Abstract
Using a newly synthesized gibberellin analog (GA3-AM) and its binding proteins, we developed a novel and efficient chemically induced dimerization (CID) system, that is completely orthogonal to the existing rapamycin-mediated protein dimerization. Combining the two systems should allow applications that were difficult or impossible with only one CID system. By using both chemical inputs (rapamycin and GA3-AM), we designed and synthesized Boolean logic gates in living mammalian cells. These gates produced output signals such as fluorescence and membrane ruffling on a timescale of seconds, a significant improvement over previous intracellular logic gates. The use of two orthogonal dimerization systems in the same cell also allows for finer modulation of protein perturbations than is possible with a single dimerizer.
We aimed to create fast-processing logic gates based on chemically inducible dimerization (CID) systems. CID has proven to be a powerful tool for inducible, rapid and specific manipulation of various signaling molecules in living cells1, 2. Rapamycin, the most commonly used chemical dimerizer, induces interaction between FK506-binding protein (FKBP) and FKBP-rapamycin-binding protein (FRB)3, 4, the system that originates from a use of FK1012 as a synthetic dimerizer in 19935. This simple principle has been deeply exploited to manipulate various aspects of cell signaling, thereby resolving fundamental biological questions that were otherwise extremely challenging6–9. Recent efforts have been made to expand the palette of CID systems10–13, aiming to control multiple signaling molecules at the same time and location, or different times and locations. However, thus far there have not been any two systems that are simultaneously orthogonal to each other and work on a rapid timescale in the context of a living cell. These conditions are prerequisite for fast-processing in vivo logic gates. Biomolecular logic gates in a cell-free system have been previously constructed utilizing nucleotides14–16 and protein enzymes17. Some of them are networked to form large-scale circuits for DNA computing18. A number of logic gates have also been constructed in living cells, generally based on protein translation as the output19, 20 and often utilizing gene circuits20–23. The CID system has also been exploited to create logic gates24. Although processing speed is a critical component of computational entities, the timescales of these logic gates in living cells were relatively slow, on the order of tens of minutes to hours. In particular, the slow response time of CID logic gates are at least partly attributed to 1. a time-consuming transcriptional process, 2. slow dimerization (except in the case of rapamycin-mediated dimerization).
In the present study, we developed a novel CID system using a plant hormone, gibberellin, a system that is completely orthogonal to the rapamycin system and that works on a timescale of seconds. Recent advances in plant biology uncovered a molecular mechanism of action by plant hormones25. Like other hormones, gibberellins regulate various aspects of plant growth and development. At a molecular level, gibberellin binds to its receptor gibberellin insensitive dwarf1 (GID1)26 and induces a conformational change. This new conformation now attracts another protein called gibberellin insensitive (GAI)27 (see Figure 1a). These binding events require a very selective gibberellin such as GA328, one of the more than one hundred gibberellin metabolites. We were able to develop and optimize a series of GID1 and GAI fusion proteins that can form a CID system activated by the compound GA3-AM, which readily enters mammalian cells and is cleaved by esterases to release active GA3. We then showed that this gibberellin-mediated CID system is fully orthogonal to rapamycin CID and can be used to induce protein translocation and to move active protein to specific subcellular locations on a timescale of seconds to minutes. Finally, by combining the gibberellin and rapamycin-based CID, we were able to generate intracellular logic gates using two distinct chemical inputs.
Figure 1.
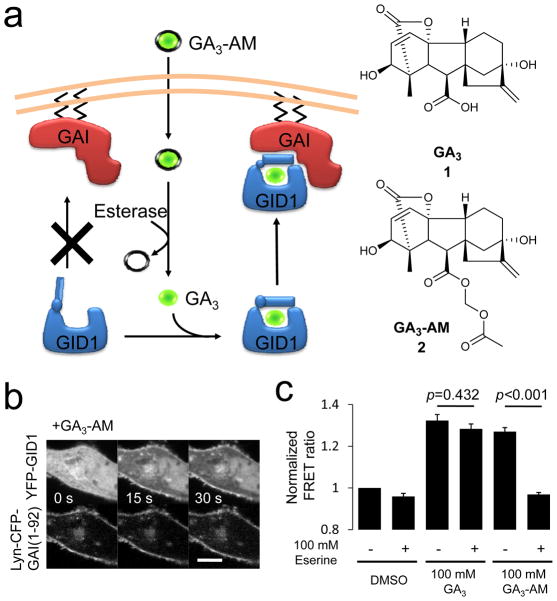
Novel CID system functioning on timescale of seconds. (a) General scheme of gibberellin-induced CID utilized in this study. GA3-AM (2) (green ball covered with a black line) is able to cross the plasma membrane of target cells, whereupon cytosolic esterase cleaves the AM group to release free GA3 (1) (green ball). GA3 then binds GID1 (blue), which induces formation of a complex between GID1 and GAI (red). (b) Time series of confocal fluorescence images of HeLa cells cotransfected with Lyn-CFP-GAI(1-92) and YFP-GID1. The cells were treated with GA3-AM (100 μM). Top row shows YFP fluorescence signal, while the bottom row shows CFP fluorescence in the same cell. Scale bar indicates 10 μm. (c) Esterase activity is required for GA3-AM dimerizing ability. Lysate was made of COS-7 cells co-transfected with CFP-GAI(1-92) and YFP-GID1, eserine (100 μM) was added and FRET measured 0 and 2 minutes after addition of DMSO, GA3, or GA3-AM as indicated. Graph represents the average of three independent experiments ± SEM. Statistical analysis was performed with an unpaired two-tailed Student’s t test assuming the two populations have the same variances.
