Summary
Adenomatous polyposis coli (APC) and End-binding protein 1 (EB1) localize to centrosomes independently of cytoplasmic microtubules (MTs) and purify with centrosomes from mammalian cell lines. Localization of EB1 to centrosomes is independent of its MT binding domain and is mediated by its C-terminus. Both APC and EB1 preferentially localize to the mother centriole and EB1 forms a cap at the end of the mother centriole that contains the subdistal appendages as defined by ε-tubulin localization. Like endogenous APC and EB1, fluorescent protein fusions of APC and EB1 localize preferentially to the mother centriole. Depletion of EB1 by RNA interference reduces MT minus-end anchoring at centrosomes and delays MT regrowth from centrosomes. In summary, our data indicate that APC and EB1 are functional components of mammalian centrosomes and that EB1 is important for anchoring cytoplasmic MT minus ends to the subdistal appendages of the mother centriole.
Keywords: Adenomatous polyposis coli, EB1, Centrosome, Mother centriole, Microtubule
Introduction
Adenomatous polyposis coli (APC) protein is the product of a tumor suppressor gene that is mutated in colorectal cancer (Groden et al., 1991) and regulates β-catenin/T-cell-factor-mediated gene expression by stimulating β-catenin degradation (Polakis, 1999). In addition to this function, there is increasing evidence for another role of APC as an organizer of the microtubule (MT) cytoskeleton. The C-terminus of APC has a MT-binding site, and APC stimulates MT assembly and bundling (Munemitsu et al., 1994; Zumbrunn et al., 2001). In epithelial cells, APC protein accumulates in cortical clusters at the tip of plasma membrane extensions (Näthke et al., 1996; Barth et al., 1997a), indicating a role for APC in promoting cell extension and migration (Barth et al., 1997b; Näthke et al., 1997; Pollack et al., 1997). A C-terminal binding partner of APC protein, End-binding protein 1 (EB1) (Su et al., 1995) localizes to polymerizing distal (plus) ends of MTs (Berrueta et al., 1998; Morrison et al., 1998; Mimori-Kiyosue et al., 2000) and can promote MT polymerization in vitro in the presence of the C-terminal binding domain of APC (Nakamura et al., 2001). The mechanism of EB1 localization to plus ends of MTs is unknown but it is independent of its association with APC (Berrueta et al., 1998; Morrison et al., 1998; Mimori-Kiyosue et al., 2000).
Although little is known about the function of the APC-EB1 complex in mammalian cells, more is understood in yeast and Drosophila. The yeast homologue of EB1, Bim1p, binds α-tubulin, localizes to the mitotic spindle and to cytoplasmic MT plus ends, and increases MT dynamicity and length (Schwartz et al., 1997; Tirnauer et al., 1999; Tirnauer and Bierer, 2000). Bim1p also localizes to both spindle poles and mediates localization of its binding partner, Kar9p, to the spindle poles (Hwang et al., 2003; Liakopoulos et al., 2003; Maekawa et al., 2003). Phosphorylation of Kar9p by Clb4/Cdc28 regulates its asymmetric enrichment at the older, bud-oriented spindle pole, from where it is loaded onto cytoplasmic MTs that are then guided in a Kar9p/myosin-dependent manner along actin cables to the bud (Hwang et al., 2003; Liakopoulos et al., 2003; Maekawa et al., 2003). Furthermore, Bim1p and Kar9p bind to the cortex at the bud tip and mediate capture of mitotic MT plus ends at that site (Korinek et al., 2000; Lee et al., 2000). These Bim1p/Kar9p-mediated processes are important for spindle orientation and movement of the nucleus to the bud neck, thereby ensuring correct chromosome segregation from mother to daughter cell (Korinek et al., 2000; Lee et al., 2000; Liakopoulos et al., 2003). In Drosophila, the APC-EB1 complex might capture mitotic MT plus ends at specific cortical sites and thereby mediate spindle orientation (Lu et al., 2001; McCartney et al., 2001).
In mammalian cells, there is evidence that the APC-EB1 complex mediates capture of mitotic MT plus ends at kinetochores, thereby ensuring proper distribution of chromosomes to daughter cells (Fodde et al., 2001; Kaplan et al., 2001). Mammalian EB1 localizes to centrosomes of interphase and mitotic cells (Berrueta et al., 1998; Morrison et al., 1998; Askham et al., 2002), and recent studies indicate that it has a role in MT growth and minus-end anchoring at centrosomes (Askham et al., 2002; Rogers et al., 2002).
Centrosomes are the major MT nucleating structures in the cell and reproduce once per cell cycle in a tightly regulated process (Fig. 1A). Centrosome duplication involves centriole duplication during S phase, centrosome maturation during G2 phase and complete separation of the two centriole pairs with associated pericentriolar material to form the spindle poles at the onset of mitosis (Stearns, 2001). At the time of centrosome separation in late G2, each centrosome contains a mother and daughter centriole (Stearns, 2001). In the single centrosome of G1 cells, MT-anchoring activity is predominantly associated with the older of the two centrioles, designated the mother centriole (Piel et al., 2000). Cytoplasmic MTs are bound and stabilized with their minus ends at specific structures, the subdistal appendages, at one end of the mother centriole (Bornens, 2002).
Fig. 1.
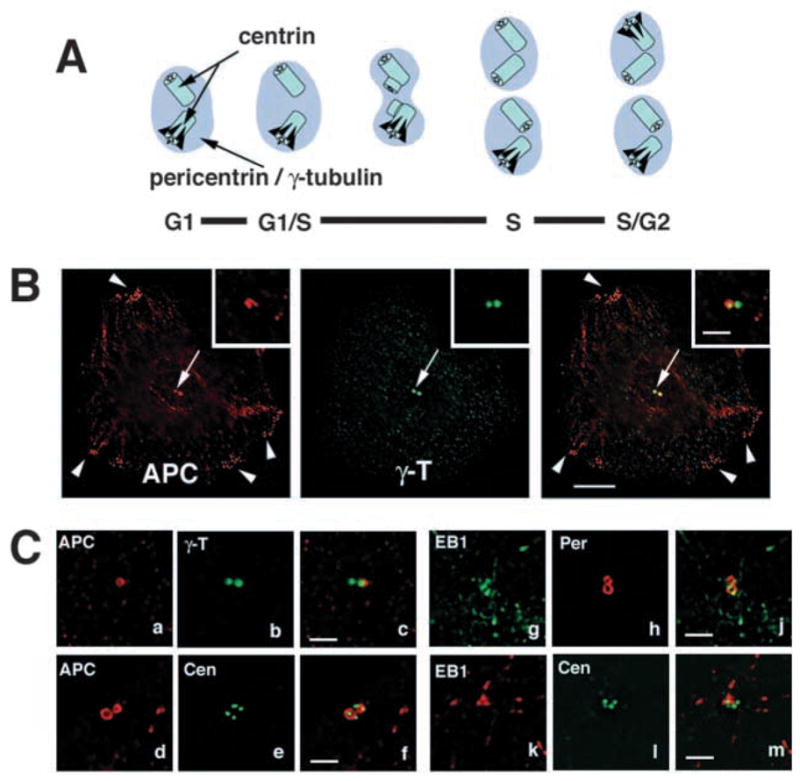
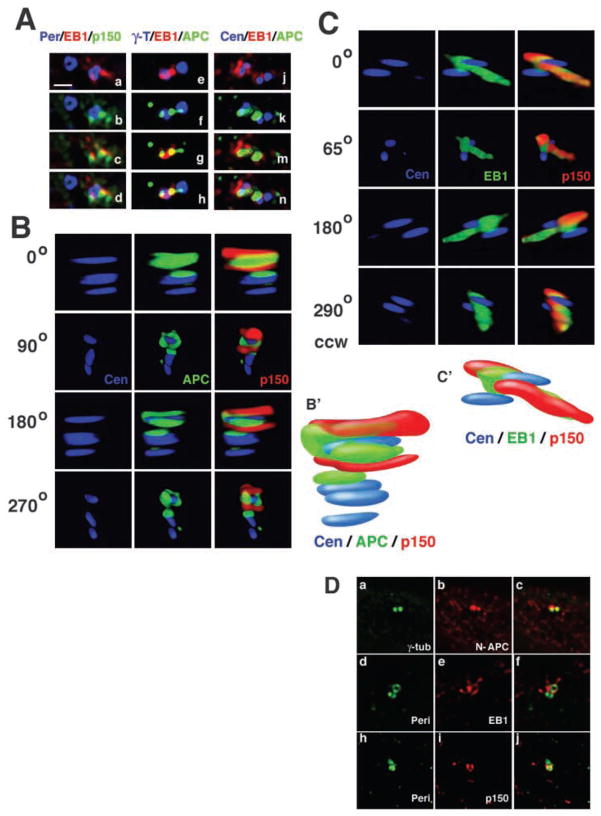
Preferential localization of APC and EB1 to a subset of centrioles. (A) Schematic representation of centrosome duplication during the cell cycle and localization of centrosome marker proteins. Pericentrin and γ-tubulin localize to the pericentriolar material (lilac area); centrin localizes to the centrioles (light blue tubes). MT-anchoring appendages on the mother centriole are marked in black. (B) Basal section of a MDCK cell co-stained for APC (red) and γ-tubulin (green). Arrowheads mark cortical APC clusters, arrow marks localization of APC with γ-tubulin. Bar, 10 μm. Insets show another example of the centrosome region of an MDCK cell at higher magnification. Bar, 2 μm. (C) Sections of U-2 OS cells showing the centrosome regions of cells immunostained for γ-tubulin (green in a–c), pericentrin (red in g–j) and centrin (green in d–f,k–m) and co-stained for APC (red in a–f) and EB1 (green in g–m). Cells with two centrosomes marked by pericentriolar pericentrin (red in g–j) or γ-tubulin (green in a–c) or with four centrioles marked by centrin (green in d–f,k–m) are also shown. APC and EB1 localize preferentially to one of two centrosomes (a–c for APC, g–j for EB1) and to two of four centrioles (d–f for APC) or to one of four centrioles (k–m for EB1). See also Movie 1 (http://jcs.biologists.org/supplemental). Bar, 2 μm.
