Abstract
The interplay among commonly used physicochemical properties in drug design was examined and utilized to create a prospective design tool focused on the alignment of key druglike attributes. Using a set of six physicochemical parameters ((a) lipophilicity, calculated partition coefficient (ClogP); (b) calculated distribution coefficient at pH = 7.4 (ClogD); (c) molecular weight (MW); (d) topological polar surface area (TPSA); (e) number of hydrogen bond donors (HBD); (f) most basic center (pKa)), a druglikeness central nervous system multiparameter optimization (CNS MPO) algorithm was built and applied to a set of marketed CNS drugs (N = 119) and Pfizer CNS candidates (N = 108), as well as to a large diversity set of Pfizer proprietary compounds (N = 11 303). The novel CNS MPO algorithm showed that 74% of marketed CNS drugs displayed a high CNS MPO score (MPO desirability score ≥ 4, using a scale of 0−6), in comparison to 60% of the Pfizer CNS candidates. This analysis suggests that this algorithm could potentially be used to identify compounds with a higher probability of successfully testing hypotheses in the clinic. In addition, a relationship between an increasing CNS MPO score and alignment of key in vitro attributes of drug discovery (favorable permeability, P-glycoprotein (P-gp) efflux, metabolic stability, and safety) was seen in the marketed CNS drug set, the Pfizer candidate set, and the Pfizer proprietary diversity set. The CNS MPO scoring function offers advantages over hard cutoffs or utilization of single parameters to optimize structure−activity relationships (SAR) by expanding medicinal chemistry design space through a holistic assessment approach. Based on six physicochemical properties commonly used by medicinal chemists, the CNS MPO function may be used prospectively at the design stage to accelerate the identification of compounds with increased probability of success.
Keywords: Multiparameter optimization (MPO), central nervous system (CNS), CNS MPO, CNS drugs, CNS candidates, lipophilicity, topological polar surface area, polarity, molecular weight, hydrogen bond donor, most basic pKa, high-throughput screening, passive permeability, Madin−Darby canine kidney, P-glycoprotein, human liver microsome stability, Unbound intrinsic clearance, drug−drug interactions, dofetilide binding, transformed human liver epithelial cells, cellular toxicity, Harrington optimization, desirability score, multivariant optimization
The high cost of drug discovery combined with high attrition rates of preclinical and clinical candidates has prompted the pharmaceutical industry to take action to improve the survival of their development candidates and increase the speed at which these candidates are identified (1). Ideally, medicinal chemists would like to increase the probability of prospectively designing molecules that survive preclinical safety studies and that possess optimal pharmacokinetic and pharmacodynamic properties to test hypotheses in the clinic. We became interested in the development of a prospective design tool based on key physicochemical properties that would enable a multiparameter optimization of druglike properties, with the goal of increasing flexibility in design and the probability of identifying candidates with optimal pharmacokinetic and safety profiles. Ultimately, it was envisioned that such a prospective design tool, together with other contemporary design concepts (2−7), would help prioritize design ideas and decrease the number of design cycles, accelerating the identification of candidates with enhanced survival.
In the preceding paper, we described a thorough analysis of 119 central nervous system (CNS) marketed drugs (drug set or drugs) and 108 Pfizer CNS candidates (candidate set or candidates) (100). We examined six fundamental physicochemical properties associated with these two sets of compounds: (a) lipophilicity, calculated partition coefficient (ClogP); (b) distribution coefficient at pH = 7.4 (ClogD); (c) molecular weight (MW); (d) topological polar surface area (TPSA) (8); (e) number of hydrogen bond donors (HBD); (f) most basic center (pKa). The CNS drug space defined by these six physicochemical properties is quite broad, but our scholarship pointed to optimum ranges for each of these properties. The analysis of the drugs and candidates clearly showed that the drug set had a high alignment of key attributes including high passive permeability (Papp), low P-glycoprotein (P-gp) efflux (9), low unbound intrinsic clearance (CLint,u) measured by human liver microsomes, (10,11) and high cell viability in a transformed human liver epithelial cell line (THLE Cv). Herein, we report our efforts to develop a CNS MPO design tool that does not focus on hard cutoffs or single end points but utilizes the above set of six fundamental physicochemical properties to, in a probabilistic manner, prospectively align druglike attributes such as high permeability, low P-gp efflux liability, low metabolic clearance, and high safety into one molecule.
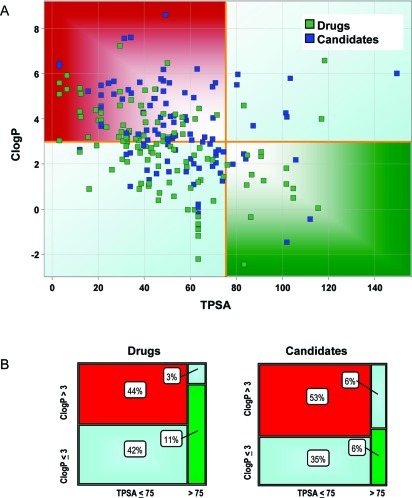
Designing for optimal pharmacokinetic and safety properties in one molecule by utilizing hard cutoffs or by focusing on a single property may restrict design space and may not align multiple attributes at once. Recently, Hughes et al. examined physicochemical properties associated with adverse events observed during in vivo toleration (IVT) studies (12). These authors concluded that compounds with both high lipophilicity (ClogP > 3) and low polarity (TPSA < 75 Å2) (3 and 75 relative risk factors) had a significantly increased relative safety risk (6:1) of showing adverse outcomes in IVT studies, in comparison to compounds that exhibited both low lipophilicity (ClogP < 3) and high polarity (TPSA > 75 Å2). An analysis of the drug set using the 3 and 75 relative risk factors (Figure 1A,B) showed that almost half of the CNS drugs (44%) reside in the higher risk space (upper left quadrant), yet these drugs overcame attrition risks and were marketed. In contrast, far fewer CNS drugs (11%) populate the lower risk space (lower right quadrant), presumably due to the challenge of routinely achieving optimal brain penetration in such a high polarity space. The plot of the candidate set is also shown for comparison in Figures 1A,B; a trend similar to that of the drug set is observed. If the ClogP > 3 and TPSA < 75 Å2 end points were to be used as strict cutoffs, the CNS chemical space that a drug designer could explore would be significantly restricted and would lead to undue hardship in the discovery of CNS drugs. Clearly, we need to understand the properties associated with CNS drugs that allow them to achieve full alignment of key druglike attributes yet overcome the safety odds in the higher risk space (ClogP > 3, TPSA < 75 Å2). An equally important objective is to successfully move CNS design into the more favorable safety risk space (ClogP < 3, TPSA > 75 Å2) while preserving CNS penetration.
Figure 1.
(A) Distribution of drugs and candidates in the ClogP and TPSA space. Orange lines represent the cutoff values for ClogP (3) and TPSA (75 Å2) relative risk factors. Compounds are colored by compound type: drugs are shown in light green and candidates in dark blue. Compounds in the lower right quadrant (ClogP < 3 and TPSA > 75 Å2) are considered to have lower risk for adverse safety findings. (B) Mosaic plots of drugs and candidates. Green boxes represent the percentage of compounds within the low-risk safety space (ClogP < 3 and TPSA > 75 Å2), and the red boxes represent the compounds in higher risk space (ClogP > 3 and TPSA < 75 Å2). The number in each box reflects the percentage of compounds in that category.