Results
Optimizing uptake of gibberellin-based dimerizer
To assess if GA3 (1) induces binding of GID1 and GAI in mammalian cells (Fig. 1a), we first established a fluorescence resonance energy transfer (FRET) assay by modifying a previously reported system28. We constructed a series of fusion proteins consisting of fluorescent proteins (CFP or YFP) and Arabidopsis thaliana GAI or GID1. First, we expressed CFP-GAI and YFP-GID1 in HeLa epithelial cells to visualize GAI-GID1 interactions upon addition of GA3 by monitoring FRET between these fusion proteins. We observed a marginal FRET increase over 10 minutes (Supplementary Figure 1). We attributed this to an inefficient membrane permeability of GA3, most likely due to a carboxylic acid group which is negatively charged at physiological pH. To improve membrane permeability of GA3, we esterified the carboxylic acid of GA3. More specifically, we used an acetoxymethyl (AM) group29 such that the negative charge of GA3 becomes masked until ambient esterases inside cells cleave the AM ester group to produce a bioactive GA3 (Fig. 1a). We named the esterified compound GA3-AM (2) (see the complete synthetic schemes in Supplementary Information). We then repeated the FRET assay with GA3-AM in replacement of GA3. GA3-AM induced a robust increase in the FRET signal on a timescale of 60 seconds (Supplementary Figure 1), indicating that GA3-AM went into cells, was converted into GA3 and then induced dimerization between CFP-GAI and YFP-GID1. Other combinations of GAI and GID1 with a different configuration (CFP-GAI + YFP-GID1, GAI-CFP + YFP-GID1) also showed a comparable FRET increase. This efficient binding may be helped by the fact that GA3 is unlikely to have any known competing proteins in mammalian cells.
Optimization of dimerizing proteins
We then determined the minimal domains of GAI and GID1. For a dimerization unit, it is better for protein components to be small in size and devoid of any regulatory domain that may affect cell signaling in an unintended manner. It has been shown that auxin-induced dimerization of two plant proteins can trigger downstream effects (i.e., protein degradation) in mammalian cells, as one of these plant proteins can signal to downstream machinery through a mammalian homolog30. Thus, we tested a series of truncated GAIs to identify a minimal GAI domain for the GA3-induced dimerization. N-terminal domains (DELLA and TVHYNP) of GAI were sufficient to interact with GID1 in yeast two hybrid assays31, while a C-terminal GRAS domain was shown to interact with GID1 once the DELLA domain binds to GID132. The GRAS domain also interacts with other proteins including SLY1/GID2 which in turn targets GAI for degradation via ubiquitin-mediated proteolysis27. We therefore constructed three truncated mutants (Supplementary Figure 2a). The first truncated mutant contains DELLA and VHYNP domains (GAI(1-92)) and the second and third mutants contain additional poly serine/threonine domains (GAI(1-151) and GAI(1-172)). In the FRET assay, we observed all these truncated GAIs bound to GID1 upon GA3-AM addition with a similar efficiency (0.0072 sec−1, 0.0095 sec−1, and 0.0106 sec−1, respectively), demonstrating that N-terminal DELLA and TVHYNP are sufficient in mammalian cells (Supplementary Figure 2b). The dynamic range of the FRET increase for the truncated and full length GAI correlated with their expression level (Supplementary Figure 3).