Here, we show that both APC and EB1 are integral components of mammalian centrosomes. Our results indicate a new, unexpected role of APC and EB1 away from the cortex or MT plus ends and at the center of the MT network, the centrosome.
Materials and Methods
Cell lines and cDNA constructs
Growth conditions for Madin-Darby canine kidney (MDCK) type II/G cells, U-2 OS human osteosarcoma cells, SW480, HeLa S3 and Cos-7 cells have been described previously (Barth et al., 1997a; Quintyne et al., 1999; Chang and Stearns, 2000; Elbashir et al., 2001). Cells were transiently transfected with expression vectors for fusion proteins between green fluorescent protein (GFP) and APC, between Discosoma sp. red fluorescent protein (DsRed) and EB1 (Barth et al., 2002) or between GFP and centrin (White et al., 2000; D’Assoro et al., 2001) using lipofectamine 2000 reagent as described by the manufacturer (Gibco BRL, Gaithersburg, MD). An MDCK cell line with stable expression of full-length EB1 fused to DsRed, used for the experiment shown in Fig. 6A, was made as described (Barth et al., 1997a).
Fig. 6.
APC localizes to the mother centriole. (A) APC co-localizes with DsRed-EB1 to the mother centriole. Sections of MDCK cells expressing DsRed-EB1 (red in a–n) showing the centrosome regions immunostained for centrosome markers pericentrin (blue in a–d), γ-tubulin (blue in e–h) or centrin (blue in j–n) and co-stained for the mother centriole marker p150Glued/dynactin (green in b–d) or APC (green in f–h,k–n). DsRed-EB1 co-localizes with p150Glued/dynactin at only one of the two centrosomes stained by pericentrin (ad). DsRed-EB1 and APC preferentially localize to the same of two centrosomes marked by γ-tubulin (e–h) and to the same two centrioles of four centrioles marked by centrin (j–n). APC precedes EB1 at the second centrosome (e–h). Only two out of 25 analysed U-2 OS and MDCK cells in asynchronous cultures had no APC at the second centrosome, whereas seven out of 28 analysed cells had no EB1 at the second centrosome, indicating that APC localizes earlier to the second mother centriole than EB1. Bar, 1 μm. (B) U-2 OS centrosome region expressing GFP-centrin as a marker for centrioles (blue) and co-immunostained for APC (green) and for the mother centriole with p150Glued/dynactin (red). Sections of this region have been deconvolved and recombined in a 3D rendering. The first column shows four centrioles in multiple angles of counterclockwise (ccw) rotation, with the two central centrioles too close to each other to be resolved as separate spots. The second and third columns show that APC tightly surrounds the mother centriole as marked by p150Glued/dynactin and extends to a second centriole. (B′) Schematic representation of 3D localization of proteins shown in B at a 205° angle. See Movie 4 (http://jcs.biologists.org/supplemental) for a 360° rotation and Materials and Methods for the resolution limits of the optical system. (C) U-2 OS centrosome region expressing GFP-centrin as a marker for centrioles (blue) and co-immunostained for EB1 (green) and for the mother centriole with p150Glued/dynactin (red). Sections of this region have been deconvolved and recombined in a 3D rendering. The first column shows two centrioles in multiple angles of ccw rotation. The second and third columns show that EB1 and p150Glued/dynactin cap one end of the mother centriole and continue in a MT-like filamentous extension from this end of the centriole. (C′) Schematic representation of 3D localization of proteins shown in C at a 0° angle. See Movie 5 (http://jcs.biologists.org/supplemental) for a 360° rotation. (D) APC-EB1 interaction is not essential for localization of APC and EB1 to centrosomes. SW480 centrosome regions were co-immunostained for the centrosomal markers γ-tubulin (green in a,c) or pericentrin (green in d,f,h,j) and APC (red in b,c), EB1 (red in e,f) or p150Glued (red in i,j). N-APC, EB1 and p150Glued localize to centrosomes in SW480 cells that express a truncated form of APC without the EB1 binding site.
Antibodies
Polyclonal rabbit antiserum was raised against a C-terminal fusion of human EB1 to maltose-binding protein (MBP). The serum was partially purified on MBP resin, tested by immunoblotting of purified EB1 and cell lysates, and used for immunofluorescence at 1:50 dilution. Affinity-purified polyclonal rabbit antiserum to a central APC domain (Näthke et al., 1996) was used at a 1:1000 dilution; mouse monoclonal antibody to EB1 (clone Ab-1; Oncogene Research Products, San Diego, CA) at 2 μg ml−1 (immunoblotting of different EB1 domains showed that this antibody recognizes the C-terminus of EB1; A. Barth, unpublished); mouse monoclonal antibody to EB1 (Beckton Dickinson Biosciences Transduction Laboratories, Lexington, KY) at 1:100 dilution; mouse monoclonal antibody to α-tubulin (clone DM1A; Sigma, St Louis, MO) at 1:200 dilution; mouse monoclonal antibody to γ-tubulin (clone GTU88; Sigma) at 1:1000 dilution; mouse monoclonal antibody to centrin at 1:200 dilution (clone 20H5; J. L. Salisbury, Mayo Clinic Foundation, Rochester, MN); rabbit polyclonal antibody to pericentrin at 1:200 dilution (T. Stearns, unpublished); and rabbit polyclonal antibody to ε-tubulin at 1:500 dilution (Chang and Stearns, 2000). Secondary antibodies against mouse, rat or rabbit IgG with minimal species cross-reactivity coupled to FITC or rhodamine were used at 1:200 dilution and coupled to Cy5 were used at 1:100 dilution (Jackson ImmunoResearch, West Grove, PA).
Centrosome purification
Centrosomes were purified from U-2 OS and MDCK cells as described elsewhere (Mitchison and Kirschner, 1986). In short, cells were treated with 10 μg ml−1 nocodazole and 5 μg ml−1 cytochalasin B for 90 minutes at 37°C, washed in PBS and lysed quickly in low ionic strength buffer (1 mM Tris-HCl, pH 8, 8 mM β-mercaptoethanol, 0.5% NP40). Centrosomes in postnuclear supernatant were concentrated by centrifugation onto a 20% Ficoll cushion and purified by fractionation in a 20–62.5% sucrose gradient. Sucrose fractions were collected and diluted in 10 mM Pipes, pH 7.2, 1 mM EDTA and 8 mM β-mercaptoethanol. Centrosomes were pelleted onto coverslips through a 20% glycerol in BRB80 (80 mM Pipes, pH 6.9, 1 mM EGTA, 1 mM MgCl2) cushion, fixed in cold methanol and assayed by immunofluorescence as described below. Sucrose gradient fractions enriched for purified centrosomes were tested for the ability to induce MT aster formation in Xenopus egg extracts. Briefly, Xenopus egg extracts were prepared as described (Murray, 1991), mixed on coverslips with aliquots of centrosome fractions and rhodamine-tubulin, and visualized directly with a Zeiss Axioplan microscope (Carl Zeiss, Thornwood, NY). Rhodamine-tubulin-labeled MT asters formed in assays containing centrosome fractions but not in control assays, containing equal volumes of fractionation buffer.
