hundreds of unannotated extensions ranging up to 18 kb. Extensive northern analyses validated the accumulation of full-length neural extended transcripts, and in situ hybridization indicated their spatial restriction to the CNS. Genes encoding RNA binding proteins (RBPs) and transcription factors were preferentially subject to 3′ UTR extensions. Motif analysis indicated enrichment of miRNA and RBP sites in the neural extensions, and their termini were enriched in canonical cis elements that promote cleavage and polyadenylation. Altogether, we reveal broad tissue-specific patterns of APA in Drosophila and transcripts with unprecedented 3′ UTR length in the nervous system.
INTRODUCTION
Alternative cleavage and polyadenylation (APA) has substantial impact on transcript diversity and function (Di Giammartino et al., 2011; Licatalosi and Darnell, 2010). APA can affect coding exons and protein sequence, but it more commonly affects the extent of 3′ untranslated region (UTR) sequence. 3′ UTRs harbor much of the cis-regulatory information for posttranscriptional regulation, including binding sites for microRNAs (miRNAs) and diverse RNA binding proteins (RBPs) (Flynt and Lai, 2008; St Johnston, 2005). Collectively, these can either positively or negatively regulate transcript stability or translational efficiency, as well as influence transcript localization. Consequently, shortening or lengthening of 3′ UTRs can substantially alter gene function across isoforms. For example, loss of distal 3′ UTR sequences allows certain oncogenes to evade repression by miRNAs, thereby potentiating their activity (Mayr and Bartel, 2009).
APA has recently been appreciated as a global phenomenon that can be broadly modulated under different cell conditions. This was originally inferred from analysis of cDNA libraries (Tian et al., 2005; Zhang et al., 2005), and examined much more deeply with the use of genome-wide techniques such as tiling microarrays (Ji et al., 2009; Ji and Tian, 2009; Sandberg et al., 2008), mRNA sequencing (RNA-seq) (Mangone et al., 2010; Ozsolak et al., 2010), and sequencing of transcript 3′ ends (Jan et al., 2011; Mangone et al., 2010; Shepard et al., 2011). Several trends emerged from such studies, including that cells proliferating upon T cell activation express shorter 3′ UTRs (Sandberg et al., 2008); that cell transformation may be correlated with 3′ UTR shortening independently of proliferation rate (Fu et al., 2011; Mayr and Bartel, 2009); that a general transition to shorter APA isoforms is observed during reprogramming of somatic cells into iPS cells (Ji and Tian, 2009); that global lengthening of 3′ UTRs occurs during mouse embryonic development and differentiation of C2C12 myocytes (Ji et al., 2009); reciprocally, that 3′ UTRs generally shorten during Caenorhabditis elegans development (Mangone et al., 2010); and that 3′ UTRs in mammalian neurons exhibit a broad trend for lengthening (Shepard et al., 2011).
To date, 3′ UTR diversity at the genome-wide scale has received relatively little attention in Drosophila melanogaster, as compared to other well-studied eukaryotes. In this study, we use strand-specific RNA-seq across a panel of dissected tissues to reveal broad usage of APA in Drosophila. Notably, we identify large cohorts of genes that exhibit 3′ UTR shortening in the testis and 3′ UTR lengthening in the central nervous system (CNS), and we provide extensive experimental confirmation of these APA events by using qPCR, northern analysis, and in situ hybridization. The usage of distal APA sites expands the transcript models of hundreds of neural genes and frequently generates extremely long 3′ UTRs, raising unanticipated complexity in their posttranscriptional regulation.
RESULTS
Analysis of D. melanogaster 3′ UTRs through Stranded mRNA Sequencing
As part of our efforts to annotate the function of every base in the Drosophila genome (modENCODE, 2010), we have conducted extensive tiling microarray and mRNA-sequencing studies (Cherbas et al., 2011; Graveley et al., 2011). Although powerful, these strategies were limited in that the transcribed strand of origin was not captured. In the current phase of the project, we switched to a stranded mRNA-seq protocol that preserves this information, and we generated libraries from 29 dissected tissues and 25 tissue culture cell lines (J.B.B. et al., unpublished data). Here, we analyze data from seven of these libraries to explore the diversity and dynamics of 3′ UTRs in Drosophila. These libraries include dissected larval and pupal CNS as well as adult female and male heads, ovaries, testis, and S2R+ cells, altogether comprising 1,071,975,003 uniquely mapped reads (Table 1).
Table 1.
Stranded RNA-Seq Library Read and Mapping Summaries
| Sample | Uniquely Mapped Reads: Replicate 1 |
Uniquely Mapped Reads: Replicate 2 |
Total Uniquely Mapped Reads |
modENCODE/NCBI-SRA Accession Numbers |
|---|---|---|---|---|
| Third instar larvae, CNS | 12,921,055 | 40,022,667 | 52,943,722 | modENCODE_3466 SRR070409, SRR070410 |
| Pupae, white pre-pupae + 2 days, CNS | 44,833,331 | 37,635,655 | 82,468,986 | modENCODE_3469 SRR100271, SRR070412 |
| Mated female, eclosion + 4 days, heads | 59,341,425 | 60,448,967 | 119,790,392 | modENCODE_3448 SRR070414, SRR070415 |
| Mated male, eclosion + 4 days, heads | 54,635,860 | 54,555,328 | 109,191,188 | modENCODE_3449 SRR070416, SRR070400 |
| Mated female, eclosion + 4 days, ovaries | 62,102,229 | 168,912,884 | 231,015,113 | modENCODE_3451 SRR070431, SRR100277, SRR100283 |
| Mated male, eclosion + 4 days, testes | 249,972,580 | 171,471,971 | 421,444,551 | modENCODE_3452 SRR070422, SRR350960, SRR070423, SRR100276, SRR350961 |
| S2-R+ | 55,121,051 | N/A | 55,121,051 | modENCODE_3487 SRR070279, SRR124149, SRR070266 |
| Total | 538,927,531 | 533,047,472 | 1,071,975,003 |
Details on RNA-seq data sets produced for this study. All the raw sequences reads and mapping files can be obtained under the designated accession numbers from the modENCODE Data Coordination Center (http://www.modencode.org/publications/dcc/index.shtml) and the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/).
