Abstract
Cadherins constitute a superfamily of cell–cell adhesion molecules expressed in many different cell types that are required for proper cellular function and maintenance of tissue architecture. Classical cadherins are the best understood class of cadherins. They are single membrane spanning proteins with a divergent extracellular domain of five repeats and a conserved cytoplasmic domain. Binding between cadherin extracellular domains is weak, but strong cell–cell adhesion develops during lateral clustering of cadherins by proteins that link the cadherin cytoplasmic domain to the actin cytoskeleton. Understanding how different regions of cadherins regulate cell–cell adhesion has been a major focus of study. Here, we examine evidence of the structure and function of the extracellular domain of classical cadherins in regard to the control of recognition and adhesive contacts between cadherins on opposing cell surfaces. Early experiments that focused on understanding the homotypic, Ca++-dependent characteristics of cadherin adhesion are discussed, and data supporting the widely accepted cis- and trans-dimer models of cadherins are analyzed.
Keywords: Cadherin, Cell–cell adhesion, Calcium, Structure, Dimer
1 Introduction
Cadherins are an important superfamily of cell–cell adhesion proteins comprising over 40 members (Takeichi 1990), and perhaps even more when the proto-cadherin family is included (Frank and Kemler 2002). Cadherins are expressed in many cells and tissues, and are evolutionarily conserved in vertebrates and invertebrates (Kemler 1992; Takeichi 1995; Gumbiner 1996). Here we focus on a subset of cadherins, the classical cadherins, that are the best described and understood. We focus on the structure and function of the extracellular domain that controls recognition and adhesive contacts between cadherins on opposing cell surfaces.
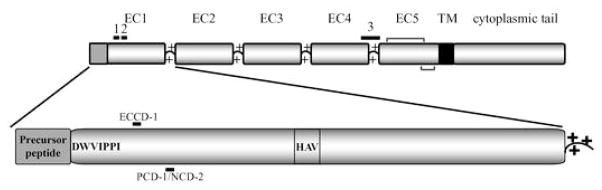
Classical cadherins, of which E-, P-, N-, and R-cadherin are members, were shown to mediate segregation of different cell populations in early studies. Segregation was based on homophilic adhesion in which cells expressing one cadherin subtype (E-cadherin for example) segregated in suspension from cell expressing a different cadherin subtype (P-cadherin). Cadherin-mediated homotypic adhesion appears to depend on binding specificity, Ca++-dependence, and molecular contacts of cadherins and cadherin–cadherin adhesion. Sequence analysis of classical cadherins reveals that they have five tandemly repeated domains in the extracellular domain, termed extracellular cadherin repeats 1–5 (EC1–5) (Fig. 1) with EC1 at the amino terminus (Hatta et al. 1988). When synthesized, cadherins contain signal and precursor peptides that are cleaved during processing and maturation of the protein in the endoplasmic reticulum (ER) and Golgi. The precursor peptide is cleaved before cell surface presentation of the cadherin, and cleavage is required for adhesive function (Ozawa and Kemler 1990).
Fig. 1.
Schematic of a classical cadherin. The schematic depicts the domain organization of a classical cadherin. Five extracellular repeats (EC1–5) are preceded by a precursor peptide which is cleaved during maturation. Ca++-binding sites (+ marks) are located at each EC junction. EC5 is followed by a single transmembrane segment and a highly conserved cytosolic domain which associates with members of the catenin protein family. Monoclonal antibody binding regions are marked with numbered black bars above the repeats and correspond to binding sites of (1) ECCD-1, (2) PCD1 and NCD1, and (3) DECMA. Disulfide bonds are shown in EC5 as gray brackets. A close view of EC1 also shows the histidine-alanine-valine (HAV) tripeptide as well as the highly conserved first seven residues of the mature protein including Trp2
How do cadherin extracellular domains interact to form cell–cell adhesions? The most prevalent models describe two cadherins within the same membrane forming a lateral or cis-dimer and that this dimer promotes adhesive dimerization (trans-dimer) of cadherins on adjacent cells (Fig. 2). The models differ in their description of the organization and mechanism of the extracellular domains in the trans-dimers. Two models describe trans-dimerization through EC1 alone while a third describes complete intercalation of cadherin extracellular domains. Each model further depicts that both cis- and trans-dimerization depend on Ca++ to bind between each EC repeat to stabilize and order extracellular domains. We will analyze the data supporting these conclusions.
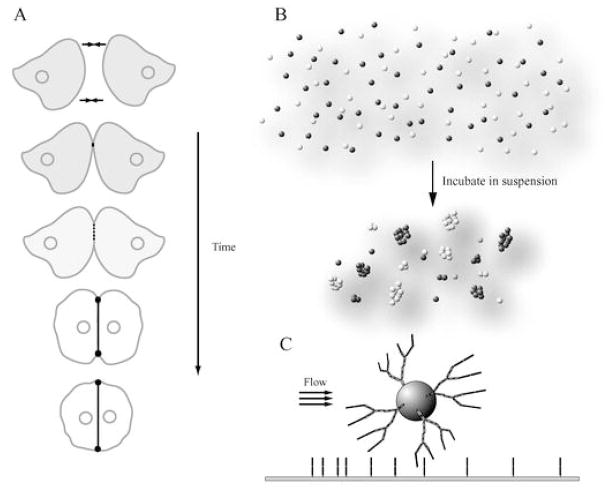
Fig. 2A–C.