Depletion of EB1 by small interfering RNA and MT regrowth after nocodazole washout
Small interfering RNAs (siRNAs) were transiently transfected into HeLa S3 cells with Oligofectamine (Invitrogen, Carlsbad, CA) as described (Elbashir et al., 2001) using 21 nucleotide duplex siRNAs directed against human EB1 (Dharmacon Research, Lafayette, CO) or a GFP control (Proligo, Boulder, CO). The EB1 siRNA was targeted against the human EB1 sequence: 5′-TTGCCTTGAAGAAAGTGAA-3′, which is identical to mouse and rat EB1, and different from human EB2 and EB3. Control cells were transfected with an siRNA targeting the GFP sequence: 5′-GCAGCACGACTTCTTCAAG-3′. 120 nM siRNA was added to each 35 mm dish with 30% confluent cultures. Medium was changed 1 day after transfection and cells were passaged onto collagen-coated coverslips 2 days after transfection. Levels of EB1 in cultures treated with siRNA were analysed by immunoblotting and by immunofluorescence 3 days after transfection. In HeLa S3 cultures treated with siRNA against EB1, more than 90% of the cells were depleted of EB1. Western blots of sodium-dodecyl-sulfate (SDS) lysates from untreated HeLa cultures or cultures treated with siRNA against EB1 or control siRNA were immunoblotted with rabbit polyclonal antiserum to EB1 C-terminus and mouse monoclonal antibody DM1A to α-tubulin (Sigma, St Louis, MO) and secondary anti-rabbit Alexa Fluor 680 (Molecular Probes, Oregon) and anti-mouse IRDye800 (Rockland, Pennsylvania). Immunoblots were scanned in a LI-COR infrared imager (LI-COR Biosciences, NE). Immunoblots were then reblotted with mouse monoclonal antibody to actin (clone 4; Chemicon, Temecula, CA) and anti-mouse IRDye800. Bands were quantified using Odyssey software (LI-COR Biosciences, NE). Measurements for EB1 and α-tubulin were normalized against actin measurements to control for gel loading and blotting differences. Three days after transfection, cells transfected with EB1- or control-siRNA were incubated with 33 μM nocodazole for 30 minutes at 4°C and 1 hour at 37°C. Cells were then cooled to room temperature. Nocodazole was washed out three times with fresh medium. After MT regrowth at room temperature for 0 minutes, 10 minutes, 20 minutes or 30 minutes, cells were washed in PBS, pH 7.4, containing 2.7 mM KCl, 1.5 mM KH2PO4, 1 mM MgCl2, 1 mM EGTA, 137 mM NaCl and 8.1 mM NaHPO4, and fixed in methanol at −20°C as described (Barth et al., 1997a). Controls were performed in which the nocodazole was not washed out or in which no nocodazole was added. Cells were immunolabeled for EB1 with purified rabbit polyclonal antiserum to EB1 C-terminus and for α-tubulin with mouse monoclonal antibody DM1A. Cells were imaged using a Zeiss Axioplan microscope with a Zeiss Plan-Neofluar 100×/1.3 oil objective (Carl Zeiss, Thornwood, NY). Images were recorded with a Zeiss MRm camera using AxioVision 3.1 acquisition software. Immunofluorescence images of EB1 were taken with identical exposure times to allow the comparison of fluorescence intensity between images. Images presented show representative cells. Adobe Photoshop version 6.0 was used to enhance the images, with linear adjustments in brightness and contrast applied uniformly and equally to EB1 images of both the EB1-siRNA-treated cells and the control cells. MT aster size was defined as the area covered by MTs that emanated from a centrosome after nocodazole removal. MTs around centrosomes were outlined by hand using the Axiovision 3.1 software’s tracing tool. MT aster size was analysed by measuring the outlined areas using Axiovision 3.1 software (Carl Zeiss, Thornwood, NY).
Cos-7 cells were treated with the same siRNA as used on HeLa cells, which is directed against a conserved region in EB1, and control siRNA targeting the following sequence from GFP: 5′-GGCTACGTCCAGGAGCGCACC-3′ (Dharmacon Research, Lafayette, CO). Cells were transfected with siRNA as described for the HeLa cells and incubated for a total of 5 days, with a second transfection performed on the third day of incubation. In Cos-7 cultures treated with siRNA against EB1, around 30% of the cells were depleted of EB1. EB1 was stained with rabbit polyclonal antiserum to EB1 C-terminus, and α-tubulin was stained with mouse monoclonal antibody DM1A (Sigma). A second experiment staining EB1 with mouse monoclonal antibody (Transduction Laboratories) and tubulin with rat monoclonal antibody YL 1/2 (Accurate Chemical & Scientific, Westbury, NY) produced similar results. Immunolabeled cells were mounted in Vectashield® with Dapi (Vector Laboratories, Burlingame, CA). Images were taken as described for the HeLa cells. MT organization at the centrosome was categorized as either ‘focused’ or ‘diffuse’. A cell in which MTs were centrally focused towards a distinct point in the nuclear periphery was classified as having a focused MT organization. Conversely, a cell that lacked this central focus point of microtubules and instead had a more uniform, broad distribution of MTs around the nuclear periphery, was classified as having a diffuse MT organization.
Immunofluorescence microscopy
2×105 MDCK, U-2 OS or 5×105 SW480 cells were seeded onto 22×22 mm collagen-coated coverslips in 35 mm tissue culture dishes and fixed 12–16 hours later. Cells were rinsed once in PBS pH 7.4 (2.7 mM KCl, 1.5 mM KH2PO4, 1.5 mM MgCl2, 1 mM EGTA, 137 mM NaCl and 8.1 mM NaHPO4), fixed 5 minutes in methanol at −20°C and then rinsed once in PBS with 0.1% Triton X-100. Cells were washed three times in PBS and blocked for 20 minutes at room temperature in PBS with 1% bovine serum albumin and 2% goat serum. Cells were labeled for immunofluorescence as described elsewhere (Barth et al., 1997b). Cells were mounted using Vectashield (Vector Laboratories, Burlingame, CA) and analysed with a Zeiss Axioplan (Carl Zeiss, Thornwood, NY) using a Zeiss Plan-Neofluar 63×/1.25 oil objective and a Zeiss AxioCam MRm camera or with a Delta Vision™ full-spectrum optical sectioning microscope system (Applied Precision, Issaquah, WA; Beckman Center Cell Sciences Imaging Facility) using Olympus PlanApo 60×/1.4 oil or UPlanApo100×/1.35 oil objectives, 1.5× auxiliary magnification and a Photometrics CH350 CCD camera (Photometrics). Optical sections were taken at 0.2 μm step size and deconvolved using constrained iterative Applied Precision SoftWoRx version 2.5 software deconvolution. Deconvolved optical sections were combined and three-dimensional (3D) reconstruction was performed through volume rendering using Volocity 2.0.1 (Improvision, Lexington, MA). 360° rotations at 5° intervals are provided online as supplementary material in form of QuickTime movies (http://jcs.biologists.org/supplemental/). Representative optical sections or different angles of the rotations are shown in Figs 1, 2, 5, 6. The 0° angles were set arbitrarily. The resolution of the optics used is about 0.23 μm in the x,y plane and about 0.7 μm in the x,z and y,z planes. Therefore, structures smaller than 0.7 μm will appear elongated in the z-axis. 3D rendered images of centrosomal proteins show their locations relative to each other but do not reflect the actual size of the structures marked by them. This is only evident in the 3D rendered images in Fig. 5A and Fig. 6B,C, and in the movies in the supplementary material.
Fig. 2.