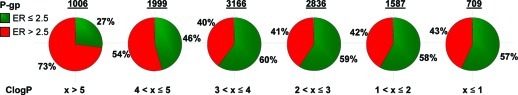
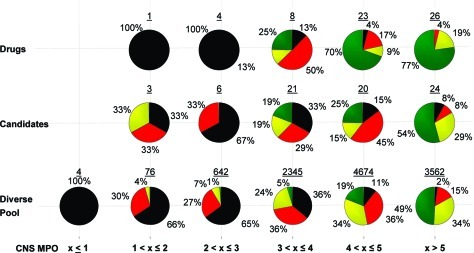
Lipophilicity as defined by ClogP has been a design component utilized by medicinal chemists for decades. The advent of the Rule of Five reinforced the importance of lipophilicity in drug design and encouraged medicinal chemists to target molecules with reduced ClogP (13). Reducing ClogP has been reported to not only improve safety outcomes (12) but also reduce human liver microsome (HLM) clearance (14). ClogP can be utilized to improve some end points quite successfully, but it falls short of aligning all of the desired ADME (absorption, distribution, metabolism, and elimination) and safety attributes. For example, with the diverse pool set of Pfizer compounds (see discussion section for details of the diverse pool), plotting P-glycoprotein (P-gp) liability versus ClogP reveals that there is little correlation between lowering ClogP and P-gp efflux liability for ClogP values ≤ 5, as shown in Figure 2. A single parameter may thus fail to simultaneously align multiple attributes, reinforcing the need to leverage multiple physicochemical properties of a molecule in order to achieve the optimum balance of ADME and safety properties.
Figure 2.
The distribution of binned P-gp efflux ratios (ER) obtained from the MDCK-MDR1 assay (low P-gp liability, ER ≤ 2.5, green; high P-gp liability, ER > 2.5, red) for the diverse pool set across a range of ClogP values. The number of compounds represented by each pie is shown above the pie graph.
Based on the above analyses, in this paper, we examine whether an increased probability of identifying aligned druglike attributes can be achieved using a multiparameter approach. First, we discuss the key components of the CNS MPO and its transformation functions based on the six fundamental physicochemical properties that are commonly utilized by medicinal chemists in compound design. Second, we examine the CNS MPO score distribution of the drug and candidate sets to determine whether the drug set can distinguish itself from the candidate set based on the MPO algorithm. Third, we look at the potential utility of the CNS MPO algorithm as a design tool. In particular, we investigate whether increasing the CNS MPO desirability score for the drugs, candidates, and a large diverse set of proprietary Pfizer compounds leads to an increase in the probability of having desired in vitro ADME and safety attributes [e.g., high passive permeability (Papp), (9) low P-gp efflux (9), low clearance (CLint,u) (10,11), high cell viability (THLE Cv) (100), or low inhibition of dofetilide binding (15)]. Fourth and last, using all three sets of compounds, we examine whether an increase in the CNS MPO desirability score results in a higher probability of identifying compounds with aligned ADME attributes in one molecule. In this section, we also assess the ability of the tool to increase design space and flexibility compared with the use of hard cutoffs for individual physicochemical properties.
Results and Discussion
A CNS Multiparameter Optimization and Desirability Score
Multiparameter optimization methods are commonly used to assess and balance the effects of several variables, weighted based on their importance to the overall objective. The term “desirability function” was first introduced by Harrington in the context of his transformation of multiple attributes into dimensionless scales, which were then arithmetically or geometrically combined into a single score (15). The technique has also been applied to library designs (16,17) and the objective function for a genetic algorithm-based molecular docking software (18).
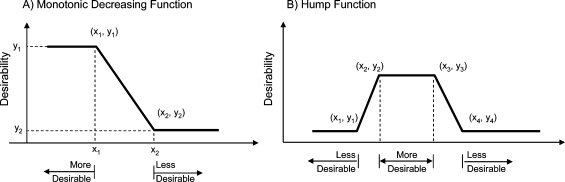
In this work, we examined a variant of Harrington’s optimization method involving a summation of the individual components to yield a composite desirability score. Each component of a desirability function is a transformed function and is usually defined by a series of inflection points delineating the desirable region(s) and undesirable region(s) of properties (x variable) with a certain desirability score (y variable) as shown in Figure 3. For example, a monotonic decreasing function is defined by a desirable region if the property x ≤ x1 and an undesirable region for x > x2. A linear transformation is applied between the two inflection points (x1 < x ≤ x2). Similarly, a hump function is defined by two undesirable regions and one desirable region, with linear transformation between the inflection points (Figure 3B). The overall desirability function is the sum of all transformed components (see Methods section). Utilization of a summation approach prevents a severe penalty in overall desirability score if one parameter is outside the desired limits set by the user, in comparison to a multiplicative approach, which could result in an overall desirability score at or near zero if one parameter was severely unfavorable. The summation approach thus avoids the hard cutoff trap and enables the expansion of desirable design space.
Figure 3.
Desirability component functions are defined by a set of inflection points; higher y values represent more desirable regions: (A) a monotonic decreasing function is defined by two inflection points; (B) a hump function is defined by four inflection points.
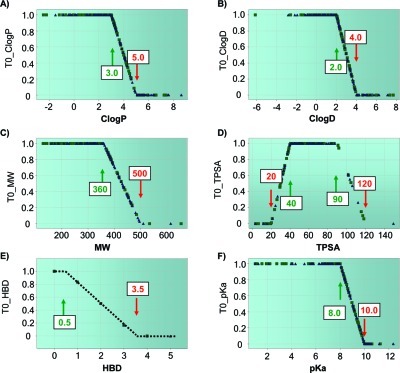
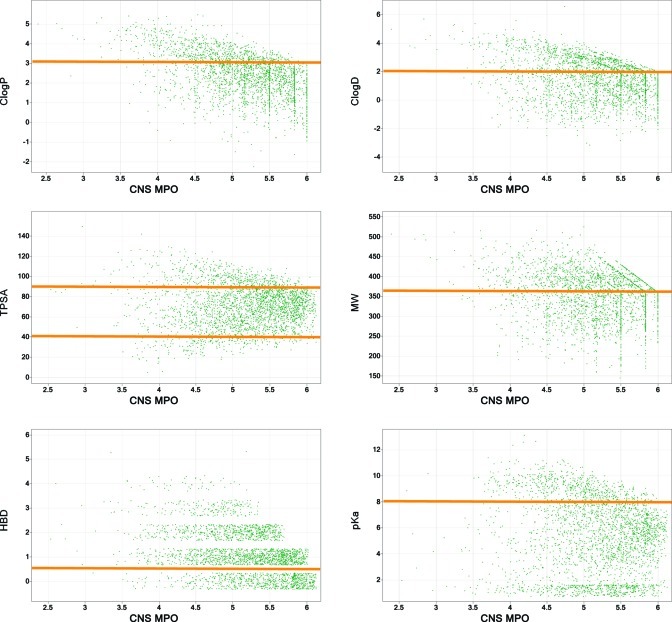
The CNS MPO score was built using six fundamental physicochemical properties (ClogP, ClogD, MW, TPSA, HBD, and pKa) commonly employed in compound design to address specific ADME and safety issues. A monotonic decreasing function was used for ClogP, ClogD, MW, HBD, and pKa, and a hump function was used for TPSA. All physicochemical properties were weighted equally with a desirability score ranging from 0.0 to 1.0 for each property. The most desirable and least desirable ranges for each physicochemical property are given in Table 1. Transformed values (T0) of the six properties were determined for each compound, and the summation of the transformed component score yielded the final “CNS MPO” desirability score, which can range from zero (0) to six (6). For each physicochemical property, the inflection points that define optimal, less optimal, and undesirable ranges were selected based on the authors’ and Pfizer scientists’ medicinal chemistry experiences (Figure 4). The inflection point selections were validated using knowledge of property distribution space for CNS drugs highlighted in the preceding publication (100) and other literature sources referenced above. Individual drugs and candidates were mapped to their corresponding property; within each of the physicochemical property plots, as can be seen in Figure 4, compounds populated most of the range of each function.