General methods to examine cadherin adhesion functions. A Schematic of cadherin distribution during epithelial cell–cell adhesion as imaged by live cell microscopy of Madin-Darby canine kidney (MDCK) cells expressing green fluorescent protein (GFP)-E-cadherin (Adams et al. 1998). B Cell segregation assay. Cells expressing one subtype of cadherin (dark spheres) are mixed with cells expressing a second subtype of cadherin (light spheres). The cells are incubated in suspension. Cadherins mediate segregation of cells expressing different cadherin subtypes (no dark and light mixed aggregates). C Adhesion flow assay. Recombinantly expressed fragments of the cadherin extracellular domain are attached to a substrate. Beads coated with the same cadherin fragment are flowed over the surface and their migration is monitored. Events in which the beads stop moving are interpreted as adhesion events
2 Ca++ and Cadherin Adhesion
Understanding the Ca++-dependence of cadherin adhesion was addressed in early experiments. Ca++ was shown to protect the cadherin extracellular domain from degradation by trypsin, suggesting a role in structural organization of the domain. However, the mechanism remains unclear. After solving the protein sequence of uvomorulin (E-cadherin), Ringwald et al. (1987) identified three internal repeats in the extracellular domain. Within these domains they identified two distinct putative Ca++-binding loops based on sequence homology to known Ca++-binding regions. Although the analysis of the extracellular domain was not exactly correct, the authors first demonstrated the basic structure of the cadherin extracellular domain, a functional domain composed of distinct, repeated units each of which are likely able to bind Ca++. A year later with the sequencing of N-cadherin and use of a different alignment program, the five internal cadherin repeats were identified (Hatta et al. 1988). The authors, however, did not comment on Ca++ binding sites. Nevertheless, sequence alignment of the known cadherins shows conservation in each of the five internal repeats of the putative Ca++-binding sites proposed by Ringwald et al. (1987).
Evidence of the functional role of Ca++ in cadherin adhesion came initially from studies on the recombinant extracellular domain of E-cadherin (Pokutta et al. 1994). Electron microscopy of the recombinant domain directly showed dependence on Ca++ for elongation and maintenance of a rigid bent rod-like structure, but the curvature was not commented upon. In the absence of Ca++, the domain was apparently disordered and not elongated, and resembled a globular structure. Ca++ binding was reversible. These conformational changes were confirmed by circular dichroism spectroscopy. A large change in the ellipticity of the CD spectrum was observed between 202–215 nm after removal of Ca++. The change in CD spectra was used to examine conformational changes at different Ca++ concentrations, and an average Kd of 42–45 μM was determined. As there were multiple Ca++ ions known to bind to the cadherin extracellular domain, it was not possible to determine whether all sites have the same Kd or the value obtained was an average. The authors also noticed an increase in the fluorescence of tryptophan upon Ca++ binding. They used this characteristic to titrate Ca++ as well and determine two different Kd values, one at 130 μM and the second at 210 μM. The results suggested that the Kd obtained from titration based on CD spectrum changes was an average, and the change in tryptophan fluorescence measured a Ca++ binding site of low affinity. Finally, Pokutta et al. (1994) measured Ca++ affinities by protection against tryptic digestion. From these experiments they observed a Kd of 24 μM, but there appeared to be cooperative binding here as well. Though unable to satisfactorily measure exact dissociation constants of the different Ca++ binding sites, the authors were able to prove the existence of high and low affinity binding sites and first observed and modeled the mechanism of Ca++-dependent adhesion whereby Ca++ acted to rigidify the extracellular domain.
The dissociation constants of Ca++ at the various binding sites were also determined using a fragment of the first two EC repeats of E-cadherin (ECAD12) (Koch et al. 1997). By monitoring the CD spectra change of ECAD12, an average Kd of 360 μM for the Ca++ binding sites between EC1 and EC2 was recorded. ECAD12 bound three Ca++ ions, and using equilibrium dialysis two Kd values of 330 μM and one Kd value of 2 mM were determined. Interestingly, when the Ca++ binding sites of the full E-cadherin extracellular domain were analyzed by equilibrium dialysis, an average Kd of only 30 μM was calculated for a total of nine Ca++ ions bound to the protein fragment.
3 Cadherin Adhesion Structure: Trans-Dimer
3.1 Early Studies
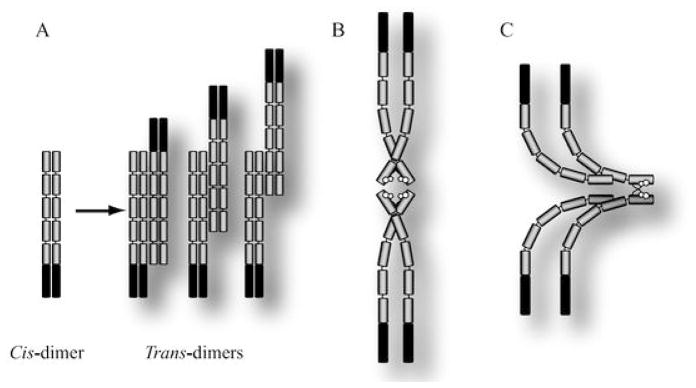
Initial studies of cadherin extracellular domain adhesion focused on determining which EC repeats are involved in adhesion and specificity. Nose et al. (1990) used a domain swapping strategy to determine the location of sites controlling cadherin subtype specificity (i.e., which domains controlled E↔E, P↔P specificity). When expressed in cadherin-deficient fibroblast L cells, wildtype E- and P-cadherins mediated sorting and segregation of cells expressing the same cadherin (Fig. 3). However, when E-cadherin EC1 was switched with EC1 of P-cadherin to generate a chimera of P-cadherin EC1 and E-cadherin EC2–5, the chimeric protein now mediated adhesion with L cells expressing P-cadherin. The authors concluded that EC1 contained sites that determine cadherin adhesion specificity. They further narrowed the region to between residues 61 and 113. However, it is noteworthy that swapping sub-regions of EC1 (residues 1–31 and 1–67) did not provide a complete switch in adhesion specificity. Rather, there was a decrease in specificity; chimeric proteins were able to mediate some adhesion between either E-cadherin- or P-cadherin-expressing cells. Thus, adhesion specificity is located in the amino terminal EC1 repeat, but it cannot be excluded that other sites outside EC1 are also involved. Nevertheless, a role for EC1 was also supported by early studies using monoclonal antibodies directed against EC1 that block the adhesion function of cadherins (Yoshida-Noro et al. 1984; Behrens et al. 1985; Gumbiner and Simons 1986; Hatta and Takeichi 1986; Nose and Takeichi 1986). However, the residues recognized by these antibodies and involved in adhesion remain unknown.
Fig. 3A–C.