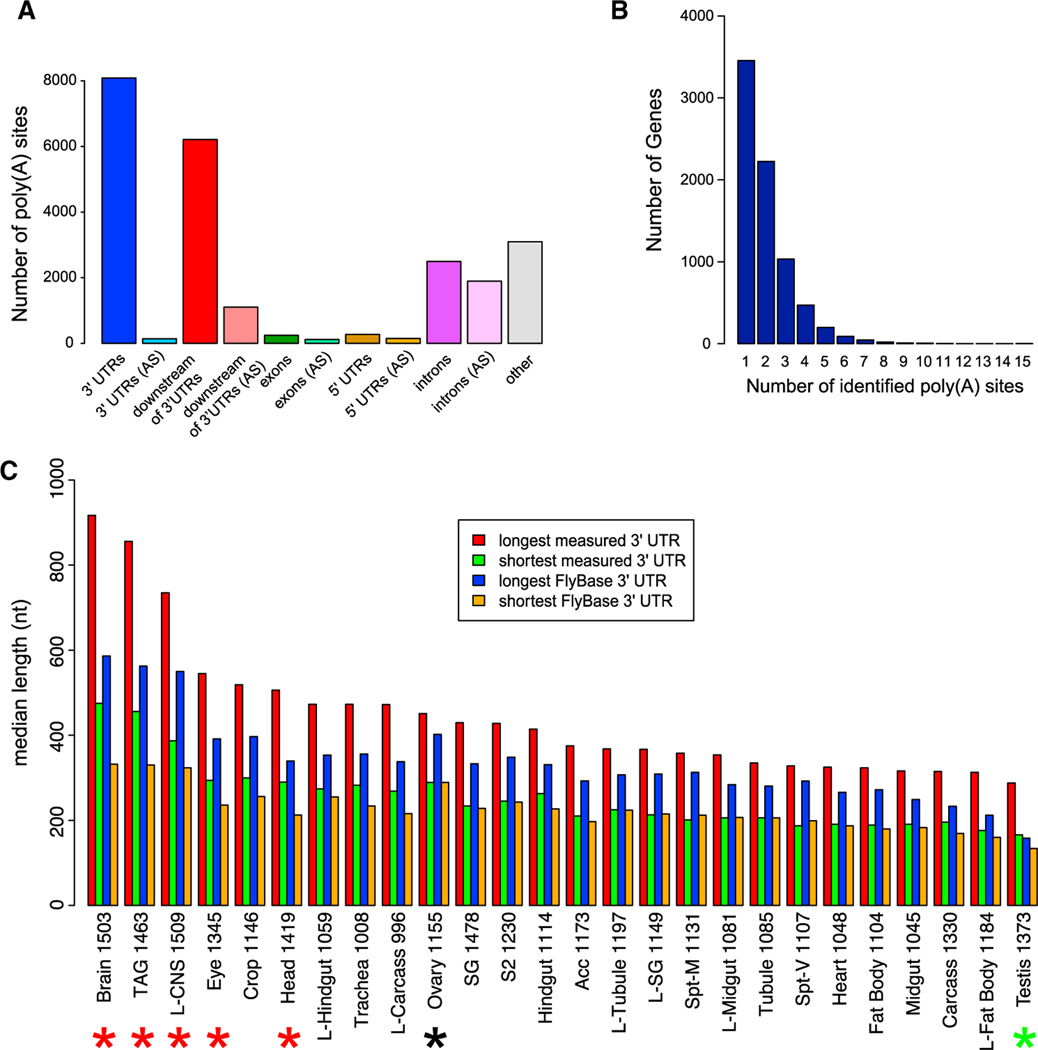
To gain further information on the precise transcript ends, we reamplified the 29 tissue libraries with the use of a 3′ primer containing six T residues and sequence complementary to the 3′ adaptor. This procedure enriched for poly(A)-spanning reads, which we initially identified as reads that terminated in ≥10 consecutive A residues that failed to align to the genome when untrimmed, yet aligned uniquely when the terminal A residues were removed. We examined these stranded RNA-seq data and the pooled poly(A)-spanning reads for evidence of APA with respect to the FlyBase gene models in release 5.32, which followed our last major transcriptome analysis (Graveley et al., 2011). After filtering out potential cases of oligo-dT priming to genomically encoded poly(A) tracts (see Experimental Procedures), we were left with 1,252,832 poly(A)-spanning reads, of which 85% were located within, or downstream of, annotated 3′ UTRs. Clusters of two or more overlapping reads were located either in or downstream of annotated 3′ UTRs ~65% of the time (Figure 1A). At this point we cannot formally attribute all of the downstream poly(A) clusters as bona fide 3′ UTR extensions; however, the strong bias for the downstream clusters to be on the sense strand of the upstream gene (Figure 1A) provided general support for this scenario. Altogether, our clusters of two or more poly(A)-spanning reads identified 14,297 putative polyadenylation sites in 7,562 genes, with 4,107 genes (54.3%) having more than one site (Figure 1B). Therefore, APA operates broadly to diversify the 3′ ends of Drosophila transcripts.
Figure 1. Poly(A)-Spanning RNA-Seq Reads Reveal Tissue-Specific Differences in 3′ UTR Length.
(A) Distribution of poly(A) sites, minimum two reads per cluster, with respect to gene annotations. AS, antisense.
(B) Distribution of 3′ UTR isoforms per gene. A cluster of two or more poly(A)-spanning reads downstream of a stop codon is classified as a poly(A)-supported 3′ end.
(C) Distribution of 3′ UTR median length (using minimum two reads per cluster) with respect to FlyAtlas gene classifications. For “longest measured 3′ UTR” and “shortest measured 3′ UTRs,” poly(A)-spanning reads downstream of annotated 3′ ends and before the adjacent gene, and reads upstream of the annotated 3′ end, but after the stop codon were attributed to that gene. Lengths of longest and shortest 3′ UTRs from the FlyBase annotations of the same genes are shown for comparison. Note that nervous system tissues (highlighted with the red asterisks) have the longest 3′ UTRs as predicted by the poly(A)-spanning reads, whereas testis has the shortest 3′ UTRs (green asterisk); ovaries (highlighted with a black asterisk) have 3′ UTRs of intermediate length. The numbers after the tissue indicate the number of genes in each category. Acc, male accessory gland; prefix “L”, larval tissue; SG, salivary gland; Spt-M, mated spermatheca; Spt-V, virgin spermatheca; S2, S2 cells; TAG, thoracicoabdominal ganglion.
We began to investigate the tissue specificity of 3′ UTR length variation and APA. We utilized the FlyAtlas expression database (Chintapalli et al., 2007) to compare 3′ UTR lengths, both for FlyBase annotations and for 3′ UTRs inferred from our poly(A)-spanning reads pooled from 29 poly(A) enriched RNA-seq libraries, of genes expressed in a variety of tissues (Figure 1C). Consistent with previous observations that annotated neural genes collectively exhibit longer 3′ UTRs relative to other Drosophila tissues (Stark et al., 2005), five out of six tissues with the longest median 3′ UTRs contained a high proportion of neurons (e.g., brain, larval CNS, and eye). In addition, data from poly(A)-spanning reads showed that these various neural tissues exhibited 3′ UTRs with median lengths that were 25%–40% longer than FlyBase gene models (Figure 1C, red asterisks). Thus, neural 3′ UTRs are in fact substantially longer than currently appreciated. Reciprocally, testis-expressed genes had the shortest median 3′ UTRs across all tissues (Figure 1C, green asterisk); ovaries expressed 3′ UTRs of intermediate length (Figure 1C, black asterisk). We selected neural tissues and testis for detailed experimental and computational analysis of tissue-specific alternative 3′ UTR patterns.
The Testis Transcriptome Is Strongly Biased for Usage of Proximal Poly(A) Sites
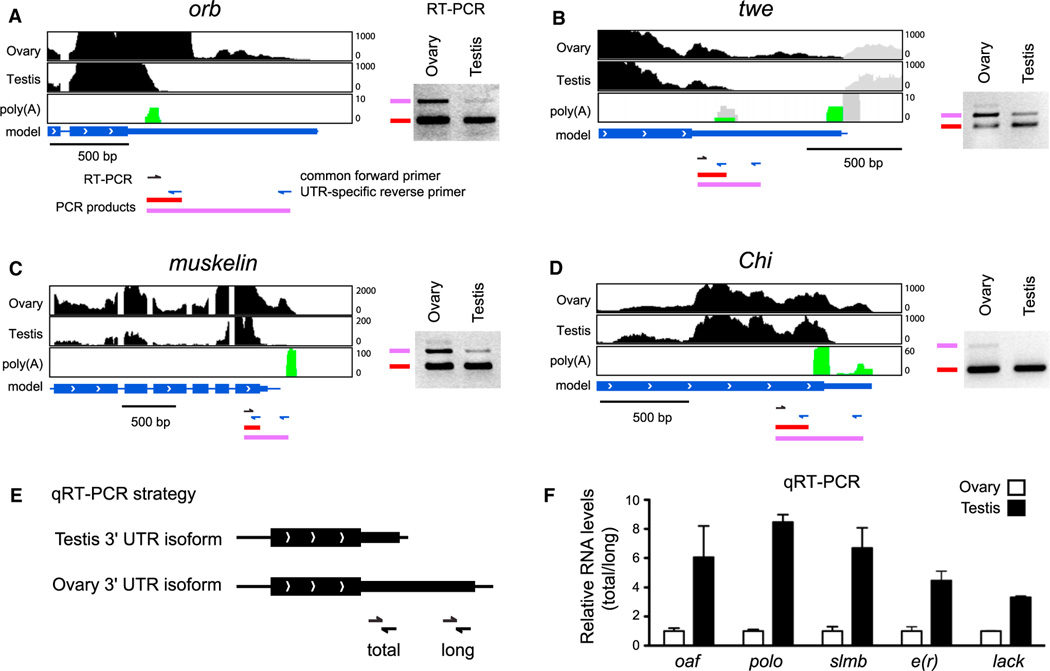
As the 3′ UTRs of testis-expressed transcripts had the shortest median length (using two or more poly(A)-spanning reads), we were interested to identify cases of APA in which a proximal site was utilized in testis. Analysis of the stranded RNA-seq data recovered 100 genes exhibiting 3′ transcript ends that were clearly shorter in testis compared to ovaries (Figure S1 and Table S1 available online). From the poly(A)-spanning reads pooled from all the tissue libraries, 47 of these genes had poly(A) support for the proximal 3′ end and 63 had poly(A) support for the distal 3′ end. Figure 2 illustrates typical examples of this phenomenon.