Gibberellin CID induces protein translocation
Next we assessed the efficacy of this new dimerization system for its ability to control protein localization. To recruit proteins from the cytoplasm to the plasma membrane, GAI(1-92) was modified with 11 amino acid residues of Lyn kinase that targets the protein to the plasma membrane33, while GID1 remained in the cytoplasm without further engineering (Figure 1a). Confocal fluorescence imaging revealed that GID1 rapidly changed its localization upon addition of GA3-AM from the cytoplasm to the plasma membrane in HeLa cells (Fig. 1b) as well as HEK293T, NIH3T3 and MCF10A cells (Supplementary Figure 4). Quantitative analysis on fluorescence intensity of cytoplasmic YFP-GID1 provided an apparent rate constant of 0.013 sec−1 (Supplementary Figure 5a). By varying the concentration of GA3-AM, we obtained dose-dependent kinetic values (Supplementary Figure 5b). We also tested GAI(1-151) and GAI(1-172) as well as full length GAI, all of which showed membrane translocation of GID1. However, the apparent rate constant of the full length GAI was slower than the other GAI truncation mutants (Supplementary Figure 5c), likely due to low protein expression. In contrast to GAI, GID1 distributes amino acids responsible for dimerization throughout the protein34, 35 which makes minimization challenging. Based on the FRET and translocation assays, we concluded that GAI(1-92) and full length GID1 are best suited as a dimerization unit induced by gibberellin.
Esterase is required for GA3-AM-induced dimerization
As our gibberellin-based chemical dimerizer is dependent on cellular esterases, questions arise regarding their expression level, abundance across cells in the population, and enzymatic efficiency. In order to evaluate these features, we measured the fluorescence intensity of calcein-AM that fluoresces upon cleavage of AM esters. These esterases are very efficient and expressed ubiquitously (Supplementary Figure 6). Consistent with this observation, AM-esterified molecular probes have been confirmed to be functional in a variety of cell types36. To address whether GA3-AM directly binds to GID1 unlike our initial prediction, we took two approaches: (1) FRET binding assays in cells using a newly synthesized non-hydrolysable GA3 analog (GA3 hydroxamate, or GA3-H (3)) (see Supplementary Information for the synthetic scheme), and (2) in vitro FRET binding assays using cell extracts containing GAI and GID1 proteins. In the first approach, we observed GA3-H inducing FRET between GID1 and GAI (Supplementary Figure 7a), but the kinetics of GA3-H were much slower than those of GA3 (Supplementary Figure 1). GA3-H was roughly 10-fold slower than GA3-AM in the plasma membrane translocation assay with an apparent rate constant of 0.00103 sec−1 at 1 mM (Supplementary Figures 5b and 7b). These results suggest that the carboxylic acid of GA3 is critical for binding GID1. In the second approach, we prepared cell extracts that contain YFP-GID1 and CFP-GAI (1-92) for the following in vitro FRET assay. We monitored FRET between the two proteins in real-time before and after addition of DMSO, GA3, or GA3-AM to the extract with or without an esterase inhibitor (eserine, 100 μM). Both GA3 and GA3-AM induced a FRET increase in the absence of eserine (Fig. 1c). However, eserine addition inhibited the FRET increase induced by GA3-AM but not by GA3 (p = 0.0003 vs p = 0.432, respectively, Fig. 1c). The inhibitory effect of eserine on the GA3-AM-induced FRET increase was concentration dependent (Supplementary Figure 8). Collectively, these results strongly support the notion that GA3, but not GA3-AM itself, is the molecule that induces dimerization of GID1 and GAI in cells.
Gibberellin CID induces localized protein activity
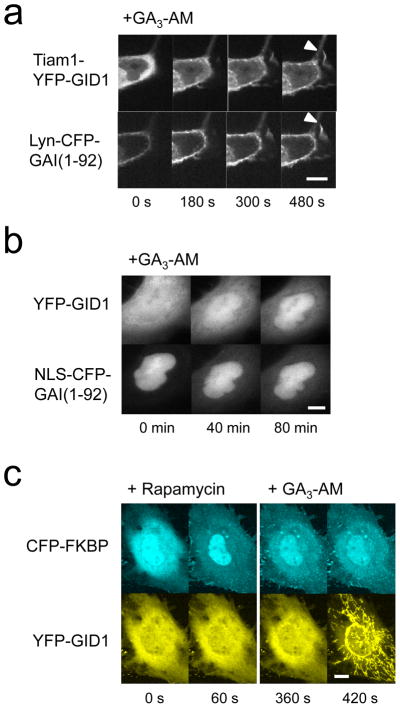
Inducible protein translocation has been successfully used to manipulate activity and levels of biomolecules1. To test if the gibberellin-induced protein translocation can be translated into changes in protein activity, we generated Tiam1-YFP-GID1. Tiam1 is a guanine nucleotide exchanger factor that activates Rac when recruited to the plasma membrane24. When the Tiam1-YFP-GID1 construct was expressed in cells with Lyn-CFP-GAI(1-92), the cells exhibited robust membrane ruffles after GA3-AM addition (Fig. 2a), indicating that Rac was inducibly activated.