Table 1. The CNS MPO Properties, Functions, Weighting, Value Range and Parameter Ranges.
| properties | transformation (T0) | weight | more desirable range (T0 = 1.0) | less desirable range (T0 = 0.0) |
|---|---|---|---|---|
| ClogP | monotonic decreasing | 1.0 | ClogP ≤ 3 | ClogP > 5 |
| ClogD | monotonic decreasing | 1.0 | ClogD ≤ 2 | ClogD > 4 |
| MW | monotonic decreasing | 1.0 | MW ≤ 360 | MW > 500 |
| TPSA | hump function | 1.0 | 40 < TPSA ≤ 90 | TPSA ≤ 20; TPSA > 120 |
| HBD | monotonic decreasing | 1.0 | HBD ≤ 0.5 | HBD > 3.5 |
| pKa | monotonic decreasing | 1.0 | pKa ≤ 8 | pKa > 10 |
Figure 4.
Each plot represents one of the six physicochemical property desirability functions used to generate the CNS MPO. Each point on a plot represents a drug or candidate: (A) ClogP; (B) ClogD; (C) MW; (D) TPSA; (E) HBD; (F) pKa. The most desirable (T0 = 1.0) and least desirable (T0 = 0.0) inflection points are marked with green and red arrows, respectively. A linear function was used to determine the desirability scores between the inflection points.
Individual component scores and overall CNS MPO desirability scores for several drugs are shown in Table 2. As an example, zolpidem’s overall CNS MPO desirability score (5.4) is a composite of optimal values for three of the six properties (MW, HBD, and pKa) and less optimal values for the remaining three properties (ClogP, ClogD, and TPSA). It is crucial to emphasize that while the overall composite CNS MPO score is important, individual transformed values (T0) are equally valuable, because they can highlight potential problems a compound may encounter. For instance, if a compound has a less optimal or undesirable T0_ClogP score, a medicinal chemist would be alerted to its potential for enhanced metabolic liability and increased risk of safety issues and could work to optimize both the holistic and individual scores. Among the 119 drugs examined, aniracetam (nootropic), caffeine (stimulant), flumazenil (benzodiazepine antagonist), and zaleplon (sedative/hypnotic) had perfect CNS MPO scores of six. For the numerical values for the full drug set, see the Supporting Information.
Table 2. CNS MPO Scores and Individual Transformed Scores (T0) for Selected Drugs.
| drugs | T0_ClogP | T0_ClogD | T0_TPSA | T0_MW | T0_HBD | T0_pKa | CNS MPO |
|---|---|---|---|---|---|---|---|
| alprazolam | 1.00 | 0.75 | 1.00 | 1.00 | 1.00 | 1.00 | 5.8 |
| zolpidem | 0.99 | 0.57 | 0.88 | 1.00 | 1.00 | 1.00 | 5.4 |
| paroxetine | 0.38 | 1.00 | 0.99 | 1.00 | 0.83 | 0.00 | 4.2 |
| risperidone | 1.00 | 0.86 | 1.00 | 0.64 | 1.00 | 1.00 | 5.5 |
| methylphenidate | 1.00 | 1.00 | 0.92 | 1.00 | 0.83 | 0.00 | 4.8 |
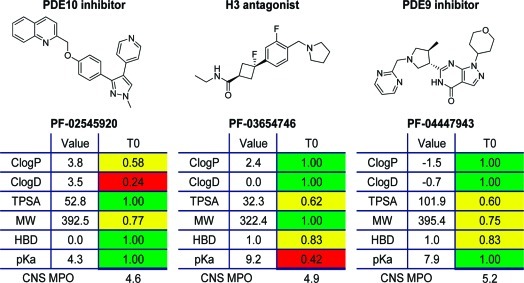
CNS MPO provides a method for balancing multiple variables without the penalty of hard cutoffs, because there are countless ways to arrive at a similar score. This is well illustrated utilizing three Pfizer CNS candidates in development: PF-02545920 (phosphodiesterase 10 (PDE10) inhibitor), PF-03654746 (histamine H3 antagonist), and PF-04447943 (phosphodiesterase 9 (PDE9) inhibitor); see Table 3. The PDE10 inhibitor has a CNS MPO score of 4.6, the H3 receptor antagonist has a CNS MPO score of 4.9, and the PDE9 inhibitor has a CNS MPO score of 5.2. All three compounds have successfully completed regulatory toxicity studies, as well as first-in-human studies, and have entered phase 2 trials, suggesting that they have suitable physicochemical properties and appropriately aligned druglike attributes to test their biological mechanisms in the clinic. All three candidates have a CNS MPO score >4.5, yet they arrive at this high CNS MPO desirability score in very different ways, reflecting the difference in characteristics for these molecules (Table 3). The PDE10 candidate has 3 out of 6 properties in optimal property space: TPSA (52.8 Å2), HBD (0), and pKa (4.3), all receiving a full property (T0) score of one. The other three properties are in less optimal space: ClogP (3.8), ClogD (3.5), and MW (392.5), with transformed values in the range of 0.2 to 0.8. Summation of the individual property values yields the overall CNS MPO desirability score of 4.6. In contrast, the H3 receptor antagonist displays optimal lipophilicity (ClogP = 2.4, ClogD = 0) and MW (322.4) properties but occupies less favorable property ranges for TPSA (32.3 Å2), HBD (1), and pKa (9.2). Finally, the PDE9 candidate resides in optimal property space for ClogP (−1.5), ClogD (−0.7), and pKa (7.9) but in slightly less optimal space with regard to TPSA (101.9 Å2), MW (395.4), and HBD (1). This triad of compounds also showcases how the CNS MPO algorithm avoids hard cutoffs: if strict limits for ClogP or ClogD had been set at ≤3.0 or design was confined to low-risk space as defined by the 3 and 75 relative risk factors (12), the PDE-10 inhibitor and the H3 antagonist, respectively, would have not been synthesized. Similarly, the PDE9 inhibitor would have not been prepared if a hard cutoff for TPSA (≤100 Å2) had been employed to increase brain penetration. The CNS MPO algorithm creates flexibility in design, expands drug design space, and may allow for the prospective design of compounds that occupy diverse property space while maintaining desirable attributes including CNS penetration.