Figure 2. The Testis Transcriptome Is Biased toward Proximal Poly(A) Site Usage.
(A–D) RNA-seq tracks for representative genes expressed in both male and female gonads, that exhibit evidence for truncated 3′ UTRs in testis compared to ovary. Additional examples can be found in Figure S1. The black regions correspond to mapped RNA-seq reads from each tissue, green indicates poly(A)-spanning reads pooled from 29 poly(A)-enriched RNA-seq libraries corresponding to the strand on which the gene of interest is expressed. The gray regions in (B) correspond to reads from the neighboring gene transcribed on the opposite strand. In (A–D), RT-PCR was performed using a common forward primer (black arrows) and UTR-specific reverse primers (blue arrows). To the right of the RNA-seq data are ethidium bromide stained gels, in which lower molecular weight bands derive from both long and short 3′ UTR isoforms, whereas higher molecular weight bands are specific to long 3′ UTR isoforms. Note the reduced levels of distal APA isoforms in testis.
(E) Schematic for quantification of alternative length 3′ UTRs in ovary and testis by qRT-PCR.
(F) qRT-PCR demonstrates 3′ UTR shortening in testis compared to ovaries for oaf, polo, slmb, e(r), and lack. Relative RNA levels are presented as ratio of total/long. Data are represented as mean ± SEM.
To confirm differential expression of 3′ UTR isoforms in gonads, we performed RT-PCR with the use of a common proximal primer and two unique distal primers for each gene. These assays indicated preferred (Figures 2A–2C) or exclusive (Figure 2D) expression of transcripts using distal poly(A) sites in ovary, relative to testis. In addition, we performed qRT-PCR experiments for five 3′ UTR shortening candidates with the use of primers that amplify all 3′ UTR species for a given gene (total) or only the longer 3′ UTR species (long) (Figure 2E). Data presented as a ratio of total/long isoforms demonstrate 3- to 8-fold increased expression of the short 3′ UTR species in testis (Figure 2F). Altogether, these analyses show a broad trend for usage of proximal poly(A) sites in the testis, resulting in shortened 3′ UTR isoforms.
The Neural Transcriptome Is Strongly Biased for Usage of Novel Distal Poly(A) Sites
We observed a strikingly converse trend in RNA-seq data from larval and pupal CNS and adult head, whose median 3′ UTR lengths were much longer than for genes expressed in all other tissues (Figure 1C). We focused our analysis on 3′ UTR extensions resulting from APA events in the 3′ UTR (UTR-APA) and not from alternative cleavage events located in internal introns or exons. We identified 66 transcripts, contained within FlyBase 5.32 gene models, for which CNS/head RNA-seq evidence clearly demonstrated usage of distal PAS relative to other tissues.
Manual browsing revealed extensive transcribed regions downstream of current gene annotations in neural tissues but not in non-neural tissues. We therefore systematically searched libraries from larval and pupal CNS and adult head for 3′ UTR extensions supported by continuous RNA-seq evidence, distal to annotated FlyBase models. This yielded 317 additional genes exhibiting UTR-APA, with longer 3′ UTR species in the nervous system (Table S1). These extensions had a significant impact on the catalog of exonic sequence in Drosophila, collectively adding >760 kb of novel sequence to the transcriptome.
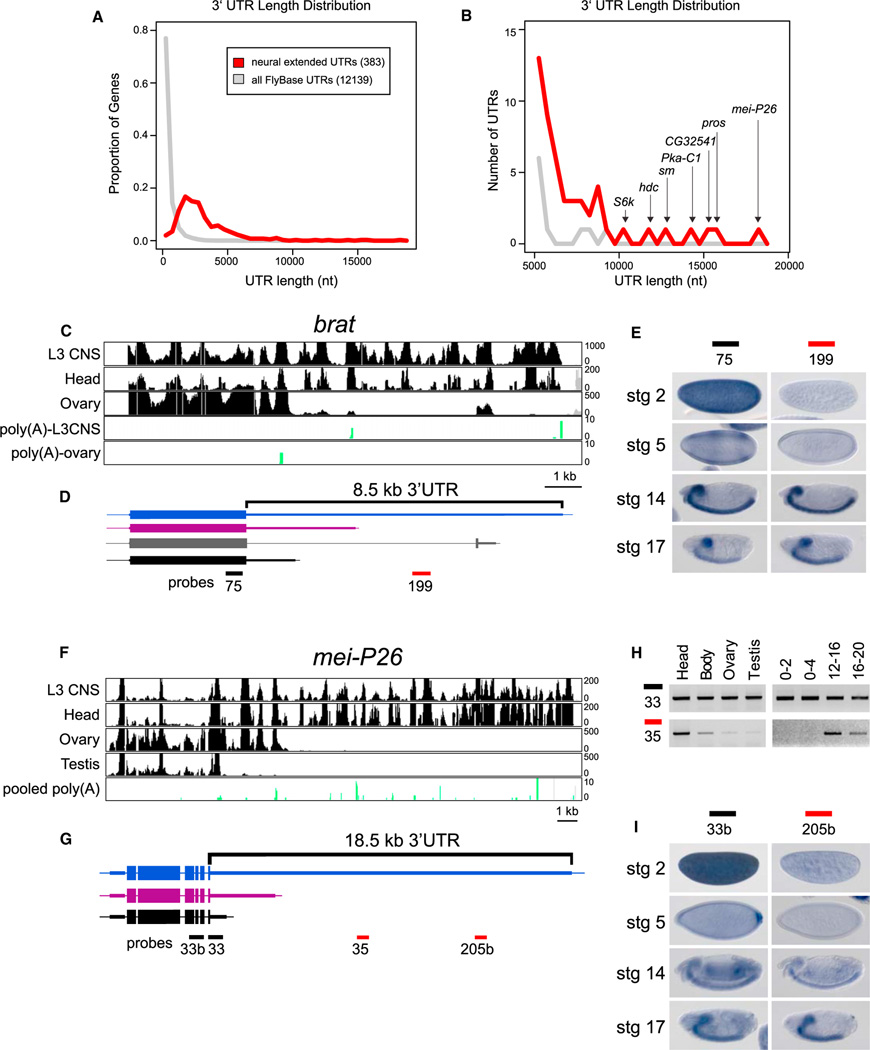
In total, we recognized 383 genes exhibiting neural-specific 3′ UTR extensions. Comparison of the 3′ UTR lengths of these 383 neural extended transcripts with FlyBase 5.32 annotations highlighted that this set of genes was rich in exceptionally long 3′ UTRs (Figures 3A and 3B). Indeed, only a handful of known Drosophila transcripts exhibit 3′ UTRs in excess of 5 kb (disregarding artifactual annotations, see Extended Experimental Procedures), whereas 51 of our newly annotated 3′ UTRs surpass this limit. We therefore sought experimental confirmation of these unusually extended 3′ UTRs.
Figure 3. Unusually Long 3′ UTR Extensions in Neural Genes.
(A) Distribution of 3′ UTR lengths of 383 transcripts that exhibit neural lengthening, relative to the longest annotated 3′ UTRs of all FlyBase 5.32 genes.