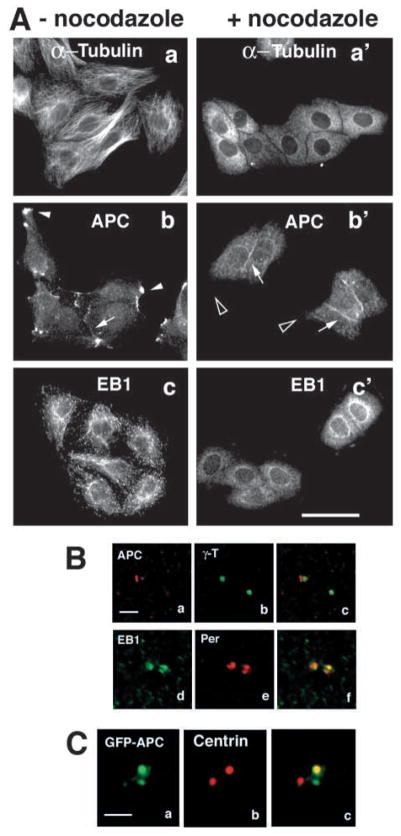
Localization of EB1 and its C-terminal binding partner APC to centrosomes is independent of intact cytoplasmic MTs. (A) MDCK cells (a–c) and MDCK cells treated with nocodazole to disrupt the MT cytoskeleton (a′-c′) were immunolabelled for α-tubulin (a,a′), APC (b,b′) or EB1 (c,c′). Filamentous MTs (a), MT-dependent cortical APC clusters (arrowheads in b) and MT plus-end localization of EB1 (c) are disrupted by nocodazole (a′, black arrowheads in b′,c′). Localization of APC to cell-cell contact sites is independent of intact MTs and seems to be enhanced in nocodazole-treated MDCK cells (arrows in b,b′). Bar, 50 μm. (B) Sections of nocodazole-treated MDCK cultures shown in (A) were stained for centrosome markers γ-tubulin (green in a–c) or pericentrin (red in d–f) and co-stained for APC (red in a–c) or EB1 (green in d–f). APC and EB1 remain localized around the centrosome in nocodazole-treated cells, whereas their MT-dependent localizations are disrupted (see A). Images in d–f show an example in which EB1 localizes to both centrosomes of a cell. Bar, 2 μm. (C) Section of a nocodazole-treated U-2 OS cell expressing GFP-APC (green in a–c) stained with the centriolar marker centrin (red in a–c). GFP-APC preferentially localizes to one of the two centrioles in this cell [Movie 2 (http://jcs.biologists.org/supplemental)]. Bar, 2 μm.
Fig. 5.
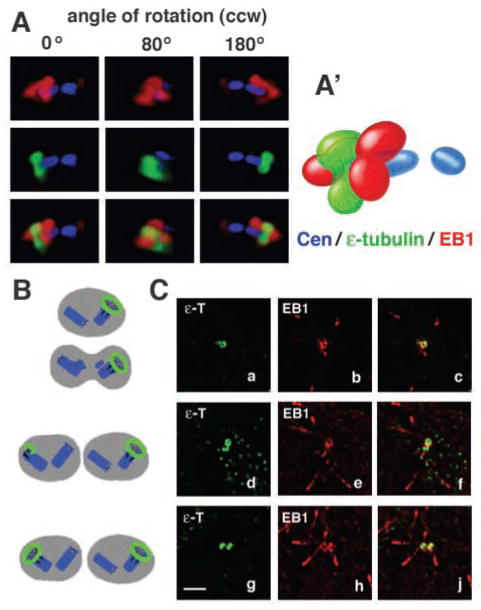
EB1 localizes to the mother centriole. (A) Centrosome region of a nocodazole-treated U-2 OS cell expressing GFP-centrin as a marker for centrioles (blue) and co-immunostained for EB1 (red) and for the mother centriole with ε-tubulin (green). Sections of this region were deconvolved and recombined in a 3D rendering. Multiple angles of a counterclockwise (ccw) rotation show that EB1 localizes with ε-tubulin to one end of the mother centriole. EB1 caps this end of the mother centriole like a wizard’s hat, whereas ε-tubulin forms a more ring-like structure around the same end. (A′) Schematic representation of 3D localization of proteins shown in A at the 0° angle. See Movie 3 (http://jcs.biologists.org/supplemental) for a 360° rotation and Materials and Methods for the resolution limits of the optical system. (B) Schematic representation of ε-tubulin distribution (green) during centrosome maturation as shown in the three panels in (C). (C) U-2 OS cell sections showing different centrosome maturation stages as marked by immunostaining for ε-tubulin (green in a,c,d,f,g,j). Sections are also stained for EB1 (red in b,c,e,f,h,j) to define EB1 localization during centrosome maturation. The first column shows increasing ε-tubulin accumulation at the second mother centriole (a,d,g). The second and third columns show that EB1 has a similar distribution pattern to ε-tubulin at different maturation stages (b–j) but that ε-tubulin precedes EB1 at the second mother centriole (c,f,j). In three out of 12 analysed cells, ε-tubulin was localized to the second centrosome, with EB1 being only at one centrosome. Bar, 2 μm.
Results
Preferential localization of APC and EB1 to a subset of centrioles
APC protein and its C-terminal binding partner EB1 preferentially localize to a subset of four centrioles in cells at the S/G2 stage of the cell cycle. MDCK cells (Fig. 1B), a well-characterized epithelial cell line, and U-2 OS cells (Fig. 1C), a cell line with well investigated centrosome structure (Chang and Stearns, 2000; Chang et al., 2003), were co-labeled with antibodies to APC and EB1, and to several centrosome markers. For every immunofluorescence experiment, a total of 10–30 cells were analysed by optical sectioning and 3D image analysis. Fig. 1 shows DeltaVision-deconvolved sections of cells in which two centrosomes are close to each other. Each centrosome is marked by either pericentrin or γ-tubulin, which label the pericentriolar material surrounding the centrioles, or by two centrioles as labeled by centriole marker protein centrin (Fig. 1A). Notice that centrosome doubling occurs by duplication of the centriole pair during S-phase of the cell cycle and so the number of centrioles indicates the cell cycle stage (Stearns, 2001). APC co-localizes with γ-tubulin near the nucleus (arrows in Fig. 1B) and localizes to cortical clusters in MDCK cells (arrowheads in Fig. 1B) as shown previously (Näthke et al., 1996; Barth et al., 1997a). APC and EB1 often localize preferentially to one of two centrosomes as marked by the pericentriolar proteins γ-tubulin (Fig. 1B insets, Fig. 1Ca–c for APC) and pericentrin (Fig. 1Cg–j for EB1), and to one or two of four centrioles in S/G2 cells (Fig. 1Cd–f for APC and Fig. 1Ck–m for EB1). 3D image analysis shows that APC wraps around most of the centriole along its length in the form of a U-shaped tube that is open at one end and more closed at the other [Movie 1 (http://jcs.biologists.org/supplemental/)]. We also observed partial alignment of APC with a second centriole along its length (Movie 1; see also Fig. 6B and Movie 4). This distribution is typical of proteins that preferentially localize to the older, mother centriole in G1 cells and are then recruited to the new mother centriole in the second centrosome in a cell cycle-dependent manner some time after centriole duplication (Mogensen et al., 2000; Piel et al., 2000; Nakagawa et al., 2001; Chang et al., 2003).
APC and EB1 are integral components of centrosomes
In order to define whether APC and EB1 localization to centrosomes is dependent on intact cytoplasmic MTs, MDCK cells (Fig. 2A,B) were treated with the MT-disrupting drug nocodazole and co-stained for MT, APC or EB1, and centrosome markers. Nocodazole treatment completely disrupted the filamentous network of cytoplasmic MTs (Fig. 2Aa,a′), MT-dependent localization of APC to cortical clusters (Fig. 2Bb,b′) and (Näthke et al., 1996), and MT plus-end localization of EB1 (Fig. 2Ac,c′). However, centrosome localization of APC and EB1 was resistant to nocodazole treatment (Fig. 2Ba–c for APC and Fig. 2Bd–f for EB1). This result shows that APC and EB1 localization to centrosomes is independent of intact cytoplasmic MTs and indicates that APC and EB1 are integral centrosome components.
As a second approach to examine APC distribution, GFP-APC was expressed in U-2 OS cells. We analysed 14 U2-OS cells expressing GFP-APC and observed preferential localization of GFP-APC to a subset of centrioles in every case. Nocodazole-resistant centrosomal GFP-APC localizes preferentially to one of two centrioles and tightly covers the centriole along its length similar to the distribution of endogenous APC [Fig. 1C, Fig. 2C; compare Movie 2 (http://jcs.biologists.org/supplemental/) to Movie 1 (http://jcs.biologists.org/supplemental/)].
We sought direct evidence that APC and EB1 are centrosome components by purifying centrosomes from nocodazole-treated MDCK cells (Fig. 3a–o) and U-2 OS cells (Fig. 3a′–o′). Purified centrosomes were active as defined by their ability to assemble rhodamine-tubulin labeled MT asters in vitro (Fig. 3o,o′) and contained centrosome markers γ-tubulin, centrin and pericentrin (Fig. 3b,b′,e,e′,h,h′,m,m′). APC co-purified with most centrosomes from MDCK cells (Fig. 3a–f, white arrowheads) and U-2 OS cells (Fig. 3a′–f′, white arrowheads); an example of a centrosome without APC is also shown (Fig. 3d–f, black arrowhead). EB1 also purified with most centrosomes from MDCK cells (Fig. 3g–j, white arrowheads) and U-2 OS cells (Fig. 3g′–j′, white arrowheads). However, we observed less co-purification of p150Glued/dynactin with centrosomes from MDCK or U-2 OS cells (Fig. 3k–n for MDCK and Fig. 3k′–n′ for U-2 OS). These results indicate that APC and EB1 are integral components of centrosomes in both cell types examined and might even be more tightly associated with centrosomes than another centrosomal protein p150Glued/ dynactin.