Figure 2.
Gibberellin-based CID can induce localized protein activity and is orthogonal to rapamycin CID. (a) Time series of confocal fluorescence images of COS-7 cells transfected with Lyn-CFP-GAI(1-92) and Tiam1-YFP-GID1 before and after addition of GA3-AM (100 μM). Arrowheads highlight ruffles. The same cell is shown in top row (imaged in YFP channel) and bottom row (CFP channel). (b) Gibberellin CID can be used to translocate proteins to the nucleus. COS-7 cells co-transfected with YFP-GID1 and NLS-CFP-GAI(1-92) were treated with GA3-AM (10 μM) and imaged. A representative cell is shown in both CFP and YFP channels. (c) Confocal fluorescence images of COS-7 cells transfected with CFP-FKBP and YFP-GID1 together with Lyn-mCherry-FRB and Tom20-mCherry-GAI(1-92). The images were taken before and after sequential addition of rapamycin and GA3-AM. Scale bars indicate 10 μm.
Rapamycin CID has also been used to control transcriptional activity by translocating a transcriptional factor into the nucleus in a rapamycin dependent manner. To see if gibberellin CID can be used in the same context, we constructed a nuclear localizing GAI (NLS-CFP-GAI(1-92)) which successfully recruited YFP-GID1 from the cytoplasm to nucleus upon addition of GA3-AM (Fig. 2b, Supplementary Figure 9), suggesting its potential usage in controlling transcriptional activity as well. YFP-GID1 was present at a low level in the nucleus even before addition of GA3-AM, which may be improved by putting a nuclear export signal sequence to the YFP-GID1 construct. Another useful application for a nuclear translocation system is manipulating signals inside the nucleus on a rapid timescale. Toward this end, we fused a YFP-GID1 to a phosphatase of phosphatidylinositol 4,5-bisphosphate (Inp54p-YFP-GID1). When expressed in cells, the protein was mostly found in the cytoplasm with very little expression in the nucleus. Subsequent GA3-AM addition induced efficient translocation into nucleus (Supplementary Figure 10).
Gibberellin and rapamycin CID systems are orthogonal
Rapamycin-induced dimerization and GA3-induced dimerization should be orthogonal to each other, as they are derived from completely different kingdoms (i.e., plant versus animal) and share no structural resemblance. To explore this, we performed a simultaneous translocation assay where both dimerization systems are introduced in the same cells. Here, we intended to recruit CFP-FKBP to the plasma membrane with rapamycin and YFP-GID1 to the mitochondria. More specifically, mCherry-FRB was anchored to the plasma membrane using a Lyn signal sequence, whereas mCherry-GAI(1-92) was anchored to the mitochondria using another signal sequence from Tom2027. These four constructs were transfected in COS-7 cells. When GA3-AM and rapamycin were added sequentially, each dimerizer induced protein translocation to their expected intracellular locations. In addition, rapamycin did not affect the gibberellin system, or vice versa, verifying their orthogonal nature (Fig. 2c, Supplementary Movie 1). In order to induce protein translocation at the same time, the chemical dimerizers were simultaneously added. This induced rapid, coinciding translocation of CFP-FKBP and YFP-GID1 to the plasma membrane and mitochondria, respectively (Supplementary Figure 11).
Excessive gibberellin induces acidification in cells
Gibberellin is reported to induce acidification in plant cells37. To test whether addition of GA3-AM induces acidification in mammalian cells, we monitored pH in cells using a Venus fluorescent protein containing one amino acid substitution (H148G) to become sensitive to proton concentration in the environment38. We co-transfected Venus(H148G) and CFP to titrate the ratio of Venus fluorescence over CFP to various extracellular pH in the presence of chemical protonophores (i.e., monensin and nigericin). At a concentration of 100 μM that we commonly used for the above mentioned experiments, GA3-AM did not induce detectable acidification in COS-7 cells (Supplementary Figure 12). 100 μM GA3-AM reduced the pH from 7.4 to 7.3 in HeLa cells. At the concentration of 100 μM, GA3 did not induce acidification in either cell type, supporting its inefficient membrane permeability. Higher concentration of GA3-AM induced a bigger shift in pH, suggesting that care should be taken for the concentration of GA3-AM. However, as shown in Supplementary Figure 5b, the EC50 is 310 nM which is much lower than 100 μM, suggesting that efficient dimerization can be induced without any acidification. Taken together, we used 10 μM GA3-AM for the following experiments.