Table 3. A CNS MPO Composite Score and Component Score Comparison of Three Pfizer CNS Candidates in Development.
 |
Drug Space
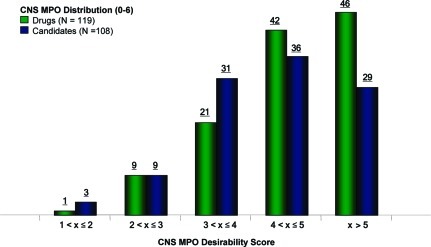
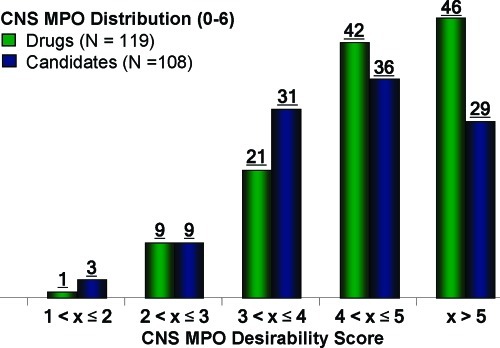
Utilizing the CNS MPO algorithm, we plotted the desirability scores for the drug and candidate sets (Figure 5). We had hypothesized that the drugs would distinguish themselves from the candidates and that these two sets of compounds would have different overall distribution of MPO scores, given that a number of the candidates had been eliminated along the drug development pathway (safety, tolerability, pharmacokinetics, etc.). Figure 5 shows that drugs are more likely distributed in higher MPO bins than candidates (p = 0.0249, one-degree freedom Mantel−Haenszel χ2 test of two by five contingency table). A significantly higher percentage of drugs (74%) had MPO scores >4 in comparison to the candidates (60%) (p = 0.0275). The CNS MPO algorithm clearly distinguished the CNS drugs from the candidate set in distribution of the desirability scores, suggesting that use of such an algorithm may increase the probability of identifying compounds with increased survival. CNS drugs with exceptionally high MPO scores (>5) were chemically diverse and represented a range of mechanistic classes including GPCRs, enzymes, ion channels, and transporters (see Supporting Information for detailed information).
Figure 5.
CNS MPO scores for drugs (green bars) and candidates (blue bars) were plotted from low to high CNS MPO score along the x-axis. The compound count for each bin appears above the bar.
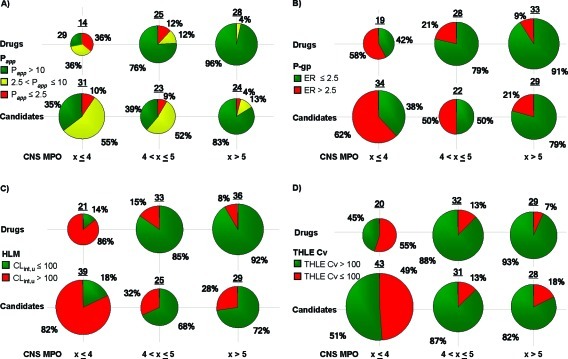
In an effort to further understand the potential utility of the CNS MPO algorithm, we sought to answer a fundamental question: “Does an increase in the CNS MPO score result in a higher probability of identifying compounds with druglike attributes?” If this proved to be true for the drug and candidate sets, then this algorithm could be useful in the prospective design of new compounds. Furthermore, medicinal chemists could use this algorithm in additional ways including the triage of high-throughput screening (HTS) hits or the evaluation of chemical matter in patents around a particular target, without running a single in vitro assay. The ADME and safety end points that we evaluated in vitro were (a) passive apparent permeability (Papp), assessed in an assay utilizing the Madin−Darby canine kidney (MDCK) cell line, (b) P-gp efflux liability, assessed in an assay utilizing an MDCK line stably transfected with the MDR1 gene, which expresses a functionally active human P-gp, (c) metabolic stability (CLint,u), assessed in a human liver microsome (HLM) stability assay, and (d) general cellular toxicity, assessed in a THLE Cv assay. For each of the end points examined, we compared CNS MPO desirability scores for the drug and candidate sets. The CNS MPO desirability scores were broken up into three different groups (≤ 4, 4−5, and >5) to evaluate the ability of the algorithm to identify compounds with the desired druglike attributes: high Papp (Figure 6A), low P-gp efflux (Figure 6B), low CLint,u (Figure 6C), and high THLE Cv (Figure 6D).
Figure 6.
Distribution of ADME and safety attributes for drugs and candidates as a function of the CNS MPO score: (A) binned values for Papp obtained from the MDCK assay, color-coded by high permeability (Papp > 10, green), moderate permeability (2.5 < Papp ≤ 10, yellow), and low permeability (Papp ≤ 2.5, red) in units of 10−6 cm/s; (B) binned values for P-gp efflux liability obtained from the MDCK-MDR1 assay, color-coded by low P-gp liability (ER ≤ 2.5, green) or high P-gp liability (ER > 2.5, red); (C) binned values for clearance (CLint,u) assessed in a human liver microsome stability assay, color-coded by low clearance (CLint,u ≤ 100 mL/(min·kg), green) and high clearance (CLint,u > 100 mL/(min·kg), red); (D) binned values for THLE Cv as measured by an ATP depletion assay, color-coded by high cell viability (IC50 > 100 μM, green) and low cell viability (IC50 ≤ 100 μM, red). Pie charts are color-coded based on the value of the bin, from desirable values (green) to undesirable values (red), and sized by the number of compounds in each pie, which is shown above each pie graph.
The first ADME end point against which the CNS MPO desirability score was evaluated was passive permeability. As the CNS MPO desirability score increased from ≤ 4 to 4−5 and to >5, so did the odds of identifying compounds with high Papp for both the drug and candidate sets (Figure 6A). From the MDCK assay data, we classified the permeability of a molecule as low, moderate, or high based on its Papp rate (expressed as 10−6 cm/s) as follows: Papp ≤ 2.5, low permeability; 2.5 < Papp ≤ 10, moderate permeability; and Papp > 10, high permeability. In the drug set, 96% of the compounds with CNS MPO score >5 had high Papp values. In contrast, only 29% of the drugs with low CNS MPO scores (CNS MPO ≤ 4) had high Papp values. Similar trends were observed for Papp values for the candidates, although the drug set generally had a higher percentage of high passively permeable compounds within each CNS MPO range examined. An increasing CNS MPO desirability score does enhance the odds of identifying compounds with the desired attribute of high passive permeability as measured by the in vitro MDCK assay.
The second ADME end point against which the CNS MPO desirability score was evaluated was P-gp liability (Figure 6B). The MDCK-MDR1 cell line is used in a bidirectional evaluation of permeability [apical to basolateral (AB) and basolateral to apical (BA)] to generate a final efflux ratio (BA/AB) value. A compound with an efflux ratio (ER) ≤ 2.5 is not considered to be a P-gp substrate and may display low P-gp efflux liability. Once more, as the CNS MPO desirability score increased so did the odds of identifying compounds with low P-gp liability. In the drug set, 91% of the compounds with CNS MPO score >5 had low P-gp liability in comparison to 42% of the compounds with scores ≤ 4. A similar trend was observed for the candidates.
The third and last ADME end point examined was metabolic stability (Figure 6C). The unbound clearance, CLint,u, is used to evaluate P450-mediated clearance; a compound is expected to have low clearance if CLint,u ≤ 100 mL/(min·kg) and high clearance if CLint,u > 100 mL/(min·kg) (100). Compounds with high CNS MPO desirability scores (>5) had significantly higher odds of displaying desired low CLint,u, 92% and 72% of the drugs and candidates, respectively. Compounds with low CNS MPO desirability scores (≤ 4) had a lower probability of displaying low CLint,u, with only 14% and 18% of the drugs and candidates, respectively, in this category.