Fig. 3.
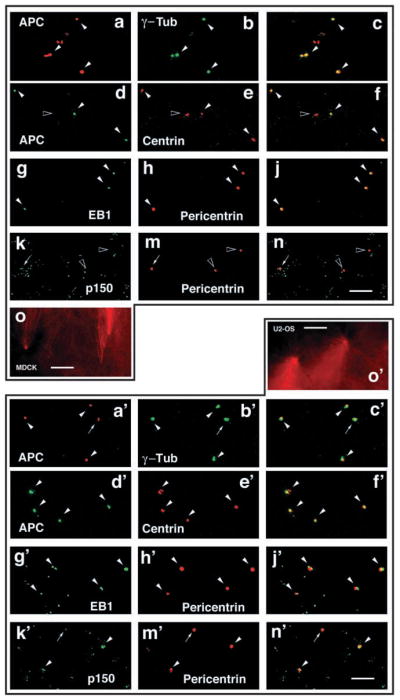
EB1 and its C-terminal binding partner APC co-purify with centrosomes. Centrosomes were purified from nocodazole-treated MDCK cells (a–o) or U-2 OS cells (a′–o′), stained for the centrosome markers γ-tubulin (green in a–c,a′–c′), centrin (red in d–f,d′–f′) or pericentrin (red in g–n,g′–n′) and co-stained for APC (red in a–c,a′–c′ and green in d–f,d′–f′), EB1 (green in g–j,g′–j′) or p150Glued/dynactin (green in k–n,k′–n′). Most of the purified centrosomes contain APC and EB1 (white arrowheads in a–j,a′–j′). An example of a centrosome without APC is shown (black arrowhead in d–f). By contrast, most of the centrosomes have little p150Glued/dynactin (arrows in k–n,k′–n′) or no p150Glued/dynactin (black arrowheads in k–n). Examples of centrosomes that contain p150Glued/dynactin are shown (white arrowheads in k′–n′). Purified MDCK (o) or U-2 OS (o′) centrosomes are functional as measured by induction of MT aster formation in vitro (o,o′). Bar, 10 μm.
The C-terminus of EB1 mediates its localization to centrosomes
Localization of EB1 to centrosomes is MT independent and so we expressed DsRed fusions of full-length EB1 and the N- and C-terminal domains of EB1 in MDCK cells to determine which domain mediates EB1 localization to centrosomes (Fig. 4). We have shown previously that the N-terminal domain (EB1NT) localizes to MTs but not to cortical APC clusters, and that the C-terminal domain (EB1CT) binds to and localizes with APC but not to MTs (Barth et al., 2002). DsRed-EB1 formed a ring around γ-tubulin and localized along MTs emanating from the centrosome (Fig. 4a–c). DsRed-EB1NT also localized along MTs emanating from the centrosome but was excluded from the centrosome area at the center of these MTs (Fig. 4d–f). We have shown previously that, with increasing levels of overexpression, the distributions of DsRed-EB1 and DsRed-EB1NT are less restricted to MT plus ends and become increasingly localized along MTs (Barth et al., 2002). However, even in cells expressing high levels, we did not detect DsRed-EB1NT around the centrosome (Fig. 4d–f). By contrast, DsRed-EB1CT, which does not co-align with MTs (Barth et al., 2002), strongly localized to the centrosome area and formed a ring around γ-tubulin (Fig. 4g–j). This result indicates that the C-terminal domain mediates EB1 localization to the centrosome and that this localization is independent of EB1’s ability to bind MTs. Notice also that DsRed-EB1 and DsRed-EB1CT, similar to endogenous EB1, often preferentially localize to one of two centrosomal γ-tubulin spots (Fig. 4a–c, insets, for DsRed-EB1 and Fig. 4g–j, insets, for DsRed-EB1CT; see also Fig. 6A for DsRed-EB1).
Fig. 4.
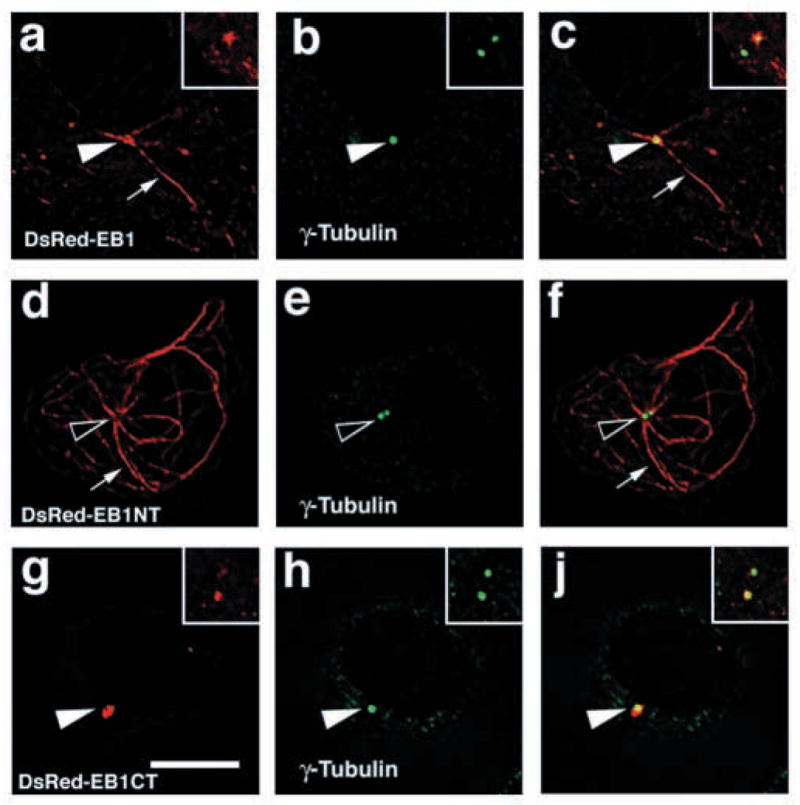
EB1 localization to the centrosome is mediated by its C-terminal domain. Sections of MDCK cells expressing full-length EB1 fused to DsRed (DsRed-EB1, red in a–c), the N-terminal domain of EB1 fused to DsRed (DsRed-EB1NT, red in d–f) or the C-terminal domain of EB1 fused to DsRed (DsRed-EB1CT, red in g–j). Cells were co-stained for γ-tubulin (green in b,c,e,f,h,j). DsRed-EB1 localizes along MTs (arrows in a,c) and forms a ring around the centrosome (white arrowheads in a–c). DsRed-EB1NT localizes along MTs (arrows in d,f) but does not localize around the centrosome (black arrowheads in d–f). DsRed-EB1CT does not localize along MTs but forms a ring around the centrosome (white arrowheads in g–j). DsRed-EB1 and DsRed-EB1CT show preferential localization to one of the two centrosomes in a cell (insets in a–c for DsRed-EB1 and insets in g–j for DsRed-EB1CT). Bar, 10 μm.
APC and EB1 localize close to the mother centriole
We examined whether the pattern of centrosomal EB1 staining in S/G2 cells is due to its localization to the mother centriole. U-2 OS cells expressing GFP-centrin as a marker for centrioles were treated with nocodazole to disrupt EB1 localization to cytoplasmic MTs and then co-labeled for the mother centriole marker ε-tubulin (Chang et al., 2003) and endogenous EB1 [Fig. 5A, Movie 3 (http://jcs.biologists.org/supplemental/)]. Although both proteins localize to the same end of the mother centriole, their distribution at this end is different. EB1 forms a cap at the end of the mother centriole and ε-tubulin folds around the centriole at the same end [Movie 3 (http://jcs.biologists.org/supplemental/)]. As determined previously by immunoelectron microscopy, ε-tubulin localizes to the subdistal appendages (Chang et al., 2003) that form a ring around one end of the mother centriole and anchor cytoplasmic MT minus ends (Bornens, 2002). EB1 capping of the mother centriole was also observed in cells with intact cytoplasmic MTs [Fig. 6C, Movie 5 (http://jcs.biologists.org/supplemental/)]. In some of these cells, additional filamentous EB1 extended out from the mother centriole and might be bound along MTs anchored to the subdistal appendages [Fig. 6C, Movie 5 (http://jcs.biologists.org/supplemental/)].