Intracellular logic gates using GA3-AM and rapamycin CID
To construct intracellular CID-based logic gates, we took advantage of our newly developed gibberellin-mediated dimerization system that works with a speed comparable to the rapamycin system. First, we created an OR gate in which the two inputs are rapamycin and GA3-AM and the output is an optical signal such as fluorescence. Our design places two dimerization units at the plasma membrane (Lyn-CFP-FRB-GAI(1-92)) and their binding partner units in the cytoplasm (FKBP-YFP-GID1). It is predicted to give rise to an increase in the FRET signal when FKBP-YFP-GID1 associates with Lyn-CFP-FRB-GAI(1-92), which should occur in the presence of rapamycin, GA3-AM or both. When these two constructs were transfected in cells, we observed the robust FRET signal under these drug treatments but not with control DMSO treatment (Supplementary Figure 13). The timescale of the process was 60 seconds.
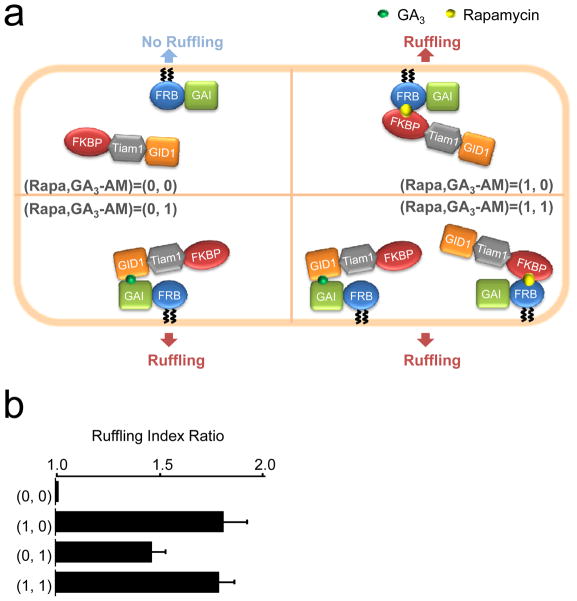
Besides speed, versatility is an additional strength of chemically-inducible dimerization systems. We and others have previously developed a variety of rapamycin-triggered molecular probes1, suggesting that the chemically-inducible logic gates have a potential to produce various signaling outputs. In pursuit of this possibility, we utilized an existing module to produce a second-messenger-mediated output. The Tiam1-based Rac activation probes were employed in our logic gates and changes in cell morphology as a result of Rac activation were visualized. For this OR gate, we transfected cells with the constructs Lyn-CFP-FRB-GAI and FKBP-YFP-Tiam1-GID1 (Fig. 3a). Membrane ruffling was observed 60–90 seconds after the addition of chemical dimerizers that induced recruitment of the Tiam1 fusion protein to the plasma membrane. Quantitative analysis indicated that the number of cells showing membrane ruffling was significantly increased after the addition of rapamycin, GA3-AM, and both (Fig. 3b). It is important to have a clear threshold for an output signal for an accurate computation, which is reasonably easy to achieve with our system due to a very low background (0,0) that is clearly distinguishable from other signals (1,0), (0,1) and (1,1).
Figure 3.
Fast processing logic gates in living cells using two CID systems. (a) Schematic diagram of an OR gate using membrane ruffling as the output signal; fluorescent reporter molecules are omitted for clarity. Membrane ruffling was measured for COS-7 cells co-transfected with Lyn-CFP-FRB-GAI(1-92) and FKBP-YFP-Tiam1-GID1 that were subject to treatment with DMSO (representing the 0,0 input), rapamycin alone (1,0), GA3-AM alone (0,1) or rapamycin and GA3-AM together (1,1). (b) Data from the OR gate shown in (a). Results represent mean ± SEM (n ≥ 20, from three independent experiments).
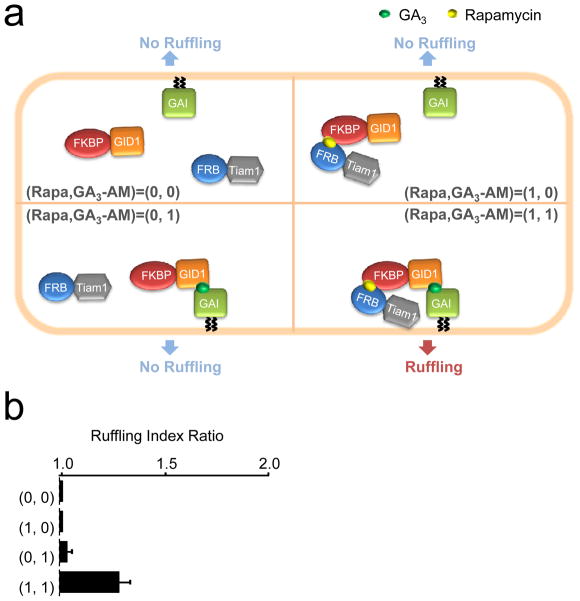
To demonstrate that other logic gates are possible with the use of orthogonal CID systems, we created an AND gate, where there should be a positive output only if both inputs are positive. Our AND gate harbors three components where GAI(1-92) is at the plasma membrane, while FKBP-GID1 and FRB-Tiam1 are in the cytoplasm (Fig. 4a); the specific constructs used were Lyn-mCherry-GAI(1-92), FKBP-YFP-GID1, and CFP-FRB-Tiam1. Indeed, when all three constructs were transfected into COS-7 cells, we observed a significant increase in membrane ruffling only when both GA3-AM and rapamycin were added, but not when either inducer was used alone (Fig. 4b).