The final end point examined versus the CNS MPO score was general cellular toxicity, measured by the THLE Cv assay (an in vitro safety assay). Cell viability was measured by ATP depletion, with the dynamic range of the assay encompassing IC50 values from single digit micromolar to ≥300 μM. (100). The higher the IC50 value, the lower the cell toxicity is and the greater the cell viability is (desired attribute). The IC50's were classified as follows: THLE Cv ≤ 100 μM (low cell viability) and THLE Cv > 100 μM (high cell viability). In the drug set, compounds with high CNS MPO scores (>5) were considerably more likely to exhibit high cell viability in comparison with compounds having low CNS MPO scores (≤ 4): 93% versus 45%, respectively. Similar findings (82% versus 51%) were obtained for the candidate set (Figure 6D).
This analysis indicates that higher CNS MPO desirability scores enhance the odds of identifying compounds in the drug and candidate sets with druglike ADME and safety attributes such as high passive permeability, low P-gp liability, low clearance, and high cellular viability.
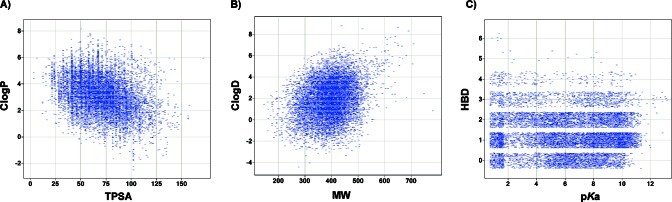
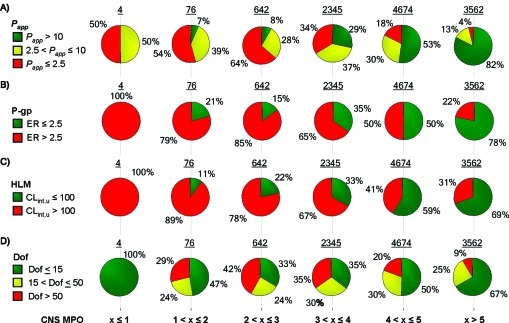
In an effort to assess the general utility of the CNS MPO algorithm, we also evaluated a large pool of proprietary Pfizer compounds, consisting of 11 303 compounds that extensively cover property space defined by ClogP, ClogD, TPSA, MW, HBD, and pKa, as shown in Figure 7A (ClogP vs TPSA), Figure 7B (ClogD vs MW), and Figure 7C (HBD vs pKa). In Figure 8, the CNS MPO desirability scores calculated for the diverse pool set are plotted from low (≤1) to high (>5) desirability for each of the following ADME and safety end points: Papp (Figure 8A), P-gp efflux (Figure 8B), CLint,u (Figure 8C), and inhibition of dofetilide binding (Dof) as a surrogate indicator of hERG potassium ion channel effects (Figure 8D). The ADME end points were assessed in vitro in the same manner as described above for the drugs and candidates. In vitro dofetilide inhibition data was obtained by measuring competitive binding to the dofetilide binding site in HEK-hERG membrane homogenates (19). The output examined was percent inhibition (% inh) of dofetilide binding, where the lower the value, the lower the risk of interference with the hERG cardiac ion channel. Results were classified as follows: % inh ≤ 15 as low-risk, 15 < % inh ≤ 50 as moderate risk, and % inh >50 as high-risk. As the CNS MPO desirability score increased from low (≤ 1) to high (>5) scores, a continuous improvement in the odds of identifying compounds with druglike attributes (high Papp, low P-gp efflux, low CLint,u, and low Dof risk) was observed. For example, compounds with a CNS MPO score of ≤ 1 had none of the most desired ADME attributes (high Papp, low P-gp liability, and low CLint,u). Compounds with CNS MPO scores in the range from 1 to ≤ 2 showed a small increase in the number of compounds with desired ADME attributes (high passive permeability 7%, low P-gp liability 21%, and low CLint,u 11%). Progressive improvement in the identification of additional compounds with the most desired attributes was observed for compounds with CNS MPO scores from 2 to 3, 3 to 4, and 4 to 5. Finally, compounds with a CNS MPO score >5 had the highest proportion of the most desired attributes: high Papp 82%, low P-gp liability 78%, low CLint,u 69%, and low Dof liability 67%. Thus, the CNS MPO desirability score does increase the probability of identifying compounds with the desired in vitro ADME and safety attributes in a large and diverse set, reinforcing the potential general utility of this tool beyond the CNS area.
Figure 7.
Plots of property space for the diverse pool set, where each square represents a compound: (A) plot of ClogP vs TPSA; (B) plot of ClogD vs MW; (C) plot of HBD vs pKa, where HBD are jittered.
Figure 8.
Distribution of ADME and safety attributes as a function of the CNS MPO desirability score for a large and diverse set of compounds. Binned values are shown for (A) passive permeability, Papp; (B) P-gp liability efflux, P-gp; (C) metabolic stability, CLint,u; and (D) inhibition of dofetilide, Dof. Pie charts are color-coded based on the value of each bin, from desirable values (green) to undesirable values (red), and the number of compounds in each pie is shown above the respective pie graph.
We then examined whether an increase in the CNS MPO desirability scores for the three sets of compounds (drugs, candidates, and diverse pool) translated to a greater likelihood of aligning desired ADME attributes (high Papp, low P-gp efflux, and low CLint,u) in one molecule. In the preceding paper, we demonstrated that drugs show a higher alignment of desired ADME (and safety) attributes in one molecule compared with candidates, suggesting that such an alignment may be a differentiating factor in survival to the market (100). Significantly, as the CNS MPO desirability score increased from low (≤ 1) to high (>5), so did the odds of identifying compounds that aligned all three ADME properties in one molecule (Figure 9). Out of the 68 drugs for which all three end points were measured, five compounds had CNS MPO desirability scores of ≤ 3 and none of these aligned all three ADME attributes. In fact, none of these compounds had any of the most desired ADME attributes. In the group of drugs with CNS MPO desirability scores from 3 to ≤ 4, 25% of the compounds attained full alignment of the three ADME attributes, with the rest displaying alignment of 2 out of 3 attributes (13%), 1 out of 3 attributes (50%), or no alignment (13%). Moving to drugs with CNS MPO desirability scores >4, a significant increase (p = 0.0002, two-tailed Fisher’s exact test) in the number of compounds with full alignment of desired ADME attributes was observed, and 77% of the drugs with CNS MPO desirability scores of >5 showed full alignment. Examination of both the candidate and general pool sets yielded similar findings: as the CNS MPO desirability score increased so did the percentage of compounds with aligned attributes. At CNS MPO scores >5, 54% and 49% of candidates and compounds in the diverse pool, respectively, showed alignment of all three attributes. Our analysis thus indicates that the probability of identifying compounds with aligned attributes increases with increasing CNS MPO desirability scores.
Figure 9.
Pie chart of binned values for alignment of desired ADME attributes: high Papp, low P-gp, and low CLint,u. Color-coding for desired ADME attributes: 3/3 (green), 2/3 (yellow), 1/3 (red), and no attributes (black). Binned CNS MPO scores are plotted along the x-axis, and drug, candidate, and diverse pool sets are plotted along the y-axis. The number of compounds in each pie is given above the pie graph.