Recruitment of ε-tubulin to the new centrosome is cell-cycle regulated and occurs only after exit from S phase (Chang et al., 2003). EB1 has a distribution similar to that of ε-tubulin at different stages of centrosome maturation (Fig. 5C) but the appearance of ε-tubulin seems to precede that of EB1 at the new centrosome (Fig. 5Cd–f). In 25% of cells analysed for ε-tubulin and EB1 localization, ε-tubulin localized to the second centrosome without EB1. However, we did not find an example in which EB1 was at the second centrosome without ε-tubulin. Furthermore, preferential localization to the mother centriole is maintained in cells overexpressing EB1. DsRed-EB1, expressed in MDCK cells, co-localized with the mother centriole marker p150Glued/dynactin (Quintyne et al., 1999; Quintyne and Schroer, 2002) to only one of two centrosomes marked by pericentrin (Fig. 6Aa–d; see also Fig. 4, insets, for DsRed-EB1 and γ-tubulin co-stain).
In order to define whether APC co-localizes with EB1 to the mother centriole, MDCK cells expressing DsRed-EB1 were co-immunolabeled for APC and γ-tubulin or centrin. APC and EB1 preferentially localize to the same centrosome marked by γ-tubulin (Fig. 6Ae–h) and to the same centrioles marked by centrin (Fig. 6Aj–n). The mother centriole localization of APC was confirmed by co-staining U-2 OS cells expressing GFP-centrin for the mother centriole marker p150Glued/dynactin (Quintyne et al., 1999; Quintyne and Schroer, 2002) and for APC [Fig. 6B, Movie 4 (http://jcs.biologists.org/supplemental/)]. APC preferentially co-localizes with p150Glued/dynactin to one of four centrioles but extends to a second centriole, indicating that APC localization to the new centrosome precedes that of p150Glued/ dynactin [Fig. 6B, Movie 4 (http://jcs.biologists.org/supplemental/)]. EB1 and p150Glued/ dynactin cap the same end of the mother centriole and, in some cells, we observed additional filamentous EB1 and p150Glued/dynactin extending out from the mother centriole that might be bound to MTs anchored to the subdistal appendages [Fig. 6C, Movie 5 (http://jcs.biologists.org/supplemental/)].
In summary, these results show that EB1 and APC preferentially localize close to the mother centriole and are recruited to the mother centriole in the new centrosome some time after centriole duplication. We analysed U-2 OS and MDCK cells by 3D reconstruction of their centrosomal regions for localization of APC or EB1 at a second of two centrosomes, as defined by γ-tubulin or pericentrin label, or a second of four centrioles, as defined by centrin label. Notice that, although there is often little APC or EB1 at the second centriole/ centrosome in S/G2 cells, we found only 8% (n=25) of cells had no APC at the second centrosome, whereas 25% (n=28) of cells had no EB1 at the second centrosome, indicating that, similar to ε-tubulin preceding EB1 at the second mother centriole, APC might precede EB1 localization to the second mother centriole (Fig. 6Ae–h).
APC-EB1 interaction is not essential for localization of APC and EB1 to centrosomes
In order to define whether APC-EB1 interaction mediates the localization of APC and/or EB1 to centrosomes, we analysed centrosome localization of endogenous truncated APC (N-APC) and EB1 in SW480 cells (Fig. 6D). The human colorectal cancer cell line SW480 expresses a stable form of APC truncated at codon 1338 (N-APC) that is recognized by APC2 antiserum (Rubinfeld et al., 1993). N-APC does not have the C-terminal microtubule and EB1 binding sites. N-APC, EB1 and p150Glued localize to centrosomes in SW480 cells, indicating that APC-EB1 interaction is not essential for the localization of these proteins to centrosomes.
MT regrowth from centrosomes is inhibited in cells depleted of EB1
To examine EB1 function at the centrosome, we analysed MT regrowth from centrosomes following nocodazole washout in HeLa S3 cells depleted of EB1. HeLa S3 cells were used because very efficient depletion of protein using siRNA has been shown before in these cells (Elbashir et al., 2001). HeLa cells incubated with siRNA against EB1 showed an eightfold reduction in the total level of EB1 compared with untreated cultures or cultures treated with control siRNA (Fig. 7A). There was no significant change in the total level of tubulin or actin in the EB1 siRNA-treated cultures.
Fig. 7.
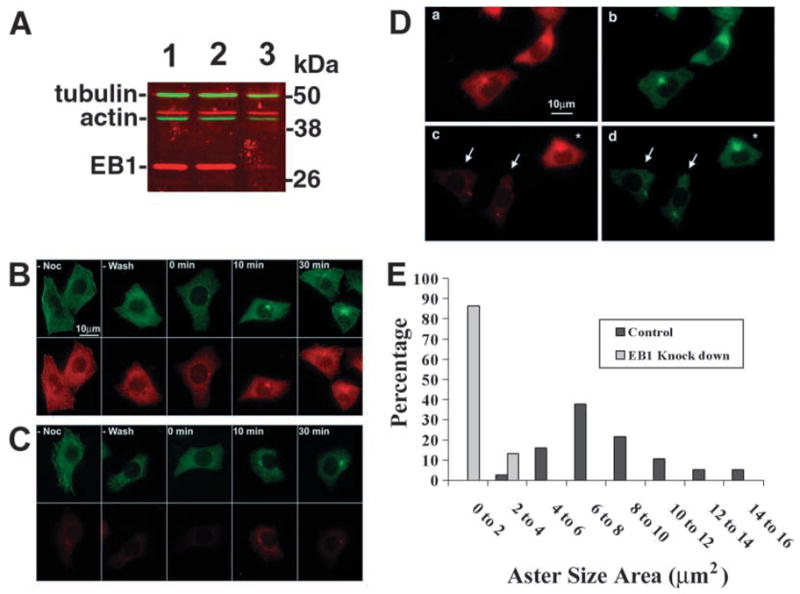
Depletion of EB1 with small interfering RNA inhibits MT regrowth from centrosomes after nocodazole washout. (A) SDS lysates from untreated HeLa cultures (1) and cultures incubated with control (2) or EB1 (3) siRNA were immunoblotted for EB1 (red), tubulin (green) and actin (green). EB1 levels were reduced in cultures treated with siRNA against EB1. Polyclonal antiserum to EB1 showed weak cross-reaction with a second slower migrating protein (red signal above actin), which was not reduced in response to EB1 siRNA. (B,C) HeLa cells incubated with siRNA against GFP as a control (B) or with siRNA against EB1 (C) were treated with nocodazole to depolymerize MTs, incubated at room temperature for the indicated times after nocodazole removal to allow regrowth of MTs and immunostained for α-tubulin (green) and EB1 (red). Some cultures were incubated with siRNA but not treated with nocodazole (–Noc) to control for preservation of MTs during fixation and, in some cultures, the nocodazole was not washed out (–wash) to control for MT regrowth during the time that was needed for the washes (compare ‘–wash’ to ‘0 minutes’). Immunofluorescence images of EB1 were taken with identical exposure times to allow comparison of fluorescence intensity between images. (D,E) Analysis of MT aster areas after 20 minutes of regrowth at room temperature. HeLa cells incubated with siRNA to GFP (a,b) or siRNA to EB1 (c,d) were immunostained for EB1 (a,c) and α-tubulin (b,d). Arrows mark cells depleted of EB1; an asterisk marks an unaffected cell with regular EB1 level in the same culture. (E) The areas covered by MT asters in EB1-depleted cells were compared with the areas covered by MT asters in cells of control cultures incubated with siRNA to GFP. Control, n=37; EB1 knock down, n=15.
Fig. 7B,C show the time course of MT regrowth at room temperature in cells incubated with control siRNA (Fig. 7B) compared with cells depleted of EB1 (Fig. 7C). 10–30 minutes after nocodazole washout, we observed pronounced MT regrowth in control cells, whereas MT asters remained small in cells depleted of EB1 (Fig. 7B,C). MT regrowth was quantified 20 minutes after nocodazole washout by measuring the area of MT asters around centrosomes (Fig. 7D,E). Cells depleted of EB1 (Fig. 7Dc–d, arrows) showed significantly smaller MT asters, covering a mean area of 1.31 μm2 (n=15) compared with 8.12 μm2 (n=37) in cells incubated with control siRNA (Fig. 7E); in a second independent experiment, the mean aster area was 1.47 μm2 (n=11) in cells depleted of EB1 compared with 17.7 μm2 (n=25) in control cells. Cells not responsive to siRNA in cultures incubated with EB1 siRNA exhibited normal MT regrowth (cell marked by asterisk in Fig. 7Dc,d). MT regrowth was delayed in EB1-depleted cells and an interphase MT network eventually forms (R. K. Louie, V. Votin and A. I. M. Barth, unpublished).