(B) Focusing on the tail of the distribution, the neural 3′ UTR extensions comprise most of the longest 3′ UTRs in the Drosophila transcriptome.
(C and D) RNA-seq tracks from indicated tissues (C) and transcript models for brat (D). A splice variant that alters the last few amino acids of Brat was apparent from RNA-seq data (gray), but this isoform was not differentially expressed.
(E) In situ hybridization revealed that both proximal and distal probes detect CNS expression in germband retracted embryos and late stage embryos, but that the proximal probe alone detects maternal deposition and blastoderm expression in two stripes along the anterior-posterior axis.
(F and G) RNA-seq tracks from indicated tissues (F) and gene models for mei-P26 (G).
(H) Semiquantitative RT-PCR indicates that RNA corresponding to the 3′ end of the coding sequence and proximal 3′ UTR can be readily detected in all tissues and embryonic time-points examined, whereas an amplicon ~8 kb into the 3′ UTR was predominantly detected in heads and late stage embryos.
(I) In situ hybridization reveals that both proximal and distal probes detect late stage expression in the brain and ventral nerve cord, but that the proximal probe alone detects maternal deposition at stage 2, posterior expression at stage 5, and ubiquitous expression at stage 14.
The TRIM-NHL family member brat exhibited extensive transcription downstream of the annotated gene model in neural tissues, comprising an ~8.5 kb 3′ UTR (Figures 3C and 3D). In addition, distinctive accumulation of poly(A)-spanning reads was observed in the larval CNS compared to ovary, with the former terminating at intermediate and distal sites and the latter at a proximal site (Figure 3C). During embryogenesis, the universal brat probe detected maternal deposition of brat and two broad stripes of zygotic expression, whereas later stage expression was confined to the brain and ventral nerve cord (VNC) (Figure 3E). In contrast, the extended isoform-specific probe failed to detect maternal or early zygotic transcripts, and hybridized exclusively to CNS transcripts at later stages (Figure 3E). Therefore, the proximal APA isoform of brat exhibits an expression pattern that is distinct from its distal APA isoform.
A more extreme example of an extended 3′ UTR is illustrated by mei-P26, which also encodes a TRIM-NHL protein. Stranded RNA-seq showed tremendous variation in tissue-specific 3′ UTR lengths, with a short 3′ UTR in testis, an intermediate 3′ UTR length in ovary and an ~18.5 kb UTR in neural samples (Figures 3F and 3G). RT-PCR analysis confirmed that the extended isoform is strongly expressed in the head samples relative to body, ovary, and testis (Figure 3H). In addition, the extended isoform appeared only in embryos 12 hr of age and older (Figure 3H). Interpretation of this temporal pattern required spatial analysis (Figure 3I). A probe detecting mei-P26 coding sequence revealed maternally deposited transcripts and posterior accumulation at stage 5. At stage 14, staining was largely ubiquitous, with enrichment in the developing brain and VNC becoming apparent by stage 17. In contrast, a probe 13 kb distal lacked maternal or early embryonic staining, and showed exclusive CNS expression in late stage embryos. Additional in situ probes designed against proximal or distal regions of the mei-P26 3′ UTR confirmed these distinctive spatial patterns (Figures S2A and S2B). Thus, the temporal accumulation of the extended mei-P26 APA isoform was a consequence of its spatial expression in the maturing nervous system. Similar results were observed with several other genes with apparent developmental lengthening (e.g., shep, heph, and msi, Figures S2C–S2J), for which a temporal trend of 3′ UTR lengthening was attributable to the expression of 3′ UTR extended isoforms in the nervous system. These data highlight the need to coordinate expression patterns determined in whole animals with knowledge of tissue-specific gene expression.
Northern Analysis Confirms Exceptionally Long APA Isoforms as Bona Fide Transcripts
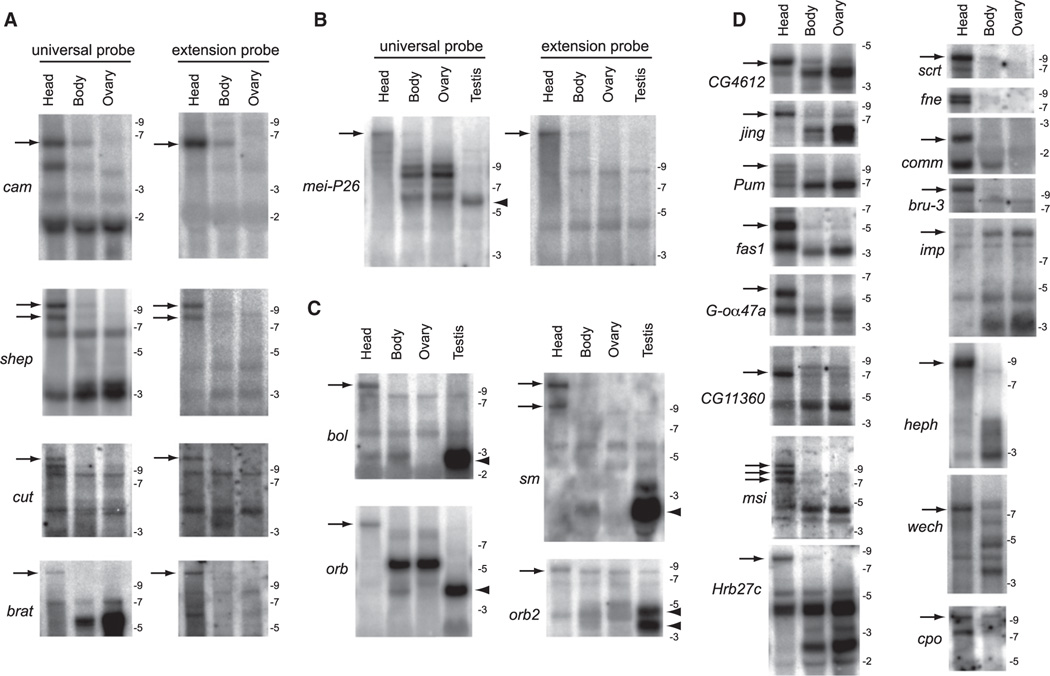
The RNA-seq, RT-PCR, and in situ data do not formally prove 3′ UTR extensions of stable transcripts, as opposed to transient or unstable RNA species, products of runaway transcription or improper termination, or distinct transcripts downstream of protein-coding genes (Mercer et al., 2011; Ponjavic et al., 2009). We distinguished these possibilities using northern analysis, which is also uniquely suited for assessing the relative accumulation of different full-length isoforms. We first compared the signals of universal probes (that should hybridize to all 3′ UTR isoforms) with extension probes specific to distal APA isoforms for cam, shep, cut, and brat (Figure 4A) and mei-P26 (Figure 4B). In all cases tested, the extension probes detected a subset of species detected by the universal probe, and these always comprised longer isoforms that were strongly enriched in heads. These data provide strict evidence of 3′ UTR extension isoforms that are contiguous with their neighboring protein-coding mRNA annotations. Many of these exceeded the longest RNA size standards, including the existence of full-length mei-P26 transcripts (Figure 4B) estimated from RNA-seq data to be some 23 kb in length, of which >18 kb was 3′ UTR.
Figure 4. Northern Analysis Validates Stable Transcripts Corresponding to Strongly Distal APA Isoforms in Drosophila Heads.
(A) Comparison of proximal “universal” probes and distal “extension” probes for a selection of exceptionally long inferred APA isoforms. In each case, a similarly sized band(s) is detected only in head samples with the paired probes (arrows), demonstrating that they connect a common 3′ UTR-extended transcript. In many cases, the sizes of these discrete lengthened transcripts far exceed the largest molecular weight marker on the RNA ladder (9 kb).
(B) Comparison of proximal universal probes and distal extension probes for mei-P26. Note the 3′ UTR lengthening in head and 3′ UTR shortening in testis relative to body/ovary samples.