Figure 4.
Fast processing logic gates in living cells using two CID systems. (a) Schematic diagram of an AND gate using membrane ruffling as the output signal. Membrane ruffling was measured for COS-7 cells co-transfected with Lyn-mCherry-GAI(1-92), FKBP-YFP-GID1, and CFP-FRB-Tiam1 that were subject to treatment with DMSO (representing the 0,0 input), rapamycin alone (1,0), GA3-AM alone (0,1) or rapamycin and GA3-AM together (1,1) (b) Data from the AND gate shown in (a). Results represent mean ± SEM (n μ 20, from three independent experiments).
Discussion
We have developed a novel, efficient protein dimerization system using chemically-modified gibberellin. With this system, we demonstrated inducible protein recruitment as well as manipulation of signaling molecules on a timescale of seconds. Furthermore, the system is completely orthogonal to the existing rapamycin-mediated dimerization system and thus suited for multivalent manipulation of different molecules at different locations. We then utilized the two orthogonal chemical dimerization systems to create representative logic gates that operate processing on a timescale of seconds. This was hundreds of times faster than the computation achieved by prevailing logic gates with genetic circuits (tens of minutes to hours) in living cells. Besides speed, logic gates need to be networked to operate higher-order computation. For this purpose, the genetic circuits are well suited because an output signal from one logic gate can be used as an input signal for another neighboring logic gate, thus readily accommodating several logic gates in a given space39, 40. In contrast, networking logic gates is challenging for chemically-inducible protein dimerization systems, as they cannot easily give out chemical dimerizers as an output signal. Non-trivial engineering would be required to make cells release particular chemical dimerizers. The knowledge obtained through the construction of logic gates using proteins may help deepen our understanding of “physiological” logic gates such as coincidence detectors (e.g. inositol trisphosphate receptor, adenylate cyclase, AP-2, N-WASP, etc.).
The use of protein signaling as an output signal has a unique advantage. Each protein has a distinctive feature so that an output signal is not limited to a binary code. In the present study, we have shown that FRET and membrane ruffling can serve as an output signal, comprising 4 bit information. We and others have developed a large number of chemically-inducible molecular probes that can be readily used to further expand the information size. It has also been reported that other plant hormones dimerize different sets of proteins in plants like gibberellin does12, 30. With multiple triggers by various plant hormones, it will be possible to diversify input signals for more complicated logic gates. Another important nature of computers is reversibility. Unfortunately, the dissociation speed of the two present chemically-inducible systems is slow compared to their association speed. Rapid reversibility will be a great asset to these logic gates toward achievement of more practical computation.
Methods
Chemical synthesis of gibberellin analogs: GA3-AM and GA3-H
All reagents and solvents were used as supplied by commercial sources without further purification. All dry solvents were purchased from Aldrich in Sure Seal bottles. Reactions involving air and/or moisture sensitive reagents were carried out under an argon atmosphere using glassware that was dried under vacuum with a heat gun. The evacuated flask was then filled with argon. Reactions were monitored by thin layer chromatography using Analtech chromatography plates (silica gel GHLF, 250 microns). Visualization was performed by staining with phosphomolybdic acid (10% PMA in ethanol) stain. Flash silica gel chromatography was performed using a Grace Reveleris flash chromatography system equipped with UV and evaporative light scattering detectors (ELSD).1H NMR (400 MHz) spectra recorded in either CD3OD or DMSO-d6 were referenced to a residual solvent peak of 3.31 ppm or 2.50 ppm, respectively. 13C NMR (100 MHz) spectra recorded in either CD3OD or DMSO-d6 were referenced to a residual solvent peak of 49.0 ppm or 39.52 ppm, respectively. HRMS mass spectra were recorded at the University of California, Riverside Mass Spectrometry Facility. A synthetic scheme of GA3-AM and GA3-H is described in more detail in Supplementary Information.
