Abstract
DNA methylation can control some CpG-poor genes but unbiased studies have not found a consistent genome-wide association with gene activity outside of CpG islands or shores possibly due to use of cell lines or limited bioinformatics analyses. We performed reduced representation bisulfite sequencing (RRBS) of rat dorsal root ganglia encompassing postmitotic primary sensory neurons (n = 5, r > 0.99; orthogonal validation p < 10−19). The rat genome suggested a dichotomy of genes previously reported in other mammals: low CpG content (< 3.2%) promoter (LCP) genes and high CpG content (≥ 3.2%) promoter (HCP) genes. A genome-wide integrated methylome-transcriptome analysis showed that LCP genes were markedly hypermethylated when repressed, and hypomethylated when active with a 40% difference in a broad region at the 5′ of the transcription start site (p < 10−87 for -6000 bp to -2000 bp, p < 10−73 for -2000 bp to +2000 bp, no difference in gene body p = 0.42). HCP genes had minimal TSS-associated methylation regardless of transcription status, but gene body methylation appeared to be lost in repressed HCP genes. Therefore, diametrically opposite methylome-transcriptome associations characterize LCP and HCP genes in postmitotic neural tissue in vivo.
Keywords: bisulfite sequencing, dorsal root ganglion, HCP promoter, high CpG content promoter genes, integrated methylome-transcriptome analysis, LCP promoter, low CpG content promoter genes, peripheral nervous system, rat
Introduction
DNA methylation is recognized as molecular mechanism in retaining cellular identity (tissue specific methylation signatures) and as a co-determinant of gene activity. In adult mammalian tissues cytosine residues preceding a guanine, the CpG dinucleotide motif, is the primary target of DNA methylation. In well-characterized examples, CpG methylation is inversely correlated with gene expression, such as in repression of transposable elements1 and tumor suppressor genes.2 Previous studies indicate that mammalian promoters can be divided into two distinct classes determined by CpG content: high CpG promoters (HCP) and low CpG promoters (LCP).3-6 Elango et al. suggested that this distinction developed during early vertebrate evolution and is a characteristic of mammalian genomes.3 Gene ontology analyses further suggested a functional bimodality whereby LCPs are strongly associated with tissue-specific genes and HCPs primarily regulate house-keeping genes.6,7 Others have used a three-tiered system of dividing promoters by CpG content, namely low, intermediate and high CpG promoters.8-10 The classic model holds that DNA methylation suppresses transcription by targeting CpG-rich regions termed CpG islands (CGI). Evidence for this focus on CpG-rich regions can be found in many older studies such as Stein et al.,11 Busslinger et al.12 or Futscher et al.13 Several recent reports on methylation in promoter regions concluded similarly that the inverse correlation between methylation and transcription is found only in promoters containing CGI, i.e., HCPs, while no correlation could be found at LCPs.9,10,14 Differing models have been presented on how methylation and transcription are related in CpG-poor regions. In a recent review, Pelizzola and Ecker concluded that DNA methylation at CpG poor promoters cannot predict expression of the downstream gene.15 Hughes et al. found that the CpG density in repressive DNA methylation is significantly lower compared with DNA methylation that is not repressive.16 Studies comparing methylation patterns in different organs found tissue differentially methylated regions (tDMR) most often outside of CGI, i.e., in CpG-poor regions.7,17-21 In cases of single genes or selected gene subgroups, an inverse correlation of LCP methylation and gene activity was shown.17,19 Raykan et al.7 provided a tDMR study employing methylated DNA immunoprecipitation (MeDIP) that included a transcriptome comparison and concluded that methylation controlled the activity of LCP genes contradicting the studies discussed above such as Weber et al.10 Considering such opposing conclusions in even the most recent literature, the question remains unsettled whether and how LCP genes are controlled by DNA methylation.
Studies mapping DNA methylation in the CNS found that the neural methylome is unique.22,23 Ghosh et al.,22 for example, showed tissue-specific CpG island methylation distinguishing neural from non-neural tissue.22 Performing restriction landmark genomic scanning (RLGS) they identified 34 differentially methylated CpG islands, which revealed neural specific CpG island hypermethylation compared with mesoderm- or endoderm-derived tissues. DNA methylation plays a critical role in neurological disorders such as Rett Syndrome24 or early life stress25 and as important regulator in the CNS, such as in memory formation.26 Most studies on the role of DNA methylation in the nervous system have analyzed the brain. No comprehensive analysis of DNA methylation is available in the peripheral nervous system (PNS).