(C) Examples of APA transcripts exhibiting neural extension (arrows) and testis shortening (arrowheads), often with intermediate-sized transcripts in body and/or ovary.
(D) A broad selection of other transcripts that exhibit 3′ UTR-extended isoforms predominantly or exclusively in head (arrows), with the exception of imp. For bands that were outside the range of the RNA ladder, the estimated size of the longest 3′ UTR isoform in head was calculated based on RNA-seq coverage from head library and existing RefSeq mRNA annotations: shep, 9.4 kb; cut, 11.6 kb; brat, 11.7 kb; mei-P26, 22.9kb; bol, 10.9 kb; orb, 8.9 kb; sm, 12.7 kb; pum, 10.1 kb; msi, 10.9 kb; scrt, 9.1 kb; wech, 7.6 kb; imp, 11.8 kb; cpo, 9.3 kb; bru-3, 9.6 kb.
Additional northern assays using universal probes provide broad support for extended APA isoforms that are enriched or indeed restricted to heads (Figures 4C and 4D). Interestingly, several genes exhibited both 3′ UTR lengthening in head and shortening in testis, relative to intermediate-sized isoforms expressed in body and/or ovary (e.g., mei-P26, bol, sm, orb, and orb2, Figures 4B and 4C). Perusal of our APA lists revealed 23 transcripts that exhibit such dual patterns of CNS and testis APA (Table S1C). Altogether, these extensive northern analyses provide a compelling view of APA dynamics leading to the accumulation of extraordinarily long APA isoforms in the Drosophila nervous system.
Predominant CNS Restriction of Neural Distal APA Isoforms
We followed up these northern analyses with additional in situ hybridization studies. As before, we compared proximal probes that would detect all 3′ UTR mRNA isoforms (universal) with extension-specific probes specific to distal APA regions. In some cases, both probes detected similar expression patterns in the CNS (e.g., khc-73, Figure 5A; CG4612 and fas1, Figure S3). However, we also observed spatially discrepant patterns of expression of paired probes. Consistent with the RNA-seq data, the extended probe often detected transcripts in a subset of the tissues for which expression was also detected for the universal probe. For example, the universal and extended probes for bru-3 detected strong expression in the brain and VNC but strong visceral muscle primordium staining was only observed with the universal probe (Figure 5B).
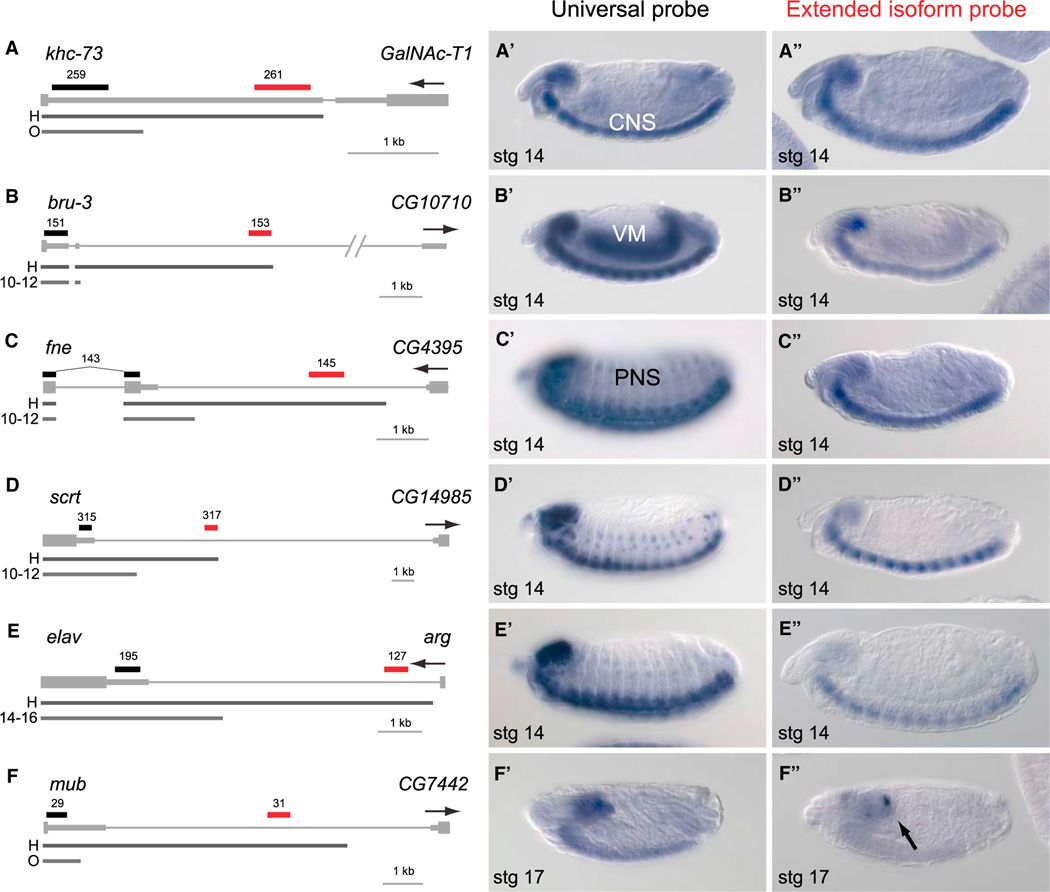
Figure 5. In Situ Hybridization Reveals Distinctive CNS Expression of Distal APA Isoforms.
Schematics to the left include gene models with 3′ UTR extensions in head RNA-seq data relative to another tissue. H, head; O, ovary; 10–12 and 12–14, 10–12 and 12–14 hr embryos. The locations of proximal (black) and distal (red) in situ probes are indicated. The gene of interest is displayed in a plus strand orientation, and the name and orientation of the neighboring downstream gene is indicated. Panels with single prime labels show in situ hybridization patterns detected with a universal probe designed to detect all transcripts from the gene of interest. Panels with double prime labels show in situ hybridization patterns of probes specific for the extended isoforms.
(A) khc-73 illustrates a gene with CNS staining detected with both universal and extended probes.
(B) The universal bru-3 probe stains visceral muscle (VM) and CNS, whereas its extension probe stains only CNS.
(C–E) fne (C), scrt (D), and elav (E) are pan-neuronally expressed in the peripheral nervous system(PNS) and CNS, yet their distal APA isoformsare specific to the CNS.
(F) mub is an example of a neural transcript where the extension probe specifically detects expression in subregions of the embryonic brain (arrow) compared to the universal probe that detects expression throughout the embryonic brain and CNS.
Curiously, in every case the distal probes detected transcripts in the CNS, and staining was often exclusive to the CNS (Figure 5 and Figure S3). This was particularly notable for a set of known pan-neural (CNS and PNS) transcripts, for which we observed distal APA isoforms largely or completely restricted to the CNS. Examples of these genes included fne, scrt, elav (Figures 5C–5E), and cut (Figure S3). In the case of the CNS-expressed gene mub, we observed that its extended 3′ UTR isoform was expressed in a subdomain of the brain (Figure 5F). The repeated observation of pan-neural APA transcripts with distinct CNS-restricted distal APA isoforms suggests that the mechanism underlying these unusually long 3′ UTR extensions is not simply neural-specific, but biased to the CNS relative to the PNS (Figure 5).
Enrichment of Nucleic Acid-Binding Proteins among Neural Genes Exhibiting Distal APA
We tested for enrichments of gene ontology annotations among the 383 neural-extended transcripts. Perhaps not surprisingly, this set of genes was highly enriched for biological processes relating to neural development or neural function (Table S2A). Strikingly though, among molecular function terms, the highest statistical enrichments observed (not including the generic categories “binding” and “protein binding”) concerned various classes of nucleic acid binding proteins. In particular, sequence-specific transcription factors were enriched at a p value of 2.68E-08, and mRNA binding was enriched at a p value of 5.77E-06. Other categories of strongly enriched molecular functions included kinases (1.96E-06) and signal transduction components (3.31E-05) (Table S2A). We did not observe a reciprocal coherence of the genes subject to utilization of proximal APA sites in testis, because no molecular function terms were enriched among the 100 transcripts in this cohort (Table S2B). In summary, we observed several classes of genes with regulatory functions preferentially subject to 3′ UTR extensions in the nervous system.