As stated in the introduction, one of our main goals was to create a new multiparameter optimization algorithm that would promote flexibility in design and expand design options beyond the use of single parameters or hard cutoffs for single or multiple parameters. To assess this design aspect, we examined the subset of compounds in the diverse set that exhibited full alignment of ADME properties (2995 out of 11 303 compounds) for their distribution against each of the six physicochemical properties in relationship to their CNS MPO desirability scores (Figure 10). As shown in each of the six plots, as the CNS MPO desirability scores increased, the density of compounds with full alignment of ADME attributes (high Papp, low P-gp liability, and low CLint,u) also increased. If hard cutoffs for one or several of the physicochemical properties had been applied at the design stage, numerous desirable compounds would have been eliminated from consideration, resulting in significant lost opportunity (Figure 10). For example, if ClogP ≤ 3 had been included as a requirement in design, approximately 30% (843 compounds) of the compounds with full ADME alignment would have not been synthesized. Overall, the CNS MPO desirability scores enable more physicochemical flexibility and expand design space, while enhancing the odds of identifying compounds with a higher probability of aligning key druglike properties as the CNS MPO scores increase. The CNS MPO algorithm may thus be a useful tool at the drug design stage to help with prioritization of ideas for compound synthesis, with the ultimate goal of decreasing the number of design cycles and accelerating the discovery of new medicines.
Figure 10.
The CNS MPO algorithm expands design space while maintaining alignment of desired attributes. Plots display the distribution of compounds from the diverse set that possess full alignment of ADME properties against each of six physicochemical properties (ClogP, ClogD, TPSA, MW, HBD, pKa) in relationship to their CNS MPO desirability scores. Orange lines represent potential hard cutoffs, where hard cutoffs in this example were defined by the CNS MPO optimal property values, for each of the physicochemical properties: ClogP = 3; ClogD = 2; TPSA, high value = 90 Å2 and low value = 40 Å2; MW = 360; HBD = 0.5; and pKa = 8.
Conclusion
Chemical space as defined by physicochemical properties is limitless, yet there are guiding principles that medicinal chemists take into consideration when designing druglike compounds (e.g., Lipinski’s Rule of Five). As part of our efforts in Neuroscience medicinal chemistry, we became interested in developing a more sophisticated understanding of the fundamental physicochemical properties that endow a compound with desirable druglike attributes. We developed a simple new multiparameter optimization design tool (CNS MPO) based on a set of physicochemical properties commonly used by medicinal chemists, with the goals of enabling greater flexibility in CNS compound design beyond the use of single parameters or hard cutoffs, expanding design space, and enhancing the odds of identifying compounds with higher probability of success. Ultimately, it was envisioned that such a design tool, together with other design principles (avoidance of toxicophores/structural alerts), could be used prospectively by medicinal chemists to decrease the number of design cycles and accelerate identification of candidates with enhanced survival.
This new CNS MPO algorithm was based on a set of six fundamental physicochemical parameters (ClogP, ClogD, MW, TPSA, HBD, and pKa) and a variation of Harrington’s optimization method. All physicochemical properties were weighted equally, with a desirability score ranging from 0.0 to 1.0 for each property and a total CNS MPO desirability score ranging from 0.0 to 6.0. For each function represented by a physicochemical property, a series of inflection points were identified that defined optimal, less optimal, and undesirable ranges for CNS agents. While these six parameters are interrelated and there is a perceptible overweighting of lipophilicity (ClogP and ClogD) and pKa (ClogD and pKa), we believe that each parameter has unique characteristics. The success of this algorithm in predicting desirable attributes demonstrated that our assumptions were reasonable. Variations in the number or weighting of properties did not lead to a more predictive algorithm. The simplicity of the CNS MPO tool makes it easy to implement using common software such as Excel. Table 4 is based on an active table, available online, that will allow rapid calculation of a CNS MPO score.
Table 4. Active CNS MPO Calculatora.
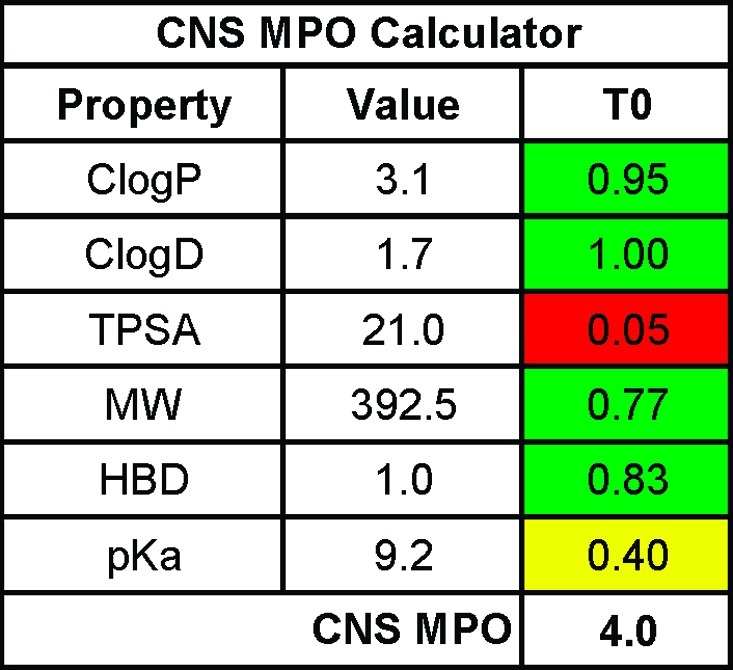 |
Access the active table online.
In order to understand the potential of the new CNS MPO algorithm as a design tool, it was applied to a set of marketed CNS drugs (N = 119) and Pfizer CNS candidates (N = 108). Our analysis showed that 74% of marketed CNS drugs were characterized by a high MPO score (MPO desirability score ≥ 4) in comparison to 60% of the Pfizer CNS candidates, suggesting that this algorithm could potentially be used to identify compounds with a higher probability of success. Our analysis also indicated that as the CNS MPO desirability scores increased, so did the odds of identifying compounds with desirable in vitro ADME and safety attributes as well as compounds where such attributes were aligned in one molecule. For example, 91−96% of the compounds in the drug set with CNS MPO scores >5 displayed high passive permeability, low P-gp liability, favorable metabolic stability, and high cellular viability, and 77% of the drugs with CNS MPO desirability scores of >5 showed full alignment of all three ADME attributes in one molecule. In our preceding paper, we showed that CNS drugs displayed a high alignment of key attributes, reinforcing the potential utility of the CNS MPO algorithm in increasing the probability of identifying compounds with enhanced survival. Our analysis thus indicates that the CNS MPO algorithm can identify compounds with aligned attributes and that the probability of identifying such compounds increases with increasing CNS MPO desirability scores.
While our efforts to develop an MPO algorithm were undertaken to prosecute the Neuroscience portfolio, we were also interested in understanding the general utility of such an algorithm. We evaluated a large pool of Pfizer compounds, consisting of 11 303 compounds that extensively covered property space. Similar trends to those observed with the CNS drugs and candidates were seen with the diverse pool set; that is, as the CNS MPO desirability scores increased so did the probability of identifying compounds with the desired in vitro ADME and safety attributes aligned in one molecule. These results reinforce the potential general utility of the CNS MPO tool across therapeutic areas beyond Neuroscience. While some physicochemical properties and inflection points are related to CNS penetration, the algorithm is not intended to be used purely as a predictor of CNS penetration. On the contrary, our analysis shows that the CNS MPO tool may find utility in other therapeutic areas as is or with appropriate modification of the function or inflection points. For example, if a therapeutic area wanted to restrict CNS penetration while maintaining druglike attributes, the following optimal inflection points could be shifted to higher values: HBD, MW, and TPSA.