The rat ranks among the most important laboratory animals but few methylome studies have been performed in this species. None has interrogated the rat genome for different gene classes that may be relevant for understanding methylome-transcriptome associations. We analyzed the distribution of the CpG motif in rat promoters and found evidence that the model of dichotomizing genes into two classes, LCP and HCP genes, should apply to the rat. Nucleotide-resolution methylation data at genome-scale is thus far not available for the rat or for the peripheral nervous system (PNS) of any other species. We performed reduced representation bisulfite sequencing (RRBS),5 a technology for interrogating hundreds of thousands of methylation sites with digital precision27,28 on the rat PNS and found characteristic and highly significant patterns relating gene methylation and activity (p≈1.0x10−74) that differed markedly between LCP and HCP genes.
Results
RRBS of genomic DNA from the L4 DRG provided information on 16x106 cytosines including 2.8x106 CpG sites. DNA methylation was found principally at cytosines in CpG sites, the known methylation motif (Fig. S1A). < 1% of cytosine methylation occurred at CpH sites with a preference for the CpA motif (Fig. S1B). Methylation levels at CpG sites followed a bimodal distribution similar to previously reported bisulfite conversion-based studies in other tissues.29-31 Over half of the CpG sites were unmethylated, a fifth appeared completely methylated, and the remaining sites were methylated at an intermediate level (Fig. S2).
RRBS quantification proved highly reproducible among five independent biological replicates. The Pearson correlation coefficient was > 0.988 for comparison of individual CpG methylation levels among all possible pairings.
An independent technology, the HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) assay, was performed as orthogonal validation. HELP detects methylation status using a pair of a methylation-sensitive- and a methylation-insensitive restriction enzyme, HpaII and MspI.32 The protocol steps of HELP and RRBS are mutually exclusive. Of 7738 sites with high and low methylation states that were unambiguously informative in both assays, 82% were concordant, which was highly significant with p < 10−19 (Table S1).
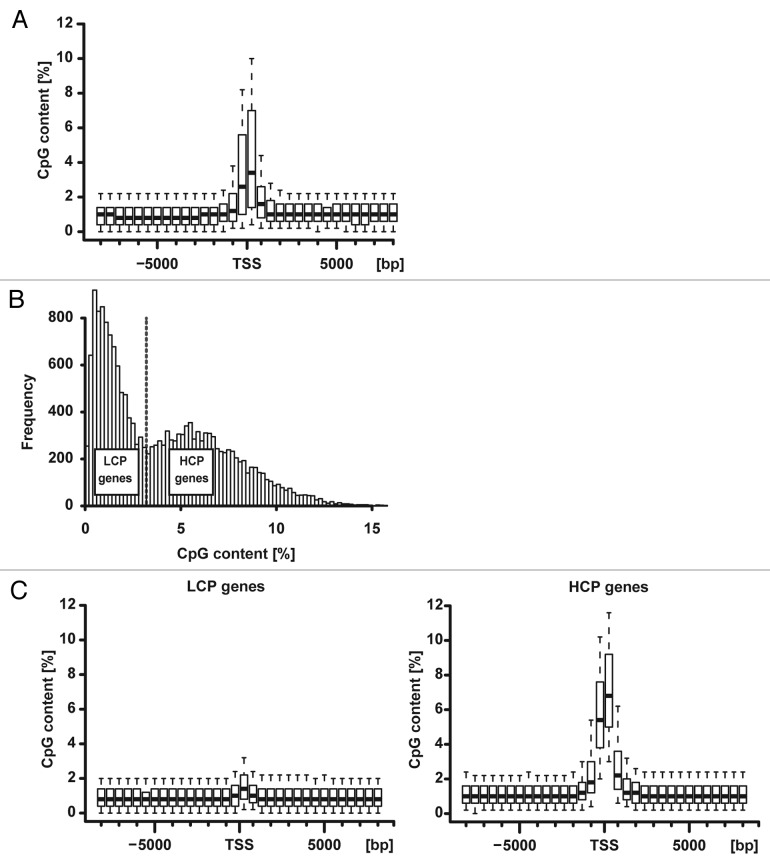
CpG dinucleotides are unevenly scattered across the genome. We found that CpG density in the rat genome peaked in the promoter region around the TSS consistent with findings in other genomes and with the observation that many promoters overlap with CGI (Fig. 1A). Next, we examined how the feature of promoter CpG density varied throughout the genome of the rat. Specifically, we wished to determine whether promoter CpG density was bimodally distributed in the rat like it is in the human genome.6 Taking the region from –500 bp to +500 bp around the TSS as a proxy, a bimodal distribution was noted, indicating two distinct groups (Fig. 1B). Dichotomizing the genes with a cutoff at a CpG density of 3.2% led to the classification of low CpG content (< 3.2%) promoter, “LCP,” genes and high CpG content (≥ 3.2%) promoter, “HCP,” genes (Figs. 1B and C). Of 17,602 protein-coding genes included in this study, 8644 were LCP genes and 8958 were HCPs.