MT minus-end-anchoring at centrosomes is inhibited in cells depleted of EB1
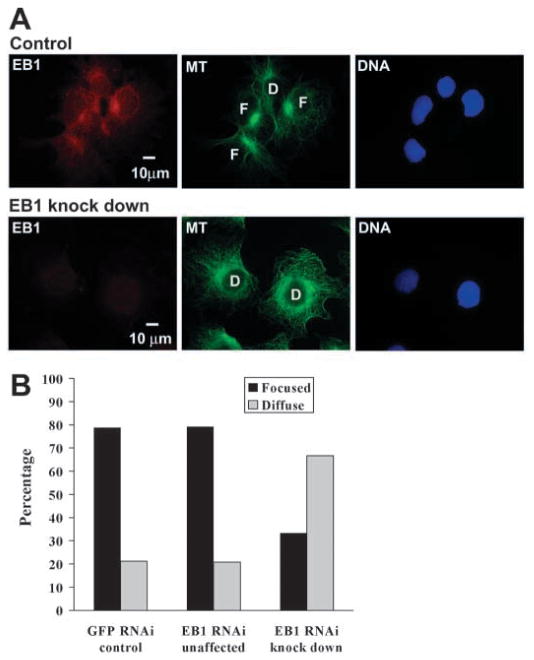
In order to analyse whether EB1 has a role in anchoring MT minus ends at the centrosome, we analysed MT minus-end focusing at centrosomes in Cos-7 cells depleted of EB1. In interphase cells of fibroblast origin, such as Cos-7 cells, most MT minus ends remain anchored at the centrosome so that the centrosome is a central focus point of cytoplasmic MTs (Quintyne et al., 1999). Therefore, MT anchoring defects are easily detectable in these cells. Cos-7 cells depleted of EB1 showed a significant increase in the number of cells in which MTs are not focused efficiently at the centrosome (Fig. 8A, compare cells marked ‘F’ for focused MTs in control-siRNA-treated cultures with cells marked ‘D’ for diffuse MTs in EB1-depleted cells). 67% of the EB1-depleted cells (n=108) showed defects in MT anchoring at the centrosome, whereas only 21% (n=101) of EB1 RNA-interference-treated cells that did not show depletion (unaffected) and 21% of cells in the control culture (n=165) had an unfocused cytoplasmic MT network (Fig. 8B); in a second, independent experiment, 89% of the EB1-depleted cells (n=47) had an unfocused MT network compared to 10% of EB1-siRNA-treated but unaffected cells (n=29) and 31% of control cells (n=118).
Fig. 8.
Depletion of EB1 with small interfering RNA reduces MT minus-end anchoring at the centrosome. (A) Cos-7 cells incubated with siRNA against GFP as a control or with siRNA against EB1 were immunostained for EB1 (red), and α-tubulin (green) and co-stained with DAPI for DNA (blue). Immunofluorescence images of EB1 were taken with identical exposure times to allow comparison of fluorescence intensity between images. Examples of cells with MT minus ends focused at the centrosome are marked as ‘F’ and examples of cells with reduced MT minus-end focus are marked as ‘D’ for diffuse. (B) The number of ‘F’ and ‘D’ type cells with EB1 knock down was quantified and compared with the number of these cell types in control cultures (‘GFP RNAi control’). Knock down was defined by reduced immunostain for EB1 (‘EB1 RNAi knock down’) and cells in the EB1 siRNA-treated cultures that did not show EB1 depletion were quantified as an internal control (‘EB1 RNAi unaffected’). Cells depleted for EB1 show reduced focus of MT minus ends at the centrosome. GFP RNAi control, n=165; EB1 RNAi unaffected, n=101; EB1 RNAi knock down, n=108.
Discussion
APC and EB1 are integral components of centrosomes
We have shown that both full-length endogenous and GFP-tagged APC and its C-terminal binding partner EB1 are retained at centrosomes after disruption of cytoplasmic MTs, and that APC and EB1 purify with functional centrosomes from mammalian cell lines. We conclude that APC and EB1 are integral components of mammalian centrosomes. Our results and the results of Tighe et al. (Tighe et al., 2001) have shown that endogenous N-APC mutant protein in colon cancer cell lines (Fig. 6D) and overexpressed N-terminal fragments of APC (Tighe et al., 2001) localize to centrosomes. Therefore, the N-terminus of APC might be involved in mediating its localization to centrosomes. In contrast to our results, Tighe et al. have not observed full-length endogenous APC at centrosomes (Tighe et al., 2001). The reason for this difference could be that full-length endogenous APC is less enriched at centrosomes than N-APC mutant protein. Furthermore, Tighe et al. might have missed its localization using conventional epifluorescence microscopy without optical sectioning (Tighe et al., 2001).
Centrosome localization of EB1 is mediated by its C-terminal domain but not by its N-terminal MT-binding domain (Fig. 4) (Askham et al., 2002). The EB1 C-terminus binds to APC and to p150Glued/dynactin, and binding of these two proteins to EB1 is mutually exclusive (Askham et al., 2002; Barth et al., 2002). Because both binding partners localize to centrosomes, either or both of them could contribute to EB1 localization at the centrosome. SW480 cells express only truncated N-APC missing the microtubule and EB1-binding sites. Analysis of N-APC, EB1 and p150Glued localization to centrosomes in these cells (Fig. 6D) shows that APC-EB1 interaction is not essential for localization of these three proteins to centrosomes.
APC and EB1 preferentially localize close to the mother centriole
Significantly, we have shown that both APC and EB1 localize close to the mother centriole at the centrosome and that this preferential localization is retained in cells overexpressing fluorescent fusion proteins of APC or EB1. APC surrounds the mother centriole like a U-shaped tube that often closes around one end of the centriole and remains open at the other. APC is more tightly associated with the centrin-labeled core structure of the mother centriole than p150Glued/dynactin. EB1 forms a cap at the end of the mother centriole that might contain the subdistal appendages as defined by its co-localization with ε-tubulin. However, further analysis at the ultrastructural level is needed to define whether EB1 localizes to the subdistal appendages.
The mother centriole is defined by specific MT-minus-end-anchoring structures termed subdistal appendages, and ε-tubulin has been localized to the subdistal appendages by immunoelectron microscopy (Chang et al., 2003). p150Glued/ dynactin is part of the dynein-dynactin complex that mediates minus-end-directed transport of cargo along MTs (Gill et al., 1991), and dynein-based MT movement is involved in centrosome-independent MT focusing (Gaglio et al., 1996). However, centrosome p150Glued/dynactin seems to be required for MT anchoring and focusing at the centrosome independent of dynein (Quintyne et al., 1999; Quintyne and Schroer, 2002). Furthermore, recent studies indicate that EB1 is important for MT minus-end anchoring at the centrosome (Askham et al., 2002; Rogers et al., 2002) and that this function might be mediated by a complex of EB1 with p150Glued/dynactin (Askham et al., 2002). We found that EB1 and p150Glued/ dynactin cap the end of the mother centriole containing the subdistal appendages as defined by ε-tubulin localization to the same end. Furthermore, in cells with intact cytoplasmic MTs, EB1 and p150Glued/dynactin extend into the cytoplasm from this end of the mother centriole most likely coating cytoplasmic MTs anchored to the mother centriole. However, we also found that EB1 localization to the mother centriole is independent of these cytoplasmic MTs because it is retained at centrosomes when cytoplasmic MTs are disrupted with nocodazole, but nocodazole treatment does not disrupt the more stable centriolar MTs.
Centrosome maturation is cell-cycle regulated and ε-tubulin is required for centriole duplication (Chang et al., 2003). Recruitment of ε-tubulin to the new centrosome occurs after exit from S phase (Chang et al., 2003). Differences in the relative amounts of APC and EB1 that accumulate at the second mother centriole indicate that APC starts to accumulate at one side of the centriole and that it precedes EB1 and p150Glued/dynactin accumulation at the second centrosome/ mother centriole. EB1 was found at the second mother centriole only when the centriole contained ε-tubulin, whereas ε-tubulin can localize to the second mother centriole without EB1, indicating that it precedes EB1 in its localization to the second mother centriole during centrosome maturation.