Our work also demonstrates that the CNS MPO algorithm offers advantages over hard cutoffs or utilization of single parameters to optimize structure−activity relationships (SAR) by providing flexibility in design and expanding medicinal chemistry design space through a holistic assessment approach. First, the CNS MPO provides a method to balance multiple variables without the penalty of strict cutoffs, because there are countless ways to arrive at a similar score. This is nicely illustrated by the three CNS candidates characterized in Table 3, the H3 antagonist and the PDE9 and PDE10 inhibitors. All of these candidates display CNS MPO desirability scores >4.5, yet they arrive at the high CNS MPO scores in very different ways. This flexibility is very important as we learn how to prospectively design for successful CNS drugs in the higher risk space (ClogP > 3, TPSA < 75 Å2) as well as the lower risk space (ClogP < 3, TPSA > 75 Å2) while preserving CNS penetration (e.g., PDE9 candidate, ClogP = −1.5, TPSA = 101.9). Second, the CNS MPO tool provides an advantage over the use of single parameters, which typically do not address all desired attributes. While ClogP is a powerful design parameter and reducing ClogP has been reported to improve metabolic stability and safety outcomes, this single property falls short of aligning all desired attributes in one molecule. For example, our analysis of a diverse set of Pfizer compounds revealed that there is little correlation between lowering ClogP and reducing P-gp efflux liability. As such, a single parameter may not simultaneously address multiple attributes, reinforcing the need to leverage multiple physicochemical properties to drive the right balance of ADME and safety properties in one molecule. Finally, the CNS MPO algorithm enables the identification of compounds with aligned desired attributes while expanding design space. As shown in Figure 10, as the CNS MPO desirability scores increased, the density of compounds with full alignment of ADME attributes also increased. If hard cutoffs for one or several of the physicochemical properties had been applied at the design stage, numerous desirable compounds would have been eliminated from consideration, resulting in significant lost opportunity and undue hardship in design. As an example, if only compounds with a ClogP ≤ 3 had been considered as part of the design criteria, approximately 30% (843 compounds) of the compounds with full ADME alignment would have not been synthesized.
It is important to emphasize that while the overall composite CNS MPO score is important, individual property desirability scores are equally valuable, because they highlight potential specific problems a compound may encounter. For example, if a compound has nonoptimal ClogP scores, a medicinal chemist would be alerted to the potential increased risk for both metabolic liability and safety issues for that compound and could work to optimize both the holistic and individual scores as necessary. Ultimately, medicinal chemists must utilize all available tools together with their judgment and experience to prioritize and make final decisions on design ideas.
In summary, the CNS MPO algorithm provides a holistic assessment of druglike ADME and safety attributes associated with a compound, and it improves the odds of identifying compounds with such attributes aligned in one molecule. The CNS MPO algorithm creates flexibility in design and expands design space, offering advantages over the use of single parameters or hard cutoffs for single or multiple parameters. We believe the algorithm will provide a new way of evaluating design ideas, impact prioritization and triage of high-throughput screening hits, and facilitate the assessment of competitive intelligence in patent and literature databases. The CNS MPO algorithm is a probabilistic tool with user-defined functions and optimal ranges that, together with other concepts, could be used prospectively in design to reduce the number of design cycles and speed the identification of compounds with enhanced survival.
Methods
Desirability Functions and MPO Score Calculation
A variant of Harrington’s optimization method, using a summation of the individual components to yield a composite desirability score, was developed. Each component of the desirability function is a transformed function, defined by a series of inflection points defining the desirable region and undesirable region(s) of properties (x variable) with a certain desirability score (y variable) as shown in Figure 3. For example, a monotonic decreasing function is defined by a desirable region if the property x ≤ x1 and an undesirable region for x > x2 (Figure 3A). A linear transformation is applied between the two inflection points (x1 < x ≤ x2). Similarly, a hump function is defined by two undesirable regions and one desirable region, with linear transformation between the inflection points (Figure 3B). In general, when a desirability component is defined by inflection points of (x1, y1), (x2, y2), ..., (xn, yn), assuming x1 < x2 < ... < xn, the score at the attribute x is determined by the piecewise linear function:
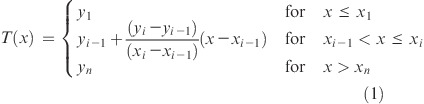 |
1 |
Once each component function is built, the overall desirability of M variables is the sum of each component as defined in eq 2 where wk is the weighting factor for attribute k(20):
| 2 |
The CNS MPO score was built based on six fundamental physicochemical properties: ClogP, ClogD, MW, TPSA, HBD, and pKa. A monotonic decreasing function was used for ClogP, ClogD, MW, HBD, and pKa, and a hump function was used for TPSA. All physicochemical properties were weighted equally, with a desirability score ranging from 0.0 to 1.0 for each property. The most desirable and least desirable ranges for each physicochemical property are listed in Table 1. Transformed values (T0) of the six properties were determined for each compound, and the summation of the transformed component score yielded the final “CNS MPO” desirability score, which can range from zero (0) to six (6). For each physicochemical property, the inflection points that define optimal, less optimal, and undesirable ranges were selected based on the authors’ medicinal chemistry experience (Figure 5). The inflection point selections were validated using knowledge of property distribution space for CNS drugs highlighted in the preceding publication (100) and other literature sources referenced above.
For the work herein, calculated physicochemical properties were obtained using standard commercial packages: Biobyte for ClogP calculations, ACD/Laboratories for ClogD at pH 7.4, and ACD/Laboratories for pKa. For calculation of TPSA, see ref (8). Statistical analyses were carried out using SAS JMP 7 statistical software (21), and the data was visualized with JMP or Spotfire Decision Site (22).
ADME Data
Data on the following in vitro ADME properties were generated in-house utilizing the following high-throughput assays: (a) passive apparent permeability, Papp, assayed utilizing the Madin−-Darby canine kidney (MDCK) cell line (9); (b) P-glycoprotein (P-gp) efflux liability, assessed via an assay utilizing the MDCK-MDR1 cell line, an MDCK line stably transfected with the MDR1 gene that expresses a functionally active human P-gp (9); (c) metabolic stability, expressed as unbound intrinsic clearance (CLint,u), calculated according to eq 3(10) using the measured intrinsic clearance (CLint), obtained via an in vitro, high-throughput human liver microsome assay, and an in silico model for free microsome fraction (cFu,mic) (11):
| 3 |
Compounds included in these studies were handled as 30 mM stock solutions generated, dispensed, and checked for purity by Pfizer’s internal sample bank and subsequently assayed in the ADME and safety assays. The data generated from these assays for the drugs and candidates are included in the previous publication (100). The same in vitro assays and in silico tool (cFu,mic) were used to generate data for the Pfizer proprietary diverse pool set.