Figure 1. Promoter CpG density defining the dichotomy of LCP vs. HCP genes in the rat genome. (A) Unselected rat genes are (in aggregate) characterized by a high density (frequency) of the CpG motif around the TSS (0 bp). Shown is the CpG density of 17,602 protein-coding genes included in the main analysis. Bins with a width of 500 nucleotides are shown. CpG density peaked in a narrow region of 1000 to 2000 nucleotides around the TSS. CpG density varied, however, considerably between genes as demonstrated by box plots indicating the 5th, 25th, 50th, 75th, 95th percentile. (B) The promoter CpG content of individual genes was bimodally distributed among the total set of genes indicating two distinct classes of promoters. The CpG content of the core promoter region was determined by choosing a 1000bp interval around the TSS (from -500bp to +500bp) as a proxy. Promoter CpG content varied among individual genes from < 0.5% to > 10%. Depiction of the promoter CpG content as histogram demonstrated two peaks at 1% and 5.5% suggesting a mixed distribution resulting from two distinct underlying populations. The position of the valley suggested a cutoff at 3.2% (vertical red line). The resulting dichotomization of genes provided a classification of “LCP” and “HCP” genes resembling that originally proposed by Saxonov et al.,6 which guided subsequent analyses. (C) CpG density in LCP genes was low not only—as expected—at the TSS but also throughout the remaining gene regions suggesting that there were no unrecognized regions of higher CpG density, CGI, farther away from the TSS. CpG density of HCP genes was high at the TSS reflecting how they were defined.
Sub-grouping genes by CpG density into more than two strata did not alter subsequent analyses further supporting the dichotomous classification. Analyzing LCP and HCP genes as two entities (rather than all genes together), however, was an important distinction.
Measurements of gene activity were available from our previous study, which had applied another massively parallel sequencing technique to the DRG, RNA-seq, validating the method extensively with qPCR.33
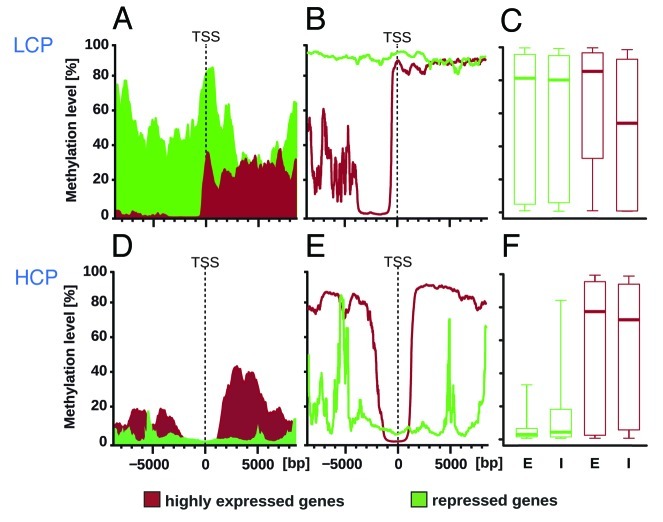
In LCP genes, transcriptional activity was linked to the methylation of CpG sites located at the TSS and within up to 8000 nucleotides 5′ of the TSS, a region commonly implicated in gene regulation (Fig. 2A). The most highly expressed genes had a mean level of CpG methylation of 0% in this upstream region, while it was > 40% for silenced genes (Fig. 2A). For this comparison, the mean % of methylation was “trimmed” by removing the highest and lowest 1/5th of values before averaging (“20% trimmed mean”), a common method in descriptive statistics to capture the middle (“central tendency”) of data sets that do not follow a normal distribution. We then performed a separate comparison of the top 1/5th of values by determining the 80th percentile rank of methylation levels. Here the difference in methylation levels between highly expressed and silenced LCP genes became even more evident: for highly expressed genes, the 80th percentile rank of methylation was < 10% in a region from -1000bp to -3000bp from the TSS, while it was > 90% for silenced genes (Fig. 2B). These findings for LCP genes are consistent with the classic model of increased methylation at promoters associated with decreased transcription. However, in the rat DRG this correlation was exclusively found in LCP genes suggesting that the repressive function of DNA methylation was most effective in shutting down genes in regions in which its target motif, the CpG dinucleotide, is sparse.