It has been suggested that the mammalian APC-EB1 complex is a functional equivalent of the yeast Kar9p-Bim1p complex (Gundersen and Bretscher, 2003). In budding yeast, the EB1 homolog Bim1p localizes to both spindle pole bodies (SPBs) and mediates localization of Kar9p to the SPBs. Preferential accumulation of Kar9p at the older, bud-oriented SPB is regulated by Clb4/Cdc28 phosphorylation of the Bim1p binding site in Kar9p. Clb4/Cdc28 localizes to the new SPB and inhibits Kar9p binding to Bim1p at the new SPB (Hwang et al., 2003; Liakopoulos et al., 2003; Maekawa et al., 2003). Kar9p at the old SPB is loaded onto cytoplasmic MTs that are then guided in a Kar9p/myosin-dependent manner along actin cables to the bud (Hwang et al., 2003; Liakopoulos et al., 2003; Maekawa et al., 2003). Our results show for the first time that mammalian APC and EB1 preferentially localize to the older, mother centriole. Interestingly, the phosphorylation site responsible for Kar9p localization to one SPB is conserved in APC and Cdc2 phosphorylation of APC reduces its affinity for EB1 (Trzepacz et al., 1997). Therefore, Cdc2 phosphorylation of APC might regulate its localization to the mother centriole but further experiments are needed to test this hypothesis.
Our results also indicate some intriguing differences between APC-EB1 localization at the mother centriole and Bim1p-Kar9p localization at the SPB. We show that, in mammalian cells, both APC and EB1 accumulate at the older, mother centriole, indicating that APC localization at centrosomes is regulated differently from that of Kar9p localization to the SPBs. Furthermore, our results indicate that APC can localize to the second mother centriole before EB1 does. There is little homology between APC and Kar9p outside their EB1-binding sites and APC is a large protein with multiple protein interaction sites. Further studies, beyond the scope of the present analysis, will be required to dissect how mother-centriole localization of APC and EB1 is regulated and to define whether APC has a similar function to Kar9p in guiding cytoplasmic MTs along actin cables towards the cortex.
Role of EB1 in MT minus-end anchoring at the centrosome
In order to investigate EB1 function at centrosomes more closely, we depleted levels of endogenous EB1 by transfection of small interfering RNA (siRNA) into HeLa S3 and Cos-7 cells. Depletion of EB1 in HeLa S3 cells strongly delayed MT regrowth from centrosomes. This delay in MT regrowth could be caused by a MT-anchoring defect resulting in dissociation of MTs from the centrosome after nucleation and/or by less-efficient MT growth from centrosomes, because EB1 is enriched at growing MT plus ends and promotes MT growth in yeast and in Xenopus extracts (Tirnauer et al., 1999; Mimori-Kiyosue et al., 2000; Nakamura et al., 2001; Tirnauer et al., 2002). The C-terminal EB1-binding domain of APC promotes the effect of EB1 on MT growth in vitro and in permeabilized mammalian cells, indicating that APC binding might be needed for this function of EB1 (Nakamura et al., 2001). Because the centrosome is the major MT-nucleating structure in the cell, a pool of centrosome APC and EB1 might be required to ensure fast, efficient outgrowth of newly nucleated MTs from the centrosome.
HeLa cells are of epithelial origin and their cytoplasmic MT network is not very strongly focused at the centrosome in interphase (Quintyne et al., 1999). Therefore, we also examined the effect of EB1 depletion in fibroblast Cos-7 cells that have a strong focus of cytoplasmic MT minus ends at the centrosome. Depletion of EB1 in these cells significantly decreased MT minus-end focus at the centrosome indicating a MT anchoring defect at the centrosome.
Taken together, our results suggest that EB1 is part of a MT-minus-end-anchoring complex at the subdistal appendages of the mother centriole. p150Glued/dynactin is probably another component of this complex (Quintyne et al., 1999; Askham et al., 2002; Quintyne and Schroer, 2002) and, because binding of EB1 to APC and p150Glued/dynactin is mutually exclusive (Askham et al., 2002), APC might regulate EB1-p150Glued/ dynactin function at the mother centriole.
Supplementary Material
Acknowledgments
We thank A. Reilein (Stanford University, Palo Alto, CA) for helpful discussions and J. L. Salisbury (Mayo Clinic Foundation, Rochester, MN) for the GFP-centrin construct and the centrin antibody. Schematic representation of 3D localization of proteins shown in Fig. 5A′ and Fig. 6B′,C′ were prepared by B. Colyear (Visual Arts Services, Stanford University). This work was supported by a grant from the NIH (NS42735) to W. J. Nelson, a Medical Scientist Training Program grant (GM07365) from the National Institute of General Medical Studies to R. K. Louie, a grant (T32GM07276-27) from the Cell and Molecular Biology Training Program to S. Bahmanyar and a grant from the NIH (GM52022) to T. Stearns.
References
- Askham JM, Vaughan KT, Goodson HV, Morrison EE. Evidence that an interaction between EB1 and p150Glued is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol Biol Cell. 2002;13:3627–3645. doi: 10.1091/mbc.E02-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AI, Pollack AL, Altschuler Y, Mostov KE, Nelson WJ. NH2-terminal deletion of beta-catenin results in stable colocalization of mutant beta-catenin with adenomatous polyposis coli protein and altered MDCK cell adhesion. J Cell Biol. 1997a;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AI, Nathke IS, Nelson WJ. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol. 1997b;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- Barth AI, Siemers KA, Nelson WJ. Dissecting interactions between EB1, microtubules and APC in cortical clusters at the plasma membrane. J Cell Sci. 2002;115:1583–1590. doi: 10.1242/jcs.115.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta L, Kraeft SK, Tirnauer JS, Schuyler SC, Chen LB, Hill DE, Pellman D, Bierer BE. The Adenomatous Polyposis Coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc Natl Acad Sci USA. 1998;95:10596–10601. doi: 10.1073/pnas.95.18.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Chang P, Stearns T. Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- Chang P, Giddings TH, Jr, Winey M, Stearns T. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat Cell Biol. 2003;5:71–76. doi: 10.1038/ncb900. [DOI] [PubMed] [Google Scholar]
- D’Assoro AB, Stivala F, Barrett S, Ferrigno G, Salisbury JL. GFP-centrin as a marker for centriole dynamics in the human breast cancer cell line MCF-7. Ital J Anat Embryol. 2001;106:103–110. [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Bretscher A. Cell biology. Microtubule asymmetry. Science. 2003;300:2040–2041. doi: 10.1126/science.1084938. [DOI] [PubMed] [Google Scholar]
- Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, McEwen DG, Grevengoed E, Maddox P, Bejsovec A, Peifer M. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat Cell Biol. 2001;3:933–938. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Isolation of mammalian centrosomes. Methods Enzymol. 1986;134:261–268. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 1998;17:3471–3477. doi: 10.1038/sj.onc.1202247. [DOI] [PubMed] [Google Scholar]
- Munemitsu S, Souza B, Muller O, Albert I, Rubinfeld B, Polakis P. The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Zhou XZ, Lu KP. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr Biol. 2001;11:1062–1067. doi: 10.1016/s0960-9822(01)00297-4. [DOI] [PubMed] [Google Scholar]
- Näthke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näthke IS, Barth AIM, Nelson WJ. Role for the tumor suppressor adenomatous polyposis coli protein in epithelial migration and adhesion: a hypothesis. In: Klymkowsky PCaMW., editor. Cytoskeletal-Membrane Interactions and Signal Transduction. 1. New York: Chapman and Hall; 1997. pp. 103–110. [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. The oncogenic activation of beta-catenin. Curr Opin Genet Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- Pollack AL, Barth AIM, Altschuler Y, Nelson WJ, Mostov KE. Dynamics of β-catenin interactions with APC protein regulate epithelial tubulogenesis. J Cell Biol. 1997;137:1651–1662. doi: 10.1083/jcb.137.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with beta-catenin. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Centrosome duplication. a centriolar pas de deux. Cell. 2001;105:417–420. doi: 10.1016/s0092-8674(01)00366-x. [DOI] [PubMed] [Google Scholar]
- Su LK, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler KW. APC binds to the novel protein EB1. Cancer Res. 1995;55:2972–2977. [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, O’Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzepacz C, Lowy AM, Kordich JJ, Groden J. Phosphorylation of the tumor suppressor adenomatous polyposis coli (APC) by the cyclin-dependent kinase p34. J Biol Chem. 1997;272:21681–21684. doi: 10.1074/jbc.272.35.21681. [DOI] [PubMed] [Google Scholar]
- White RA, Pan Z, Salisbury JL. GFP-centrin as a marker for centriole dynamics in living cells. Microsc Res Tech. 2000;49:451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zumbrunn J, Kinoshita K, Hyman AA, Näthke IS. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK-3β phosphorylation. Curr Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.