Safety Data
Data for the following in vitro safety end points were generated in house via high-throughput assays according to reported methods: (a) Cell viability, measured as activity in an in vitro cellular toxicity assay as a surrogate for acute in vivo toxicity (24). The cellular toxicity data was generated using a transformed human liver epithelial cell line where ATP levels were detected using a bioluminescent end point (100). This assay utilized luciferase to catalyze the formation of light from ATP and luciferin. (b) hERG liability, assessed via inhibition of dofetilide binding (19) as a surrogate indicator of hERG potassium ion channel effects (blocking of hERG may result in prolongation of the QT interval of cardiac rhythm (25)).
Acknowledgments
The authors thank Brian Bronk, Shibing Deng, David Gray, Chris Helal, Spiros Liras, Scot Mente, and Stefan Steyn for helpful discussion. The authors also thank Katherine Brighty for her insightful comments on this manuscript.
Note Added after ASAP Publication
This paper was published on the Web on March 25, 2010, with an incorrect WEO file. The corrected version was reposted on March 30, 2010.
Supporting Information Available
CNS MPO component and desirability scores for drugs (119) and candidates (108) and CNS MPO desirability distribution plot for drug with target class labeled. This material is available free of charge via the Internet at http://pubs.acs.org.
These authors contributed equally to this work.
Supplementary Material
References
- Kola I.; Landis J. (2004) Opinion: Can the pharmaceutical industry reduce attrition rates?. Nat. Rev. Drug Discovery 3, 711–716. [DOI] [PubMed] [Google Scholar]
- Leeson P. D.; Springthorpe B. (2007) The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discovery 6, 881–890. [DOI] [PubMed] [Google Scholar]
- Abad-Zapatero C. (2007) Ligand efficiency indices for effective drug discovery. Expert Opin. Drug Discovery 2, 469–488. [DOI] [PubMed] [Google Scholar]
- Keserue G. M.; Makara G. M. (2009) The influence of lead discovery strategies on the properties of drug candidates. Nat. Rev. Drug Discovery 8, 203–212. [DOI] [PubMed] [Google Scholar]
- Kalgutkar A. S. (2008) Role of bioactivation in idiosyncratic drug toxicity: Structure-toxicity relationships. Biotechnol.: Pharm. Aspects 9, 27–55. [Google Scholar]
- Kalgutkar A. S.; Gardner I.; Obach R. S.; Shaffer C. L.; Callegari E.; Henne K. R.; Mutlib A. E.; Dalvie D. K.; Lee J. S.; Nakai Y.; O'Donnell J. P.; Boer J.; Harriman S. P. (2005) A comprehensive listing of bioactivation pathways of organic functional groups. Curr. Drug Metab. 6, 161–225. [DOI] [PubMed] [Google Scholar]
- Kalgutkar A. S.; Soglia J. R. (2005) Minimising the potential for metabolic activation in drug discovery. Expert Opin. Drug Metab. Toxicol. 1, 91–142. [DOI] [PubMed] [Google Scholar]
- Wager T. T., Chandrasekaran R. Y., Hou X., Troutman M. D., Verhoesf P. R., Villalobos A., Will Y.. Defining desirable central nervous system drug space through the alignment of molecular properties, in vitro ADME and safety attributes. ACS Chem. Neurosci. 2010, 1, DOI: 10.1021/cn100007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P.; Rohde B.; Selzer P. (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its applications to the prediction of drug transport properties. J. Med. Chem. 43, 3714–3717. [DOI] [PubMed] [Google Scholar]
- Feng B.; Mills Jessica B.; Davidson Ralph E.; Mireles Rouchelle J.; Janiszewski John S.; Troutman Matthew D.; de Morais Sonia M. (2008) In vitro P-glycoprotein assays to predict the in vivo interactions of P-glycoprotein with drugs in the central nervous system. Drug Metab. Dispos. 36, 268–275. [DOI] [PubMed] [Google Scholar]
- Hosea N. A.; Collard W. T.; Cole S.; Maurer T. S.; Fang R. X.; Jones H.; Kakar S. M.; Nakai Y.; Smith B. J.; Webster R.; Beaumont K. (2009) Prediction of human pharmacokinetics from preclinical information: Comparative accuracy of quantitative prediction approaches. J. Clin. Pharmacol. 49, 513–533. [DOI] [PubMed] [Google Scholar]
- Gao H.; Yao L.; Mathieu H. W.; Zhang Y.; Maurer T. S.; Troutman M. D.; Scott D. O.; Ruggeri R. B.; Lin J. (2008) In silico modeling of nonspecific binding to human liver microsomes. Drug Metab. Dispos. 36, 2130–2135. [DOI] [PubMed] [Google Scholar]
- Hughes J. D.; Blagg J.; Price D. A.; Bailey S.; DeCrescenzo G. A.; Devraj R. V.; Ellsworth E.; Fobian Y. M.; Gibbs M. E.; Gilles R. W.; Greene N.; Huang E.; Krieger-Burke T.; Loesel J.; Wager T.; Whiteley L.; Zhang Y. (2008) Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 18, 4872–4875. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. (2004) Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technol. 1, 337–341. [DOI] [PubMed] [Google Scholar]
- Waterhouse Rikki N. (2003) Determination of lipophilicity and its use as a predictor of blood-brain barrier penetration of molecular imaging agents. Mol. Imaging Biol. 5, 376–389. [DOI] [PubMed] [Google Scholar]
- E. C. Harrington J. (1965) The desirability function. Ind. Qual. Control 21, 494–498. [Google Scholar]
- Le Bailly de Tilleghem C.; Beck B.; Boulanger B.; Govaerts B. (2005) A fast exchange algorithm for designing focused libraries in lead optimization. J. Chem. Inf. Model. 45, 758–767. [DOI] [PubMed] [Google Scholar]
- Mandal A.; Johnson K.; Wu C. F. J.; Bornemeier D. (2007) Identifying promising compounds in drug discovery: Genetic algorithms and some new statistical techniques. J. Chem. Inf. Model. 47, 981–988. [DOI] [PubMed] [Google Scholar]
- Gehlhaar D. K.; Verkhivker G. M.; Rejto P. A.; Sherman C. J.; Fogel D. B.; Fogel L. J.; Freer S. T. (1995) Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 2, 317–324. [DOI] [PubMed] [Google Scholar]
- Deacon M.; Singleton D.; Szalkai N.; Pasieczny R.; Peacock C.; Price D.; Boyd J.; Boyd H.; Steidl-Nichols J. V.; Williams C. (2007) Early evaluation of compound QT prolongation effects: A predictive 384-well fluorescence polarization binding assay for measuring hERG blockade. J. Pharmacol. Toxicol. Methods 55, 238–247. [DOI] [PubMed] [Google Scholar]
- Lowe C. W. (1967) A report on a simplex evolutionary operation for multiple responses. Trans. Inst. Chem. Eng. 45, T3–T7. [Google Scholar]
- JMP. (1989−2007) SAS JMP 7, SAS Institute, Inc., Cary, NC.
- Spotfire. (1996−2007) Spotfire Decision Site 9, TIBCO Softrware Inc., Somerville, MA.
- Walum E.; Hedander J.; Garberg P. (2005) Research perspectives for pre-screening alternatives to animal experimentation. Toxicol. Appl. Pharmacol. 207, S393–S397. [DOI] [PubMed] [Google Scholar]
- Fermini B.; Fossa A. A. (2003) The impact of drug-induced QT interval prolongation on drug discovery and development. Nat. Rev. Drug Discovery 2, 439–447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.