Figure 2. Diametric methylome-transcriptome relationships in LCP vs. HCP genes. (A) LCP genes (20% trimmed mean): Mean CpG methylation levels are shown for highly expressed (red) and repressed (green) LCP genes (low CpG content promoter genes containing < 3.2% CpG). Shown is the 20% trimmed mean in a 1000bp-wide moving window. CpG sites located at the TSS and within several thousand nucleotides 5′ of the TSS differed markedly between highly expressed and repressed LCP genes. Differences were highly significant in the region -6000bp to -2000bp with p≈3.7x10−88 and in the region -2000bp to +2000bp with p≈1.0x10−74. In the region +2000bp to +6000bp there was no significant difference with p = 0.42. (B) LCP genes (80th percentile): The 80th percentile rank of CpG methylation levels supported the same observation demonstrating hypomethylation of highly active genes and hypermethylation of silenced genes 5′ of the TSS. Methylation downstream of the TSS was high in LCP genes regardless of gene activity. (C) Gene body methylation in LCP genes: CpG methylation was similar in the gene bodies of highly expressed and silent LCP genes. Shown are boxplots for exons (E) and introns (I) indicating the 10th, 25th, 50th, 75th, and 90th percentile rank of methylation levels for each gene group. (D) HCP genes (20% trimmed mean): Mean CpG methylation levels of highly expressed and repressed HCP genes showed the characteristic deep valley of hypomethylation around the TSS, which is the region of high CpG motif density defining the HCP gene group. Methylation of silenced genes appeared to be only minimally higher at the TSS. (E) HCP genes (80th percentile): The 80th percentile rank of CpG methylation levels further supported the observation that the TSS of HCP genes remained poorly methylated regardless of the level of gene activity. Highly expressed HCP genes were marked by methylation outside of the TSS while the silenced HCP genes appeared to be relatively hypomethylated throughout the whole gene. (F) Gene body methylation in HCP genes: CpG methylation differed between the gene bodies of highly expressed and silent HCP genes. Differences were highly statistically significant with p≈2.6x10−156 for exons and p≈1.2x10−75 for introns.
LCP genes appeared highly methylated downstream of the TSS, i.e., in the region of the gene body whether or not they were expressed. Because a positive correlation of gene body methylation and expression was reported by others, we addressed this question further in a complementary analysis binning CpG methylation level data for exons and introns (first exon excluded from the exon bin because it overlaps the TSS). This comparison confirmed the above by showing that in LCP genes methylation of introns and exons was high regardless of expression levels (Fig. 2C).
In HCP genes the TSS was only minimally affected by methylation, remaining essentially unmethylated regardless of the level of gene expression (Figs. 2D and E). Outside of the TSS region highly expressed HCP genes were highly methylated and repressed HCP genes were hypomethylated. High methylation levels extended several thousand nucleotides on both sides of the TSS including the gene body. As with the LCP genes above, the main findings for HCP genes were apparent by comparing the two subgroups of genes (highly expressed and silent) using the trimmed mean % of methylation (Fig. 2D). The subgroup differences appeared even more clearly when the top 1/5th of methylation values was compared using the 80th percentile rank (Fig. 2E).
However, in HCP genes differences in gene body methylation were linked to expression. Introns and exons were poorly methylated in repressed genes and highly methylated in active genes (Fig. 2F), a relationship consistent with the hypermethylation of transcribed sequences reported previously.8,29,30,34
Discussion
A sequencing-based integrated, genome-wide methylome-transcriptome analysis was performed in the DRG, a postmitotic neural tissue harvested ex vivo without subsequent in vitro culture. Tissue was from the rat, an important laboratory animal for which a genome-wide DNA methylation map with nucleotide resolution was previously lacking. We used RRBS, a technique that was validated by several others35 and may represent the state of the art in the DNA methylation field. We added our own orthogonal validation with another genome-wide methylome analysis tool, the HELP assay, proving highly significant agreement for CpG sites covered in both assays (p < 10−19). While many other RRBS studies were limited to two replicates per tissue we analyzed 5 biological replicates demonstrating very high reproducibility. Taken together, the reported data therefore represents a technically faithful representation of the methylation levels of over 2.8x106 CpG corresponding to approximately 6% of such sites in the rat genome (CpG-level data will be available through the online rat genome database RGD as described in methods).