Cadherin adhesion models. A Trans-dimerization model of interdigitated cadherin domains by Sivasankar et al. (2001). Cis-dimerized cadherins in opposing membranes are able to bind through several different mechanisms. One mechanism involves complete interdigitation of cadherin extracellular domains. A second binding complex occurs through overlap of EC1–4, and a third occurs between EC1 and 2 alone. B Cis- and trans-dimerization model proposed by Pertz et al. (1999). W2 acts as an allosteric activator to allow EC1 domains of opposing cadherins to interact through an unknown mechanism. Ca++-coordination first allows cadherins to cis-dimerize, but only at high Ca++-concentrations (>1 mM) will W2 dock in the hydrophobic pocket. C Cis- and trans-dimer model proposed by Bogon et al. (2002). Cis-dimers form through an interaction of EC1 in one cadherin with EC2 and some of EC3 of a second cadherin. These cis-dimers would organize into a semi-crystalline array forming trans-dimers through exchange of W2 via the strand dimer. Of note is the fact that only one cadherin of a cis-dimer interacts with an opposing cis-dimer on a neighboring cell. In all schematics, only cadherins involved in trans-adhesion show W2 docking
Tripeptides based on protein–protein interfaces have been useful in examining adhesion of integrins to extracellular matrix, and a similar strategy has been used with cadherins. Blashuck et al. (1990) hypothesized a tripeptide adhesion sequence was present in cadherins. Sequence analysis identified several potential tripeptide adhesion sequences, and they tested whether these peptides blocked normal cadherin–cadherin adhesion, aggregation, and blastocyst compaction. The authors showed that only the histidine-alanine-valine (HAV) sequence from the EC1 repeat of all classical cadherins is likely important in adhesion. Mouse blastocysts incubated with a decapeptide derived from N-cadherin containing the HAV sequence failed to compact. Furthermore, rat dorsal root ganglia did not extend neurites over astrocytes in the presence of the HAV-containing peptide. Both blastocyst compaction and neurite extension are mediated by E-cadherin and N-cadherin, respectively. Because the HAV sequence is found in the EC1 of all classical cadherins, this evidence gives further support to the developing model of EC1 involvement in mediating both adhesion and specificity of cadherins.
The HAV sequence has continued to be studied as a potential mediator and recognition sequence of cadherin adhesion. Based on the inhibitory aspect of the HAV-containing decapeptide, Williams et al. (2000) showed that cyclic peptides containing HAV were better than linear peptides in inhibiting N-cadherin-mediated neurite extension of cerebellar neurons over N-cadherin-expressing 3T3 cells. In addition, they found that incorporation into the cyclic peptides of specific residues flanking the HAV sequence in N-cadherin increased their inhibitory activity. Interestingly, if they incorporated flanking residues from E-cadherin rather than N-cadherin, the cyclic peptides did not inhibit N-cadherin adhesion. It should be noted that several of the peptides derived from N-cadherin sequence failed to inhibit N-cadherin adhesion. It would have been more informative if the E-cadherin-derived cyclic peptides were shown to inhibit E-cadherin, but not N-cadherin adhesion. Further evidence for the involvement of EC1 in cadherin adhesion was revealed in a study from the same group on small peptide agonists of cadherin adhesion (Williams et al. 2002). The authors demonstrated that a recombinant N-cadherin EC1 domain was able to inhibit neurite outgrowth from cerebellar neurons over 3T3 cells expressing N-cadherin. Together, these results demonstrate that residues flanking the HAV sequence are potential mediators of cadherin adhesion specificity.
Despite strong evidence, using a variety of experimental approaches, for a role of EC1 in adhesion, some evidence points to a model in which additional EC repeats are required for adhesion. The monoclonal antibody DECMA-1 blocks E-cadherin adhesion (Vestweber and Kemler 1985). Mapping of the epitope showed that it is directed against the EC4/EC5 boundary rather than EC1, as shown for other inhibitory antibodies (see above and Fig. 1) (Ozawa et al. 1990). Ozawa et al. (1990) showed also that E-cadherin contains at least one disulfide bond, and that this bond is important, but not required, for cadherin–cadherin adhesion. Sequence analysis showed only EC5 to have potential disulfide bonds [confirmed by crystal structure (Boggon et al. 2002)] adding further support to some role of EC4–5 in cadherin adhesion.
Analysis of crystal structures of cadherin extracellular domains provided new information on molecular interactions between cadherin EC domains. Because EC1 was suspect in adhesive interactions, the initial focus of crystal structure studies was EC1. The first crystal of cadherin EC1 was from N-cadherin (Shapiro et al. 1995). It raised more questions than answers about past evidence of EC1. First, three different crystal forms were observed, one contained a single molecule in the asymmetric unit of the crystal lattice, and the other two contained two molecules per asymmetric unit in different orientations. The molecular interactions described (see below) were observed in all the crystal forms, but mostly only as crystal packing interactions. Second, the structure revealed that the HAV tripeptide, though on the surface, is partially buried and obscured, and a potential dimer could not be assigned to correlate with the HAV sequence (Fig. 4A and 4A′). The structure did help explain why a single linear sequence of the protein could not be identified as the determinant of cadherin specificity.
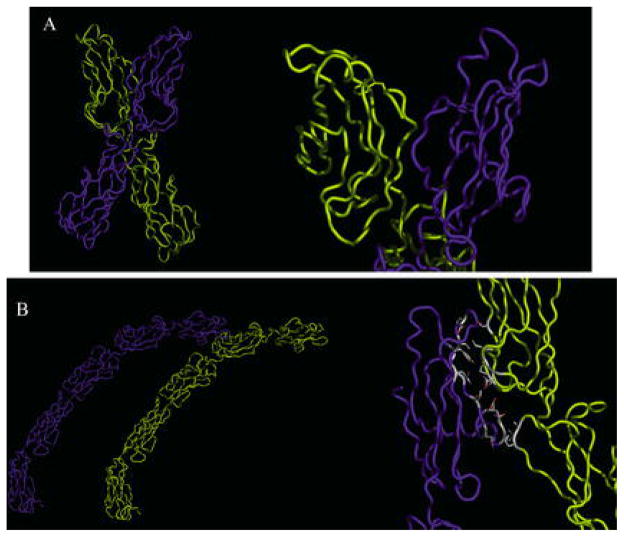
Fig. 4A–C.