Quality of Poly(A) Sites and Local Conservation of Alternative Transcript Termini Analysis
We performed motif analysis on sequences surrounding alternative 3′ ends of CNS APA (Figure 6A) and testis APA (Figure 6B) transcripts, for which poly(A)-spanning reads pooled from all 29 samples described above definitively marked the site of cleavage and polyadenylation. The precise demarcation of transcript ends using the poly(A)-spanning reads enabled us to search for cis elements potentially interacting with the poly(A) machinery. De-novo searches identified the canonical AAUAAA PAS upstream of poly(A) sites, and a G/U rich sequence downstream of (the distal) poly(A) sites (Figure S4) that resembled the GU-rich downstream sequence element (DSE), known to increase the efficiency of 3′ end processing in mammalian cells via interaction with CstF-64 (MacDonald et al., 1994). We also observed a degenerate A-rich motif enriched mostly upstream of poly(A) sites (Figure S4). Because no motifs were found that were clearly distinct from known poly(A)-associated elements (Hu et al., 2005; Ozsolak et al., 2010), we proceeded to analyze the canonical and variant polyadenylation signals and the inferred DSE motif in more detail.
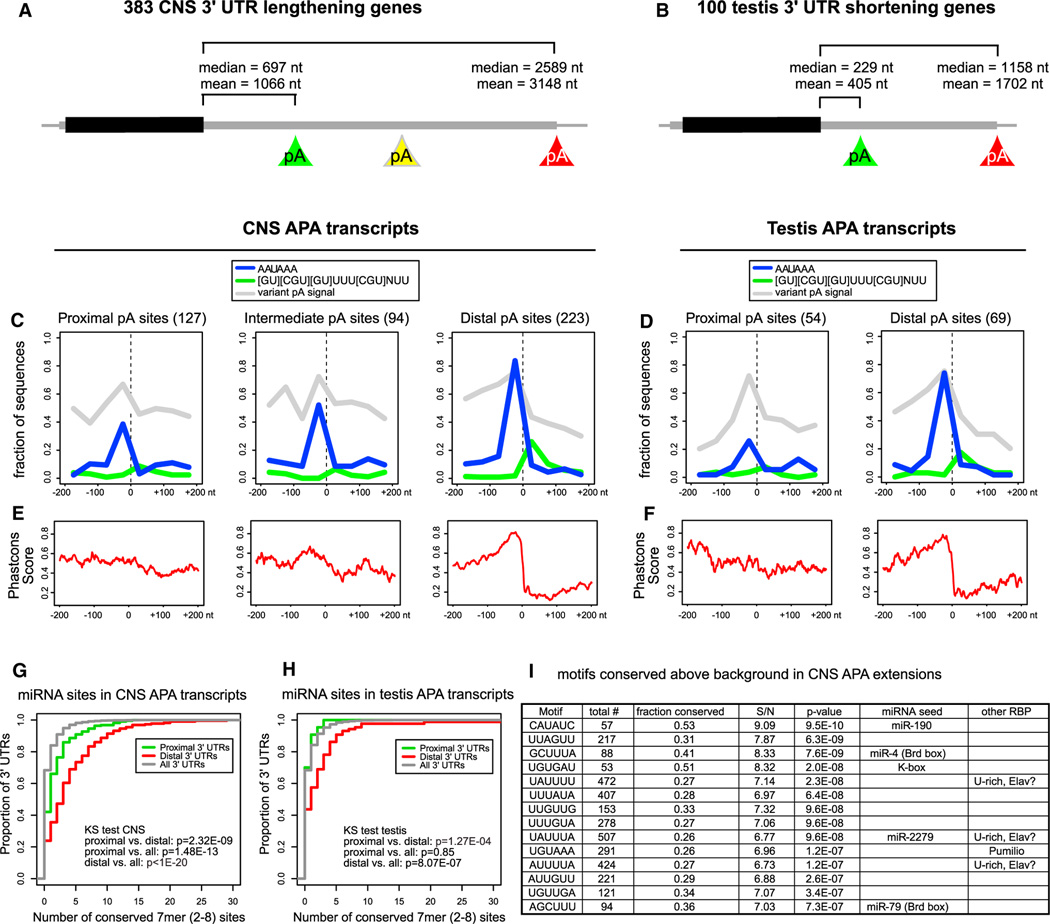
Figure 6. Quality and Conservation of Alternative Poly(A) Sites and miRNA Sites.
(A and B) Average 3′ UTR lengths of the proximal and distal transcript isoforms for genes exhibiting CNS 3′ UTR lengthening (A) and testis 3′ UTR shortening (B).
(C and D) Sequence motifs involved in polyadenylation around poly(A) sites identified by poly(A)-spanning reads: Canonical PAS (in blue), GU-rich DSE (in green) and variant PAS (in gray). (C) For CNS extended genes, distal sites are enriched for the canonical PAS compared to proximal sites (Fisher’s exact test p value = 4.9E-16), whereas the frequency at the intermediate poly(A) sites lies in between. The distal poly(A) sites are also enriched for the DSE motif, compared to the proximal and intermediate poly(A) sites (Fisher’s exact test p value = 7.2E-4). All poly(A) sites show similar collective levels of variant PAS; see also Figure S5 for analysis of individual PAS variants. (D) Testis APA transcripts display similar patterns of motif occurrence and conservation as the CNS transcripts (enrichment of AAUAAA in distal versus proximal sites, Fisher’s exact test p value = 2.6E-4).
(E) PhastCons conservation scores in the vicinity of poly(A) sites. Proximal and intermediate CNS poly(A) sites have intermediate conservation levels whereas distal sites have a region of high conservation just upstream of the poly(A) site and a region of low conservation downstream of the poly(A) site.
(F) Proximal poly(A) sites in testis transcripts have intermediate conservation levels whereas the distal sites have a region of high conservation just upstream of the poly(A) site and region of low conservation downstream of the poly(A) site.
(G) Cumulative distribution of numbers of miRNA target sites across all Drosophila 3′ UTRs, compared with the proximal and distal APA 3′ UTR variants of the 383 transcripts exhibiting neural lengthening. The distal extensions bear significantly more miRNA binding sites than Drosophila 3′ UTRs do in general.
(H) Cumulative distribution of miRNA target site numbers in the 100 testis APA transcripts.
(I) Motifs conserved above background in 3′ UTR extensions of neural transcripts. Shown are the top 6-mer motifs conserved above background in greater than or equal to seven Drosophilid species among the 383 3′ UTR extensions of neural transcripts. Many sites correspond to miRNA seeds or neural RBP sites (e.g., Pumilio and U-rich sequences that may potentially include Elav binding sites). A complete list of 6-mer and 7-mer motifs conserved above background are shown in Table S3. S/N, signal/noise ratio.
Upon comparing proximal, intermediate, and distal isoforms of our collection of genes with neural extensions, we observed a progressively increasing fraction bearing the canonical AAUAAA polyadenylation signal just upstream of poly(A) sites (Figure 6C). In contrast, variant PAS (Figure S5) were collectively equally represented upstream of these various cohorts of transcript ends (Figure 6C). We observed that distal neural poly(A) sites contained substantially higher frequency of DSE motifs, relative to intermediate or proximal isoforms. We observed a similar trend in the 3′ termini of genes that exhibit testis shortening, with the distal poly(A) sites exhibiting a much higher frequency of canonical AAUAAA PAS and DSE motifs (Figure 6D).