Live-cell confocal and epifluorescence microscopy
The majority of live cell dual color or tri-color images were performed on a spinning-disc confocal microscope. CFP and YFP excitations were conducted with a helium-cadmium laser and argon laser (CVI-Melles Griot), respectively. mCherry excitation was conducted with the argon laser. The two lasers were fiber-coupled (OZ optics) to the spinning disk confocal unit (CSU10; Yokogawa) mounted with dual CFP-YFP dichroic mirrors (Semrock). The lasers were processed with appropriate filter sets for CFP, YPF, and mCherry (Chroma Technology) to capture fluorescence images with a CCD camera (Orca ER, Hamamatsu Photonics), driven by Metamorph 7.5 imaging software (Molecular Devices). Images were taken using a 40x objective (Zeiss) mounted on an inverted Axiovert 200 microscope (Zeiss). Some additional imaging was done on an epifluorescence microscope. CFP and YFP excitation were performed by an X-Cite Series 120Q mercury vapor lamp and processed through appropriate filter cubes. Images were taken using a 63x objective (Zeiss) mounted on an inverted Axiovert 135 TV microscope (Zeiss) and captured by a QIClick CCD camera (QImaging). Time-lapse live cell imaging was collected mostly every 15 seconds and performed at room temperature.
Cytosolic pH measurements in live cells
Intracellular pH change upon addition of 100 (or 333) μM gibberellin derivatives (GA3 or GA3-AM) was evaluated by using fluorescent protein variant (Venus(H148G)) and ECFP. This H148G mutant is optimal for measuring cytosolic pH because it has a pKa of around 8. CFP is an internal reference as it has a pKa around 4.5. Between 1 and 2 days after transfection, cells were imaged at room temperature. Calibration of fluorescence intensity of (Venus(H148G) and ECFP) in living cell was accomplished with 5 μM of each ionophore nigericin and monensin in media over the pH range 5–9.
FRET measurement in live cells
Fluorescence images were taken every 15 seconds as described above. Images were normally collected for two minutes prior to addition of GA3, GA3-AM, or rapamycin dissolved in DMSO (0.1%), then collected for eight minutes after addition of drug. CFP/YFP FRET was normalized to the average of the reading for the five timepoints immediately prior to addition of drug. Graphs represent the average from three independent experiments of 10–15 cells per experiment.
In vitro FRET binding assay
Transient co-transfection of CFP-GAI(1-92) and YFP-GID1 was carried out using FuGeneHD (Promega) in Cos-7 cells. 24 hours after transfection, the cells were lysed in lysis buffer containing 20 mM Tris-HCl, pH 7.5, 120 mM NaCl, 1 mM DTT, 1 mM PMSF, 1 mM EDTA, 5 mM MgCl2, Protease inhibitor cocktail (Roche), Phosphatase inhibitor cocktail 2 (Sigma-Aldrich), and 1% NP-40, and cleared lysates were subjected to the in vitro FRET assay. Using an EPI fluorescence microscope, fluorescence images were taken 0 and 2 minutes after addition of either DMSO (control), GA3, or GA3-AM. The ratio of CFP-YFP FRET between these two time points was calculated, and then normalized to the value obtained from DMSO treatment. When used, eserine (Sigma) was added to the lysate prior to the start of the assay.
Membrane translocation assay
Fluorescence images were taken every 15 seconds during the translocation assay. Membrane translocation induced by GA3-AM dissolved in DMSO (0.1%) was evaluated by fitting the initial part of the normalized time course of the decrease in cytoplasmic fluorescence signal intensity to the exponential function e−rt, where r is the rate constant used as an index of membrane translocation.
Quantification of ruffling
Membrane ruffles were defined as undulating membrane protrusions, folding back and transported forward, that fail to adhere. Membrane ruffles can be distinguished from lamellipodia by observing the dorsal plane. The extent of ruffling of each cell was scored using a scale of 1–3, where 1 indicates that no ruffles were present, 2 indicates that ruffling was confined to isolated regions covering no more than 25% of the peripheral area, and 3 indicates that extensive ruffles were present covering more than 25% of the peripheral area (Supplementary reference 11). Cells showing a score of 3 prior to drug addition were excluded from analysis. The ratio of the ruffling index (post-stimulus/pre-stimulus) for more than 20 cells from 3 independent experiments was calculated.
Visualization of esterase activity in living cells
Calcein-AM (ANASPEC) was added to live Cos-7 cells at 100 μM for 30 seconds immediately followed by washout of extracellular media in prior to time-series fluorescence imaging on an EPI microscope. Cells were pre-stained with Cholera toxin subunit B labelled with a dye (CTB AlexaFluor 555 from Invitrogen) to ensure the contour of cells.