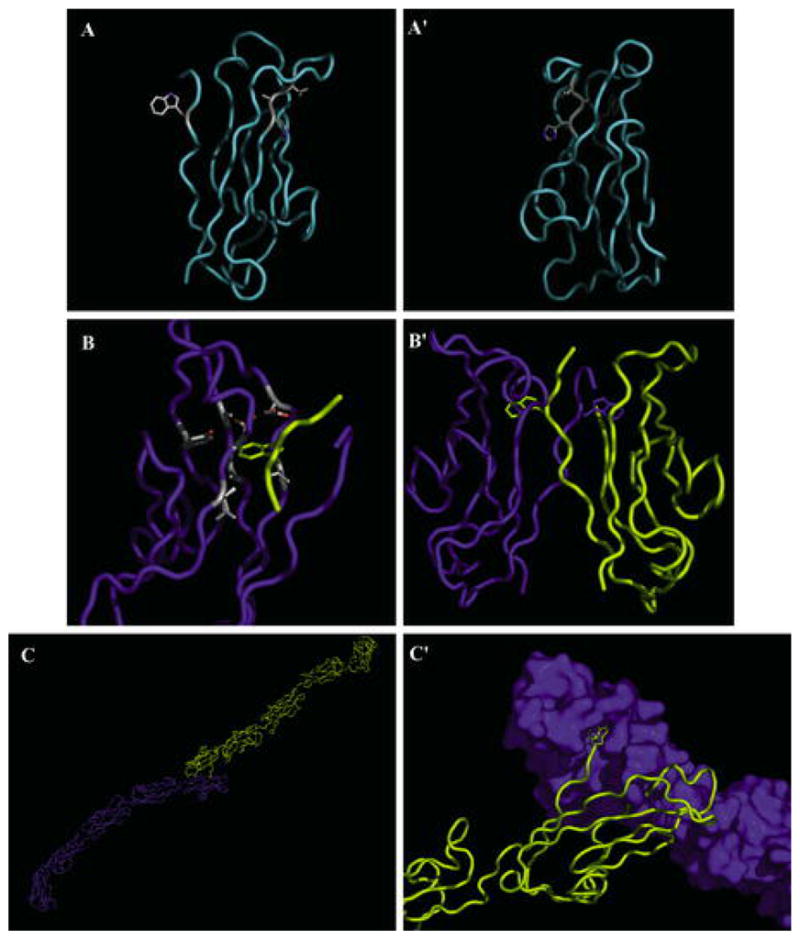
Structure of EC1 and strand dimer. A N-cadherin EC1 is shown in a ribbon with the HAV sequence and W2 side chains. A′ Ninety degree rotation of the structure in A. B Close-up of W2 docking in the hydrophobic pocket in the strand dimer. W2 of one cadherin (yellow) is docked in a pocket of an opposing cadherin (blue). Side chains of residues composing the surface of the pocket are shown. B′ EC1 domains participating in the strand dimer exchange of W2. C Strand dimer of the complete C-cadherin extracellular domain. C′ A close-up of EC1 domains in the C-cadherin strand dimer. A surface model of cadherin (blue) is shown against a ribbon model (yellow). [A, A′, B, and B′ are from Protein Data Bank (PDB) accession 1NCI (Shapiro et al. 1995), and C and C′ from PDB accession 1L3 W (Boggon et al. 2002)]
The shape of the cadherin repeat is a β-barrel structure, and residues in close proximity on the surface of the protein are not necessarily close in the primary structure of the protein. An intriguing interaction is between two domains with their long axes aligned in a roughly parallel, not antiparallel, orientation. The authors suggested that this interaction might be a putative lateral or cis-dimer (see below for discussion of cis-dimers). Additionally, tryptophan 2 (W2) of each domain was inserted into a hydrophobic pocket of the adjacent cadherin in what was termed a “strand dimer” (Fig. 4). The strand dimer was the major characteristic of lateral cadherin dimers. Interestingly, the hydrophobic pocket accepting the side chain of W2 is composed, in part, of the alanine from the HAV tripeptide.
3.2 Recent Studies, Evolving Models
The first crystal structure of the full cadherin extracellular domain showed that the strand dimer, inferred from early structures to represent a cis-dimer, may in fact be representative of the association of cadherins on neighboring cells (Boggon et al. 2002). The structure shows the characteristic strand dimer with the EC1 domains of the two molecules in a roughly parallel orientation; however, the rest of the extracellular domain adopts a curved structure rather than the assumed rigid straight structure (Fig. 4C). The curvature of the structure is such that the long axis of EC1 is roughly perpendicular to the long axis of EC5. Based on this new evidence, the authors proposed a model that the strand dimer mediated trans-dimerization and that cis-dimerization occurred through a previously undescribed interaction.
The key characteristic of the strand dimer is intercalation of W2 into the hydrophobic pocket of an opposing cadherin. Experiments focusing on W2 and the hydrophobic pocket demonstrate their importance (Tamura et al. 1998; Pertz et al. 1999; Ahrens et al. 2002; Perret et al. 2002). Mutation of W2, A78, or A80 (the alanines comprising parts of the hydrophobic pocket) inhibits cell aggregation, bead aggregation, and cell-bead binding. Of particular note are the studies by Pertz et al. (1999) which used a chimeric protein of the E-cadherin extracellular domain fused to the coiled-coil pentamerization domain of cartilage oligomatrix protein (ECADCOMP). Using electron microscopy to examine structure and interactions, Pertz et al. (1999) showed that ECADCOMP forms a pentamer in solution. In the presence of Ca++, the E-cadherin extracellular domain adopts a bent rod structure, two of which in a pentamer can form a ring-like structure, inferred as a putative cis-interaction (Fig. 5). There are instances in which two ring-like structures are in contact in a putative trans-interaction. An ECADCOMP carrying the W2A mutation, while still able to adopt a ring structure, is never seen in association with a second ring. While all other data cannot distinguish between a role in cis- or trans-dimerization, these electron microscopy studies support a model in which W2 docking in the hydrophobic pocket is required for trans-dimerization.
Fig. 5.