We also assessed the levels of conservation at the various categories of poly(A) sites, using PhastCons scores (http://genome.ucsc.edu). This revealed an intermediate level of conservation (0.5–0.6) in the proximity of proximal poly(A) sites, that was relatively similar upstream and downstream of the poly(A) sites. In contrast, the levels of conservation in the vicinity of distal neural poly(A) sites rose sharply in the preceding ~50 bp, peaking at ~0.8 at the poly(A) sites, and then rapidly dropped to background levels of 0.2 (Figure 6E). This suggests that the regions in the immediate upstream vicinity of distal neural poly(A) sites are selected for sequences that mediate highly efficient cleavage and polyadenylation. A similar trend was observed in the vicinity of distal poly(A) sites of transcripts that were preferentially shortened in testis (Figure 6F).
Impact of Neural 3′ UTR Extensions on Posttranscriptional Regulation
We sought to infer the functional impact of tissue-biased patterns of APA in Drosophila. One strategy was to investigate the frequency of miRNA binding sites in the proximal versus distal portions of APA regulated transcripts. For each category of 3′ UTR we cataloged the number of conserved seed matches (two to eight 7-mer sites) for miRNAs conserved between D. melanogaster and D. pseudoobscura (Ruby et al., 2007). Cumulative distribution plots showed that proximal regions of CNS APA transcripts carried higher numbers of conserved miRNA sites than observed with all Drosophila 3′ UTRs (Figure 6G). Kolmogorov-Smirnov (KS) tests demonstrated significantly higher numbers of miRNA binding sites in distal versus proximal regions of the CNS APA transcripts. Analysis of the testis APA transcripts showed that the proximal 3′ UTRs utilized in testis had similar numbers of miRNA binding sites relative to all Drosophila 3′ UTRs, but the distal regions of these APA transcripts exhibited significantly more miRNA binding sites (Figure 6H). A simple interpretation is that APA brings neural transcripts with extended 3′ UTRs under posttranscriptional control unique to the CNS, whereas APA spares testis transcripts with shortened 3′ UTRs from posttranscriptional control that applies to longer isoforms expressed outside the testis.
We next employed an unbiased strategy to ask what motifs are most preferentially conserved in the extended portions of neural distal APA transcripts. We assessed all 6-mers or 7-mers for conservation in the 383 neural 3′ UTR extensions, above a background binomial distribution of control motifs with similar occurrence and GC content. Interestingly, many of the most-conserved motifs corresponded to miRNA binding sites, including those of miR-190, K box miRNAs (miR-2/11/13, etc.), and Brd box miRNAs (miR-4/79). Another highly conserved motif corresponds to the binding site for Pumilio, and many conserved U-rich motifs potentially including Elav binding sites (Figure 6I). Additional miRNA seeds were observed among less-conserved (but still significantly-conserved) motifs (Table S3).
Overall, the logic of our observed UTR-APA examples appears to bring neural extended transcripts under the control of neural post-transcriptional regulatory machinery, because miR-190 is strongly enriched in adult head relative to body (Ruby et al., 2007), a cluster of miR-2/13 miRNAs is specifically expressed in the CNS (Aboobaker et al., 2005), Brd box miRNAs are known to regulate nervous system development (Lai and Posakony, 1997; Lai et al., 2005), and Pumilio and Elav are well known as regulatory RNA binding proteins in the nervous system (Menon et al., 2004; Soller and White, 2003).
DISCUSSION
Toward a More Complete Description of the Drosophila Transcriptome
One of the charges of the modENCODE project is the comprehensive characterization of the fly and worm transcriptomes. Although we recently analyzed 3.5 billion RNA-seq reads spanning Drosophila development (Graveley et al., 2011), our new libraries add substantially to the catalog of genic regions in this species. We confidently identify 317 transcripts with previously unannotated 3′ UTR extensions in the nervous system, comprising >760 kb of novel 3′ UTR sequence. The large number of transcript models affected was surprising, given that the nervous system is historically one of the more well-studied tissues in Drosophila. Equally unexpected was the sheer length of many neural distal APA isoforms. A number of them extend more than 10 kb, longer than virtually all other known Drosophila transcripts, and we validated many of these as stable full-length transcripts using northern analysis.
These comprise a conservative annotation of 3′ UTR variation, as there were additional instances of extended transcription for which the bounds and continuity could not be confidently judged. In addition, we observed orphan poly(A) sites downstream of 3′ UTRs that were not clearly associated with RNA-seq evidence. These may include transcripts with low or restricted expression in the nervous system. Reciprocally, our analysis of 3′ UTR shortening focused on APA that differed between male and female gonads, and did not include genes that were not expressed in both testis and ovary.
Beyond these APA isoforms, the datasets described herein comprise a valuable resource for furthering the annotation of the transcriptome. In particular, the dissected tissues enrich for transcripts that are rare in whole animals, and the stranded nature of the data help distinguish closely apposed transcription units, especially when they are produced from opposite strands. A fuller accounting of novel transcribed regions revealed by these data will be reported elsewhere (J.B.B. et al., unpublished data).
Tissue-Biased Features of Drosophila APA and Comparison with Other Model Systems
Our collection of >1 million poly(A)-spanning reads provides the largest resource of alternative 3′ ends in Drosophila to date, and provides direct evidence for APA events in more than 50% of detected Drosophila genes. Among this broad palette of APA transcripts, we discovered trends for shortening in the testis and lengthening in the CNS. Combined approaches of tissue-specific RNA-seq and in situ hybridization were important for interpreting these phenomena. For example, we observed an apparent trend for 3′ UTR lengthening during development in Drosophila (e.g., Figure S2), possibly concordant with the lengthening of 3′ UTRs observed during mouse embryonic development (Ji et al., 2009). However, our analysis reveals that such distal APA usage is broadly accounted for by the CNS 3′ UTR extensions. Therefore in Drosophila, the apparent developmental regulation of APA is actually due to the tissue specificity of this process, with the nervous system being present only in later but not earlier embryonic development. This highlights that interpretation of trends from temporal progression should be coordinated with knowledge of tissue development.
While this work was in preparation, Hilgers et al., (2011) used tiling microarrays to report on developmentally regulated 3′ UTR lengthening in Drosophila. Their analysis identified 30 genes with long zygotic 3′ UTR extensions, 15 of which were not annotated, and several of which were shown to be neural-specific. Our tissue-specific RNA-seq data broadly extend these findings to 383 genes with 3′ UTR extensions in head versus other tissues (28 of which were also identified by Hilgers et al. [2011]). As well, our northern data provide first evidence that these constitute bona fide 3′ UTR extensions of upstream coding sequences that accumulate as stable full-length transcripts.
These findings in the Drosophila CNS raise comparisons with a recent report that many mammalian long noncoding RNAs map downstream of neural transcripts, comprising coexpressed pairs in the brain (Ponjavic et al., 2009). In that study, the existence of 3′ UTR and downstream CAGE tags were taken as part of the evidence of independently transcribed ncRNAs, and RT-PCR tests yielded negative evidence of connectivity between the annotated neural mRNAs and their downstream ncRNAs. Such 3′ UTR CAGE tags have also been observed in Drosophila (Hoskins et al., 2011; Mercer et al., 2011), but their functional significance is not yet known. Indeed, many of the genes that we analyzed are associated with such 3′ UTR CAGE tags, yet northern analysis did not reveal the stable accumulation of any as distinct transcripts. Instead, in all cases we observed only transcripts corresponding to 3′ UTR extensions of upstream mRNAs (Figure 4 and Figure S6). It may be informative to assay for the existence of longer transcripts contiguous with protein coding mRNAs in the mammalian brain using northern analysis.
Our observations in Drosophila build upon other reports of analogous phenomena in vertebrates. For example, analysis of murine ESTs showed a trend of shortened 3′ UTRs in spermatogenesis that correlated with reduced usage of canonical AAUAAA signals (Liu et al., 2007). Reciprocally, microarray analysis and deep sequencing of 3′ ends in mouse and human tissues showed that brain and nervous system were among the tissues that tended to favor distal PAS usage (Sandberg et al., 2008; Shepard et al., 2011; Zhang et al., 2005). The mechanistic bases of these tissue-specific APA trends are poorly understood, but our finding that these trends are conserved in Drosophila indicates that this genetic system will be valuable for dissecting these processes.