Statistical analysis
Statistical analysis was performed with an unpaired two-tailed Student’s t test assuming the two populations have the same variances.
Supplementary Material
Acknowledgments
This study was supported in part by the National Institute of Health (NIH) (GM092930 and DK090868 to T.I., and NS072241 to M.J.W), the National Science Foundation (IOS-0641548 and MCB-0923723 to T.-p.S.), and the National Center for Research Resources of the NIH and NIH Roadmap for Medical Research (UL1 RR 025005 to C.M., D.J.M). TU is a recipient of a fellowship from the Japanese Society for the Promotion of Science. MC is a recipient of the Provost’s Undergraduate Research Award.
Footnotes
Author Contribution
T.M., A.S., M.C. and T.I. generated DNA constructs and T.M., R.D., T.U., A.S. and T.I. performed cell biology experiments. T.-p.S. advised on design of the gibberellin system. T.M. conducted biochemical experiments under supervision of M.J.W. C.M. and D.M. synthesized GA3-AM and GA3-H. T.I. conceived original ideas. R.D. and T.I. wrote the paper.
Competing Financial Interest
There is a pending patent associated with the gibberellin-induced dimerization system.
References
- 1.Fegan A, White B, Carlson JC, Wagner CR. Chemically controlled protein assembly: techniques and applications. Chem Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber S, Kapoor TM, Wess G. Chemical biology : from small molecules to systems biology and drug design. Wiley-VCH; Weinheim: 2007. [Google Scholar]
- 3.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 4.Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 5.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu T, et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korzeniowski MK, Manjarres IM, Varnai P, Balla T. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci Signal. 2010;3:ra82. doi: 10.1126/scisignal.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno T, Falkenburger BH, Pohlmeyer C, Inoue T. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Sci Signal. 2011;4:ra87. doi: 10.1126/scisignal.2002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayle JH, et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Czlapinski JL, et al. Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. J Am Chem Soc. 2008;130:13186–13187. doi: 10.1021/ja8037728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang FS, Ho WQ, Crabtree GR. Engineering the ABA Plant Stress Pathway for Regulation of Induced Proximity. Sci Signal. 2011;4:rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci U S A. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 15.Stojanovic MN. Molecular computing with deoxyribozymes. Prog Nucleic Acid Res Mol Biol. 2008;82:199–217. doi: 10.1016/S0079-6603(08)00006-8. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida W, Yokobayashi Y. Photonic Boolean logic gates based on DNA aptamers. Chem Commun (Camb) 2007:195–197. doi: 10.1039/b613201d. [DOI] [PubMed] [Google Scholar]
- 17.Katz E, Privman V. Enzyme-based logic systems for information processing. Chem Soc Rev. 2010;39:1835–1857. doi: 10.1039/b806038j. [DOI] [PubMed] [Google Scholar]
- 18.Qian L, Winfree E. A simple DNA gate motif for synthesizing large-scale circuits. J R Soc Interface. 2011;8:1281–1297. doi: 10.1098/rsif.2010.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. J Am Chem Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 20.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 23.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 2006;4:e45. doi: 10.1371/journal.pbio.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronson JE, Mazur WW, Cornish VW. Transcription factor logic using chemical complementation. Mol Biosyst. 2008;4:56–58. doi: 10.1039/b713852k. [DOI] [PubMed] [Google Scholar]
- 25.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 26.Ueguchi-Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 27.Hirano K, Ueguchi-Tanaka M, Matsuoka M. GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 2008;13:192–199. doi: 10.1016/j.tplants.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Ueguchi-Tanaka M, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsien RY. A non-disruptive technique for loading calcium buffers and indicators into cells. Nature. 1981;290:527–528. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirano K, et al. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell. 2010;22:2680–2696. doi: 10.1105/tpc.110.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 35.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 36.Malgaroli A, Milani D, Meldolesi J, Pozzan T. Fura-2 measurement of cytosolic free Ca2+ in monolayers and suspensions of various types of animal cells. J Cell Biol. 1987;105:2145–2155. doi: 10.1083/jcb.105.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson SJ, Jones RL. Gibberellic Acid Induces Vacuolar Acidification in Barley Aleurone. Plant Cell. 1996;8:2211–2221. doi: 10.1105/tpc.8.12.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tojima T, et al. Attractive axon guidance involves asymmetric membrane transport and exocytosis in the growth cone. Nat Neurosci. 2007;10:58–66. doi: 10.1038/nn1814. [DOI] [PubMed] [Google Scholar]
- 39.Regot S, et al. Distributed biological computation with multicellular engineered networks. Nature. 2010;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 40.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2010;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.