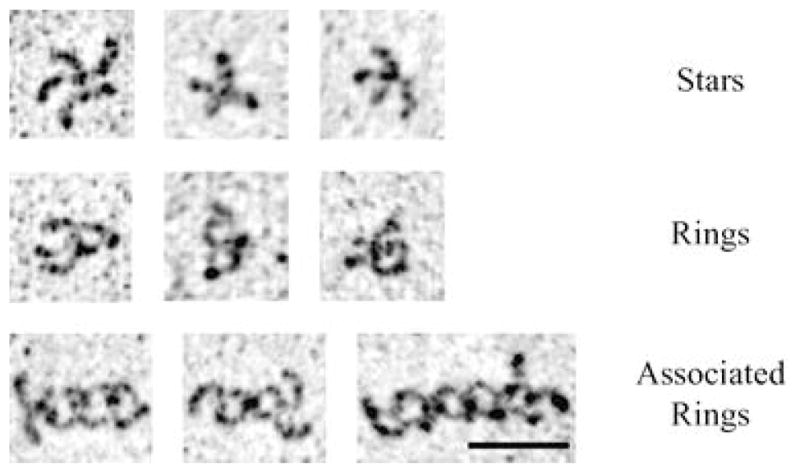
ECADCOMP conformations. ECADCOMP visualized by rotary shadowing electron microscopy. The top row shows non-dimerized pentamers in a star-like pattern. The middle row shows pentamers in which two or four E-cadherin extracellular domains have formed a ring-like structure. The bottom row shows multimerized pentamers with a concentric ring orientation. The ring structures are proposed cis-dimers, and the concentric rings are proposed trans-dimers of cis-dimerized cadherins. Used by permission from Pertz et al. (1999)
A second model has been proposed in which the strand dimer is not directly involved in trans-dimerization (Koch et al. 1999; Pertz et al. 1999). Crystal studies of EC1 and 2 of N-cadherin and E-cadherin showed an apparent cis-dimer lacking the strand dimer. In two of the crystals, the amino terminus was disordered and could not be resolved (Nagar et al. 1996; Tamura et al. 1998). The third crystal showed W2 docking into the hydrophobic pocket of its own protein (Pertz et al. 1999). Pertz et al. (1999) proposed that W2 is not involved in strand exchange and direct intermolecular interactions, but rather is required as an allosteric activator for trans-dimerization. The conclusion was based on the assumption that W2 is required for cis-dimerization, an assumption which the authors proved wrong. Note that ECADCOMP bearing a W2A mutation was seen by electron microscopy to form ring-like structures in an apparent cis-dimerization but was unable to oligomerize into concentric ring structures.
Other studies continue to support a model of W2 as an allosteric effector rather than a direct mediator of trans-adhesion. Nuclear magnetic resonance (NMR) analysis of Ca++-dependent dimers and oligomers of E-cadherin EC1 and 2 [the same construct used by Pertz et al. (1999) for crystallographic studies] suggested that W2 is buried in the hydrophobic pocket of its own molecule (Haussinger et al. 2002). Analysis of {1H} (15N) shifts of W2 indole group revealed no significant change in the orientation of W2 during Ca++-dependent dimerization and oligomerization of E-cadherin EC1 and 2. Additionally, the {1H}-15N nuclear Overhauser enhancements (NOEs) of the W2 indole group in the presence and absence of 600 μM Ca++ indicate considerable flexibility of the residue in both monomeric and aggregated states that would not be expected if it were fixed in the hydrophobic pocket of a neighboring molecule as depicted in the strand dimer model. One caveat of this study, though, is the presence of an N-terminal methionine as a cloning and protein expression artifact. Previous studies showed that precise cleavage of the precursor peptide of cadherins is required for full adhesive functionality (Ozawa and Kemler 1990).
3.3 Further Functional Studies of EC Domains in Adhesion
EC1 was first modeled as the adhesive domain because it was found to be involved in specificity, contained the HAV adhesion sequence, and N-cadherin EC1 packed in an antiparallel orientation as if in a trans-dimer in one crystal form (Blaschuk et al. 1990; Nose et al. 1990; Shapiro et al. 1995). However, the precise orientation of EC1 in the trans-dimer remains unclear. Nevertheless, adhesion through EC1 remains the model best fitting all data, and additional studies have been presented which strengthen this view.
First, further work on ECADCOMP showed that adhesive pentamers oligomerized through the N-terminal regions of E-cadherin (Tomschy et al. 1996; Koch et al. 1999; Pertz et al. 1999). It is unclear, however, exactly which parts of the N-terminus are involved in adhesion. Second, the studies of Perret et al. (2002) demonstrated adhesive events between cadherin fragments of just EC1 and 2. To investigate kinetics of cadherin trans adhesion, they constructed an E-cadherin extracellular fragment consisting of EC1 and 2 with a C-terminal His tag (E-cad1/2). Beads were coated with an antibody against the His tag, and E-cad1/2 was added to them. A mica surface was prepared by adsorbing Ni++ to the surface and then chelating the E-cad1/2 His tag directly. The authors visualized the cadherin-coated beads as they rolled across the cadherin-coated surface and measured the duration and frequency of stop events interpreted as cadherin adhesion between the bead and surface (Fig. 3). An approximately fivefold decrease in the frequency of binding events was observed when the tryptophan analog I3A or an E-cad1/2 with the W2A mutation was used, thereby further supporting a role of EC1 and W2 in adhesion.
These results, however, are different from those of Chappuis-Flament et al. (2001). In similar adhesion flow experiments, the authors used various C-cadherin fragments, missing one or several of the EC repeats, fused to the Fc domain of IgG. The cadherin fragments were bound directly to protein A-coated beads through the Fc domain, and their function was tested by Ca++-dependent bead aggregation. In these experiments, at least three cadherin EC repeats were required for strong Ca++-dependent adhesion. The authors were unable to measure adhesion by bead aggregation or in a flow assay of a cadherin fragment of EC1 and 2 alone. However, EC1 and 2 were shown to be required in all the assays performed, while the additional repeat could be either EC3, EC4, or EC5. The third domain had to be an EC repeat, though, as fibronectin repeats fused to only EC1 and 2 failed to mediate bead aggregation. The differences in experimental design and aims between these experiments and those of Perret et al. (2002), specifically attachment of the cadherin fragment to the beads and analysis of single adhesion events versus bulk adhesion properties, could explain the different conclusions arrived by the two sets of experiments. However, it is clear from both studies that EC1 and 2 are required for adhesion.