Biological Significance of Tissue-Specific APA in Drosophila
In the nervous system, the expression of 3′ UTR lengthened isoforms subjects these genes to spatially restricted posttranscriptional control. We show this to apply to many hundreds of transcripts, and find that binding sites for neural miRNAs and neural RNA binding proteins are among the most highly conserved motifs within these extensions. Certainly miRNA-mediated control might serve to restrict transcript function, potentially in the context of local translation or transcript recycling in response to environmental cues and neural activity. However, neural 3′ UTR extensions may not solely confer downregulation. A distal APA variant of mammalian brain-derived neurotrophic factor (BDNF), but not its proximal APA variant, localizes to dendrites where it plays a role in long term potentiation (An et al., 2008). Moreover, Drosophila polo undergoes APA, in which the distal isoform is required to support efficient Polo translation (Pinto et al., 2011); thus, 3′ UTR extensions can promote translation. Finally, it is worth considering whether the considerable real estate within these neural 3′ UTR extensions may serve structural or scaffolding functions, or perhaps act as “sponges” that attract miRNA or RBP complexes.
We are intrigued by the fact that among the network of Drosophila genes exhibiting 3′ UTR lengthening in the nervous system, the top-enriched molecular functions are transcription factors and RBPs. This may imply special needs to regulate these key regulatory molecules in the nervous system. Interestingly, several of the RBPs that are subject to extraordinarily lengthened 3′ UTRs are themselves involved in 3′ UTR determination and/or miRNA-mediated regulation (e.g., elav, mei-P26, brat, pumilio, ago1). This raises the possibility of a complex network of auto- and cross-regulating posttranscriptional regulatory factors in the CNS. Recent technical advances in identifying RBP interactions with RNA elements such as PAR-CLIP and HITS-CLIP (Hafner et al., 2010; Licatalosi et al., 2008) enable genome-wide examination of RBP-target interactions, and should prove useful for elucidating these complex networks.
EXPERIMENTAL PROCEDURES
Sample Preparation and RNA-seq
Total RNA was isolated from tissues dissected from Oregon R animals in biological duplicates or from cultured S2R+ cells. Strand-specific RNA-seq libraries were prepared using prerelease Directional mRNA-seq Library Kits (Illumina). Briefly, poly(A)+ RNA was isolated by oligo-dT selection, fragmented, and treated with phosphatase and polynucleotide kinase to repair the ends. RNA adapters (3′ and 5′ ) were then sequentially ligated to the RNA fragments and reverse transcribed using a primer complementary to the 3′ linker. The libraries were then PCR amplified and sequenced on either an Illumina GAIIx using paired-end 76 bp chemistry or a HiSeq2000 using paired-end 100 bp reads. Reads were simultaneously aligned to the genome and splice junctions using Bowtie (Langmead, 2010) and SPA to report uniquely aligned reads as described (Graveley et al., 2011). The raw fastq RNA-seq data were deposited at the NCBI Short Read Archive, and the processed bam files were deposited at the modENCODE Data Coordination Center; the accession numbers are summarized in Table 1.
Poly(A) Enrichment
One nanogram of the strand-specific RNA-Seq libraries were reamplified by PCR using a primer complementary to the 5′ adaptor and a second primer complementary to the 3′ adaptor with six T residues at the 3′ end. After 10 rounds of amplification, the 3′ primer with the T extension was replaced with a 3′ primer complementary to the adaptor with a 5′ extension containing a 6 nt index sequence and a sequence complementary to the flow cell primer. After an additional 15 rounds of amplication, the libraries were quantitated, 10–12 libraries were pooled together and sequenced on an Illumina HiSeq2000 using paired-end 100 bp and 6 bp index read chemistry. Reads were split into the respective samples using the index sequence and aligned as described above. All of the raw fastq data and alignments of poly(A)-spanning reads from the poly(A)-enriched libraries were deposited at NCBI Gene Expression Omnibus under Series GSE3390.
Experimental Analysis of Gene Expression
In situ hybridization was performed on mixed stage Canton S embryos according to the BDGP 96-well plate in situ hybridization procedure (Tomancak et al., 2002), with modifications to utilize 1.7 ml tubes. Quantitative RT-PCR was performed using SYBR green PCR mastermix (QIAGEN) on a CFX96 real-time system (BioRad). Northern analysis of was performed as previously described (Mayr and Bartel, 2009) using 2–3 mg of poly(A)+ RNA or 8–12 mg of total RNA per lane. Detailed experimental procedures are provided in the Extended Experimental Procedures. Oligo sequences used to generate in situ probes, northern probes and qPCR amplicons are listed in Table S4.
Analysis of PAS Defined by Poly(A)-Spanning Reads
Read pairs that failed to align to the genome were examined to identify cases where the first read containedR10 A residues at the 3′ end or the second read contained ≥ 10 T residues at the 5′ end. Terminal A or T residues were trimmed from the reads and uniquely aligned poly(A)-spanning reads identified. These were filtered for instances of oligo dT priming to potentially genomically encoded poly(A) stretches by removing reads where at least eight out of the ten nucleotides downstream of the matching regions were adenines. Remaining reads were clustered so that all reads mapping to the same strand and ending within ten nucleotides were collapsed into a cluster. Clusters supported by at least two reads were considered for further analysis.
3′ UTR Analysis
A combination of bioinformatic searches and manual browsing was performed to identify genes with neural 3′ UTR extensions. This was necessitated by the discontinuous nature of stranded RNA-seq data. The computational scan was based on the number of reads proximal and distal to annotated 3′ ends in head and ovary libraries, assuming that genes that displayed a higher ratio of distal/proximal reads in head samples compared to ovary samples could be mRNAs with neural 3′ UTR extensions. To identify genes with 3′ UTR truncations in testis, we performed a computational search for an increased ratio of proximal/distal reads between testis and ovary. Manual browsing to validate transcript models was performed using Jbrowse (jbrowse.org) and Integrated Genome Viewer (www.broadinstitute.org/igv) for the entire Drosophila genome with tracks loaded from multiple Drosophila tissues.
Motif Analysis
Regions around the polyadenylation sites (±50 nt) were scanned for sequence motifs using MEME (Bailey et al., 2006) and Weeder (Pavesi et al., 2001). We also performed directed searches for the canonical PAS AAUAAA and known variants (Retelska et al., 2006). The neural extended and testis shortened portions of the 3′ UTRs were scanned for conserved 6-mers and 7-mers using the approach described in (Xie et al., 2005). We also searched for known miRNA seeds (Ruby et al., 2007) within annotated and extended 3′ UTRs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shujie Xiao for help with dissections, David Miller for help with fly preparation, Chris Streck for providing the RNA-seq library kits, and the UCHC Translational Genomics Core facility for use of the Illumina GAIIx and HiSeq2000. P.M. was supported by a fellowship from the Canadian Institutes of Health Research, and J.O.W. was supported by a fellowship from the Swedish Research Council. This work was funded by an award from the National Human Genome Research Institute modENCODE project (U01-HB004271) to S.E.C. (Principal Investigator), P.C., and B.R.G. (co-Principal Investigators) under Department of Energy contract DE-AC02-05CH11231. Work in E.C.L.’s group was supported by R01-GM083300, U01-HG004261, and RC2-HG005639.
Footnotes
ACCESSION NUMBERS
Raw fastq RNA-seq data have been deposited at the NCBI Short Read Archive, and the processed .bam files have been deposited at the modENCODE Data Coordination Center. Accession numbers for these data are summarized in Table 1.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, four tables, and one data set and can be found with this article online at doi:10.1016/j.celrep.2012.01.001.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported License (CC-BY; http://creativecommons.org/licenses/by/3.0/legalcode).
REFERENCES
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. USA. 2011;108:15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Chapter 11, Unit 11.17. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of Celegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;17(Suppl 1):S207–S214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, Moreira A. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:research0088.1–research0088.14. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.