A different experimental design has been used to examine a role of additional EC repeats, i.e., other than just EC1 in adhesion. Sivasankar and colleagues, using a surface force apparatus, demonstrated that the strongest adhesive interaction between cadherins on two surfaces occurred at a minimum distance of about 25 nm (Sivasankar et al. 1999; Leckband and Sivasankar 2000; Sivasankar et al. 2001). Interestingly, this is the approximate length of a cadherin extracellular domain if all the repeats would be in a straight line, one after the other. However, as noted earlier, electron microscopy of the extracellular domain of E-cadherin and the crystal structure of the full-length extracellular domain of C-cadherin depict the cadherin extracellular domain as a bent rod (Pokutta et al. 1994; Tomschy et al. 1996; Ahrens et al. 2002; Boggon et al. 2002). Due to the curve of the extracellular domain, the trans-adhesion dimer proposed by Boggon et al. (2002) would occur between surfaces approximately 25 nm apart. Additionally, analysis of oligomerized dimers of E-cadherin, using the c-Jun/c-Fos dimerization system, reveals a measurement of adhesive dimerization between EC1 of approximately 25 nm. If EC5 of each extracellular domain is oriented perpendicular to the surface and each cadherin is interacting with the cadherin on the opposite side (left–right, right–left), then the calculated distance between two theoretical surfaces bearing the E-cadherin c-Jun/c-Fos dimers is approximately 28 nm. In each of these models, however, a single cadherin itself would stand only 10–15 nm from the plasma membrane surface. This conclusion conflicts with measurements of the cadherin extracellular domain height analyzed with the surface force apparatus (Sivasankar et al. 2001). These measurements showed that at least some cadherins can extend up to 20 nm from a surface. Furthermore, adhesive interactions were detected with the surface force apparatus even if the cadherin surfaces were only allowed to approach to 30 and 40 nm of each other. This result suggests that adhesive interactions can occur between cadherins that are not in a bent configuration. Since the structure of the cadherin extracellular domain is potentially a bent rod, measurements with the surface force apparatus do support a model in which the N-terminal EC1 repeat is solely involved in trans-adhesion if the cadherin extracellular domain is able to adopt multiple rigid orientations, but they also support a model in which multiple EC repeats intercalate during cadherin adhesion.
Taken together, we know that EC1 is required for adhesion and at least partly is responsible for homotypic specificity. Multiple EC repeats are likely involved in adhesion as well, but it is unclear whether they directly form adhesive contacts with an opposing cadherin or simply correctly present EC1 to an opposing cadherin. All evidence supports a model in which EC1 alone interacts in adhesion, but further experiments are required: functional adhesion must be verified while identifying or knowing the orientation of the cadherin extracellular domains. The W2 residue plays a role in adhesion, but the exact mechanism remains to be determined. Experiments to date have not been able to distinguish between W2 as an allosteric activator of trans-adhesion or as a direct mediator of trans-adhesion through the strand dimer observed in crystal structures. Finally, though the HAV sequence does appear to have a function in adhesion (mutation of the alanine and use of HAV-containing peptides), the mechanism is unknown. Mutation studies of the other residues in the tripeptide may help to explain the function of HAV. Additionally, structural studies with HAV peptides may help to explain their ability to inhibit adhesion. The precise determination of all residues involved and required for adhesion would help in determining the true molecular interactions of cadherin adhesion.
4 Cadherin Adhesion Structure: Cis-Dimer
Formation of lateral cadherin dimers, referred to as cis-dimers, was first proposed based on crystal structures of EC1 of N-cadherin (Shapiro et al. 1995). This conclusion was based on the observation that individual EC1 domains packed in a parallel orientation, representative of proteins that had originated from the same cell membrane.
The first functional evidence for cis-dimerization of cadherins came from studies using purified C-cadherin extracellular domain (Brieher et al. 1996). C-cadherin extracellular domain separated into two peaks through a gel filtration column. Crosslinking of protein fractions showed that the higher molecular weight peak corresponded to dimer and the lower molecular weight peak corresponded to monomer. The putative dimer from the high molecular weight fractions was shown to have a higher adhesive potential in a cell adhesion assay than the low molecular weight monomer fractions. The authors concluded that these results confirm lateral dimerization of the cadherin extracellular domain-promoted adhesive dimerization. However, whether the extracellular domains in the dimer fractions were in a parallel (cis-dimer) or antiparallel (trans-dimer) orientation was not determined.
A key experiment on lateral dimerization of cadherins was performed in vivo (Takeda et al. 1999). Using cadherin-deficient L cells, full-length, wildtype E-cadherin or a chimeric E-cadherin fused directly to the actin-binding domain of α-catenin (Eαcat) was ectopically expressed. When cadherin-expressing cells were treated with the crosslinking agent 3,3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP) and solubilized, monomers and dimers of E-cadherin were identified. These dimers could arise from either lateral or adhesive trans-interactions. To test this, cells expressing E-cadherin and cells expressing Eαcat were cocultured. The cocultures were crosslinked and then analyzed by immunoblot for α-catenin. Only monomers and homodimers of Eαcat were observed. If the dimers were adhesive dimers, heterodimers of E-cadherin and Eαcat would also be expected. The authors additionally showed that these lateral dimers were only found in adherent cells; if cells were grown in the presence of ethyleneglycoltetraacetic acid (EGTA), low-Ca++ medium, or cadherin-inhibiting antibodies, lateral dimers could not be crosslinked. The conclusion drawn was that lateral cadherin dimers are a functional unit for cadherin adhesion. However, a second conclusion is also supported by the data, that cadherins are only able to be crosslinked into lateral dimers because of the increase in local concentration of cadherins on the cell surface during cadherin-mediated adhesion.
Cis-dimer formation has been further investigated by immunoprecipitation experiments without first crosslinking the proteins (Chitaev and Troyanovsky 1998; Klingelhofer et al. 2000; Shan et al. 2000). Immunoprecipitations showed complexes of cadherins believed to represent cis-dimers isolated from cell lysates. Chitaev and Troyanovsky (1998) provide evidence that E-cadherin forms lateral dimers through a Ca++-independent/W2-dependent mechanism. However, in a following study from the same group, Klingelhofer et al. (2000) suggest E-cadherin and P-cadherin can form hetero cis-dimers through multiple mechanisms. They present data suggesting heterocomplex formation in the presence or absence of Ca++ and also dependent and independent of W2. The W2-independent mechanism required the absence of Ca++. Additionally, Shan et al. (2000) found R-cadherin/N-cadherin heterocomplexes could be coimmunoprecipitated from cells expressing both cadherin subtypes. They showed also that the R-cadherin/N-cadherin interaction appears to be real because E-cadherin, if coexpressed with R-cadherin, is not coimmunoprecipitated with R-cadherin. The fact that these lateral cadherin interactions are Ca++-independent is in direct conflict with the data of Takeda et al. (1999) in which the lateral cadherin dimers they could crosslink were Ca++-dependent. The data from these immunoprecipitation experiments are very difficult to interpret in light of the various models of W2 involvement in trans- and cis-dimerization, and different methods are required to fully address the formation of lateral cadherin complexes in vivo.
Cis-dimerization has also been examined using peptides (Williams et al. 2002). Short peptide HAV-containing antagonists were dimerized so that two adhesion sites were on the same molecule. These peptides were found to act as agonists for neurite outgrowth, a cadherin-mediated adhesion response, on 3T3 cells lacking N-cadherin expression. Dimeric peptides of a second putative adhesion site containing INPISG also activated neurite outgrowth in the cerebellar neuron system. Cyclic monomeric peptides were able to block the activation by these dimeric peptides. It can be concluded that lateral dimerization or clustering of cadherins is able to mediate cellular response. It is not clear, however, if the dimeric peptides promoted adhesion (no assays were performed to test this), and they could act independently of adhesion since N-cadherin binds and activates fibroblast growth factor (FGF) receptors in the neurons (Williams et al. 1994; Saffell et al. 1997; Williams et al. 2001).
Crystal structures of the first two EC repeats show possible lateral dimer organization. Two different crystals have been presented of E-cadherin EC1 and EC2 (Nagar et al. 1996; Pertz et al. 1999). Each shows the two domain fragments aligned lengthwise in a parallel orientation. The region of closest contact is around the Ca++ binding sites between EC1 and 2 such that the proteins together adopt a sort of twisted “X” configuration (Fig. 6). The first crystal structure showed a less likely dimerization interaction as it was mediated by several water molecules and the interface was relatively small (Nagar et al. 1996). The second published structure was able to resolve more of the protein to show W2 docking into the hydrophobic pocket of its own molecule (Pertz et al. 1999). Additionally, the authors observed a lateral dimer they believed to be more stable also in the shape of an intertwisted “X”. There have been no biological assays done to conclusively confirm either interaction. The N-cadherin EC1/2 structure showed what appeared to be a lateral dimer, but no strand dimer was detected as previously described for the N-cadherin EC1 domain alone (Tamura et al. 1998). Tamura et al. (1998) mention a crystal packing interface possibly involved in trans adhesion, but they allude to results suggesting it is not a real interface and the one described in earlier work of N-cadherin EC1 (Shapiro et al. 1995) is more likely correct.
Fig. 6.
Structures of the cadherin cis-dimer. A Structure from the E-cadherin crystal by Pertz et al (1999). The overall alignment of the two Ecad12 fragments is a slightly twisted “X”. A close view of W2 in each EC1 reveals it is docking in the hydrophobic pocket of its own EC1. B Structure of the C-cadherin extracellular domain in a proposed cis-dimer. A groove in EC1 of one cadherin binds over a bulge in EC2 of a lateral cadherin. [A created from PDB accession 1FF5 (Pertz et al. 1999), and B created from PDB accession 1L3 W (Boggon et al. 2002)]
With the crystallization of the complete cadherin extracellular domain and the discovery that the strand dimer possibly mediated trans-, not cis-, dimerization, Boggon et al. (2002) had to formulate a new molecular model for the cis-dimer. The authors described a putative cis-dimer interaction based on the crystal packing interactions between a groove in EC1 of one cadherin and a bulge on EC2 of a second cadherin (Fig. 6). The proposed interaction showed two adjacent cadherins aligned with their N-termini pointed in the same direction. This interaction would allow cadherins within the same membrane to organize in a continuous linear array. Interestingly, the surface of the groove in EC1 is composed in small part by the histidine and valine of the HAV tripeptide. Functional significance has not been verified.
Electron microscopy analysis of ECADCOMP supports a different cis-dimer model. In their studies on Ca++-dependent adhesion, Tomschy et al. (1996) and Pertz et al. (1999) describe association of E-cadherin extracellular domains within a single pentamer before extracellular domains in different pentamers adhere. They base this on the observation that ring-like structures are found in some isolated pentamers at high Ca++ concentrations, but they are always seen as the adherent cadherins when two or more pentamers associate. In contrast to the model proposed by Boggon et al. (2002), the cadherins in this cis-dimer would bend toward each other and the N-termini of the extracellular domains would point in opposite directions. The conclusion that these data represent a real cis interaction and that this interaction is required before trans adhesion is valid. However, there is another equally possible explanation: Because the extracellular domain is a curved rod and the extracellular domains are constrained by the pentamerization domain, they simply lay on the surface in an orientation that appears like a cis interaction. When two pentamers associate through a single extracellular domain, a second is brought into the structure quickly because the effective concentration of extracellular domains is very high, and the two adherent pairs simply lay flat to appear as two adjacent ring-like structures, again because of the constraint imposed by the pentamerization domain.
The cis-dimer model requires more conclusive validation. The fact that lateral complexes of cadherin can form in vivo is certain, but the question remains concerning whether these complexes serve a specific role in adhesion or if they are simply the result of cadherin clustering by its anchor to the actin cytoskeleton. Data should be obtained that show a distinct difference in the adhesive properties of known cadherin cis-dimers versus single cadherin extracellular domains. All experiments to date have either not shown a difference in adhesion due to cis-dimerization or have not determined the specific orientation of the dimers. In addition, the molecular interactions of cadherin cis-dimers must be determined. Models based on the packing in crystal structures continue to change and offer conflicting views. As remains with the trans-dimer model, residues involved in cis-dimerization must be determined.
Acknowledgments
Work from the Nelson Laboratory is supported by NIH GM55227, and T.D.P. is also supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
References
- Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–19. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens T, Pertz O, Haussinger D, Fauser C, Schulthess T, Engel J. Analysis of heterophilic and homophilic interactions of cadherins using the c-Jun/c-Fos dimerization domains. J Biol Chem. 2002;277:19455–60. doi: 10.1074/jbc.M200606200. [DOI] [PubMed] [Google Scholar]
- Behrens J, Birchmeier W, Goodman SL, Imhof BA. Dissociation of Madin-Darby canine kidney epithelial cells by the monoclonal antibody anti-arc-1: mechanistic aspects and identification of the antigen as a component related to uvomorulin. J Cell Biol. 1985;101:1307–15. doi: 10.1083/jcb.101.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschuk OW, Sullivan R, David S, Pouliot Y. Identification of a cadherin cell adhesion recognition sequence. Dev Biol. 1990;139:227–9. doi: 10.1016/0012-1606(90)90290-y. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–13. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Yap AS, Gumbiner BM. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol. 1996;135:487–96. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev NA, Troyanovsky SM. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J Cell Biol. 1998;142:837–46. doi: 10.1083/jcb.142.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–62. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- Gumbiner B, Simons K. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J Cell Biol. 1986;102:457–68. doi: 10.1083/jcb.102.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–57. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–9. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- Hatta K, Nose A, Nagafuchi A, Takeichi M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion molecule: its identity in the cadherin gene family. J Cell Biol. 1988;106:873–81. doi: 10.1083/jcb.106.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussinger D, Ahrens T, Sass HJ, Pertz O, Engel J, Grzesiek S. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–39. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- Kemler R. Classical cadherins. Semin Cell Biol. 1992;3:149–55. doi: 10.1016/s1043-4682(10)80011-x. [DOI] [PubMed] [Google Scholar]
- Klingelhofer J, Troyanovsky RB, Laur OY, Troyanovsky S. Amino-terminal domain of classic cadherins determines the specificity of the adhesive interactions. J Cell Sci. 2000;113 (Pt 16):2829–36. doi: 10.1242/jcs.113.16.2829. [DOI] [PubMed] [Google Scholar]
- Koch AW, Pokutta S, Lustig A, Engel J. Calcium binding and homoassociation of E-cadherin domains. Biochemistry. 1997;36:7697–705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- Koch AW, Bozic D, Pertz O, Engel J. Homophilic adhesion by cadherins. Curr Opin Struct Biol. 1999;9:275–81. doi: 10.1016/S0959-440X(99)80038-4. [DOI] [PubMed] [Google Scholar]
- Leckband D, Sivasankar S. Mechanism of homophilic cadherin adhesion. Curr Opin Cell Biol. 2000;12:587–92. doi: 10.1016/s0955-0674(00)00136-8. [DOI] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–4. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nose A, Takeichi M. A novel cadherin cell adhesion molecule: its expression patterns associated with implantation and organogenesis of mouse embryos. J Cell Biol. 1986;103:2649–58. doi: 10.1083/jcb.103.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–55. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Kemler R. Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol. 1990;111:1645–50. doi: 10.1083/jcb.111.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Hoschutzky H, Herrenknecht K, Kemler R. A possible new adhesive site in the cell-adhesion molecule uvomorulin. Mech Dev. 1990;33:49–56. doi: 10.1016/0925-4773(90)90134-8. [DOI] [PubMed] [Google Scholar]
- Perret E, Benoliel AM, Nassoy P, Pierres A, Delmas V, Thiery JP, Bongrand P, Feracci H. Fast dissociation kinetics between individual E-cadherin fragments revealed by flow chamber analysis. EMBO J. 2002;21:2537–46. doi: 10.1093/emboj/21.11.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–47. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- Ringwald M, Schuh R, Vestweber D, Eistetter H, Lottspeich F, Engel J, Dolz R, Jahnig F, Epplen J, Mayer S, et al. The structure of cell adhesion molecule uvomorulin. Insights into the molecular mechanism of Ca2+-dependent cell adhesion. EMBO J. 1987;6:3647–53. doi: 10.1002/j.1460-2075.1987.tb02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffell JL, Williams EJ, Mason IJ, Walsh FS, Doherty P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron. 1997;18:231–42. doi: 10.1016/s0896-6273(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–90. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA. Structural basis of cell–cell adhesion by cadherins. Nature. 1995;374:327–37. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Sivasankar S, Brieher W, Lavrik N, Gumbiner B, Leckband D. Direct molecular force measurements of multiple adhesive interactions between cadherin ectodomains. Proc Natl Acad Sci U S A. 1999;96:11820–4. doi: 10.1073/pnas.96.21.11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Gumbiner B, Leckband D. Direct measurements of multiple adhesive alignments and unbinding trajectories between cadherin extracellular domains. Biophys J. 2001;80:1758–68. doi: 10.1016/S0006-3495(01)76146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S. E-cadherin functions as a cis-dimer at the cell–cell adhesive interface in vivo. Nat Struct Biol. 1999;6:310–2. doi: 10.1038/7542. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: a molecular family important in selective cell–cell adhesion. Annu Rev Biochem. 1990;59:237–52. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–63. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–14. [PMC free article] [PubMed] [Google Scholar]
- Vestweber D, Kemler R. Identification of a putative cell adhesion domain of uvomorulin. EMBO J. 1985;4:3393–8. doi: 10.1002/j.1460-2075.1985.tb04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJ, Furness J, Walsh FS, Doherty P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron. 1994;13:583–94. doi: 10.1016/0896-6273(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Williams E, Williams G, Gour BJ, Blaschuk OW, Doherty P. A novel family of cyclic peptide antagonists suggests that N-cadherin specificity is determined by amino acids that flank the HAV motif. J Biol Chem. 2000;275:4007–12. doi: 10.1074/jbc.275.6.4007. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Williams G, Howell FV, Skaper SD, Walsh FS, Doherty P. Identification of an N-cadherin motif that can interact with the fibroblast growth factor receptor and is required for axonal growth. J Biol Chem. 2001;276:43879–86. doi: 10.1074/jbc.M105876200. [DOI] [PubMed] [Google Scholar]
- Williams G, Williams EJ, Doherty P. Dimeric versions of two short N-cadherin binding motifs (HAVDI and INPISG) function as N-cadherin agonists. J Biol Chem. 2002;277:4361–7. doi: 10.1074/jbc.M109185200. [DOI] [PubMed] [Google Scholar]
- Yoshida-Noro C, Suzuki N, Takeichi M. Molecular nature of the calcium-dependent cell–cell adhesion system in mouse teratocarcinoma and embryonic cells studied with a monoclonal antibody. Dev Biol. 1984;101:19–27. doi: 10.1016/0012-1606(84)90112-x. [DOI] [PubMed] [Google Scholar]