Examination of the rat genome sequence suggested two promoter types defined by CpG density: LCP and HCP. The distinction between LCP and HCP genes based on the bimodality of the underlying distribution of promoter CpG density was originally proposed for the human genome by Saxonov et al.6 and has been used recently in the analysis of genome-wide studies of human promoter control by histone modification.36
LCP and HCP genes are similarly frequent in the mammalian genome. Consequently, mechanisms affecting only one group may be diluted by noise from the other or will be cancelled out entirely, if all genes are analyzed as a single universe. Another incremental improvement to the analysis was the use of a trimmed mean and percentile ranks when comparing aggregate methylation levels between two groups of genes. In our case, the analysis of all genes combined using the untrimmed mean (average) showed methylation effects at the TSS (Fig.S3) resembling the depictions in other recent reports8,29,30,37 while obscuring important insights. However, analyzing LCP and HCP genes as distinct entities demonstrated different associations between gene methylation and activity in each group (Fig. 2). The main findings of our study therefore include not only biological observations but also suggestions toward refining standard analyses of genome-wide methylome-transcriptome data sets. For LCP genes our data clearly showed that there was an inverse correlation between promoter methylation and gene activity. Our findings contribute to a body of literature, which has not found agreement to date on the issue whether or not DNA methylation acts on CpG-poor promoters. Our findings agree with studies by Eckhardt et al.,17 Han et al.,19 Hughes et al.16Eckhardt et al.17 that showed an inverse correlation between methylation and transcription for a single low CpG promoter gene, oncostatin. Han et al.19 showed this relationship for the two tissue-specific CpG poor promoters, LAMB3 and RUNX3.19 Our findings for LCPs are also in alignment with studies focusing on t-DMRs, which found that hypermethylated t-DMRs associated with CpG poor regions (many but not all at promoters) are linked to gene repression. A comprehensive RRBS study by Meissner et al. included data on LCP promoters showing that in some cases methylation changed during tissue differentiation correlating with activating or repressing histone marks.36 On the other hand, many reports found that LCP promoters are not targeted or not affected by DNA methylation, e.g., Weber et al.10 and Koga et al.14 reported that LCP genes were expressed regardless of promoter methylation and a recent review article reached a similar conclusion.15 Some of the discrepancies in the literature may be related to experimental design, e.g., both of the above studies relied on a lower-resolution assay, MeDIP, which performs less sensitively in CpG-poor regions. Weber et al.10 used RNAP II occupancy as proxy for expression, which may be problematic because RNAP II can also bind to inactive genes. Our study is not affected by either of these potential pitfalls. Our experiments provide nucleotide resolution data on more CpG sites in LCP genes than any of the above-cited reports. By design our study was not limited to selected LCP genes but executed an unbiased and genome-wide analysis strengthening the case that gene methylation and activity are linked at LCP promoters.
Our findings for HCPs are consistent with the observation in many prior studies that the TSS (of HCPs) is typically unmethylated even in inactive genes. We studied a healthy, postmitotic tissue. Therefore, our findings do not disagree with the many well-established cases of HCP gene shutdown by promoter hypermethylation occurring in cancer or other diseases (as discussed in the introduction). We found that there was minimal if any difference in methylation in HCPs in the TSS region in the rat DRG. At the same time our data suggests a positive correlation between gene activity and the methylation of the body region of HCP genes, i.e., exons (p ≈ 2.6x10−156) and introns (p ≈ 1.2x10−75). This is consistent with the original report of this observation by Zilberman et al.34 in A. thaliana and subsequent studies in a variety of human embryonic and adult cell types.8,29,30 In our study, the observation did not extend to LCP genes. This dissimilarity between LCP and HCP genes appears not related to CpG density in the gene body, which is similar in both gene groups as shown in Figure 1C.
Furthermore, close examination of our data on HCP genes suggested the possibility that there might also be a positive correlation between gene activity and DNA methylation of a several-thousand nucleotide wide region upstream of the TSS, a topic that may warrant further exploration.
The main finding of our study was that the LCP vs. HCP distinction is strongly predictive of different methylome-transcriptome relationships. It is important to note, that similar to other studies emphasizing a genome-wide, agnostic approach our data demonstrates correlations between gene methylation and activity but does not establish causal relationships. At the same time, other genome biology studies suggest that the methylome differences observed here between HCP and LCP may indeed be mechanistically linked to transcription through control of chromatin. A recent report on another epigenetic mechanism, histone post-translational modification, found that different histone modifications predicted the activity of LCP and HCP genes (H3K4me3 and H3K79me1 vs. H3K27ac and H4K20me1).36 Taken together, these and our results support the model that alternate regulatory mechanisms are active in LCP and HCP genes.
How DNA methylation (co-)determines (or is associated with) gene activity may also depend on the tissue or experimental system under study. For instance shutdown of HCP promoters by DNA methylation is common in tumors but (as discussed above) may not be operative in normal tissues such as the PNS investigated here. Furthermore, it has been shown that in vitro culture of cells for as few as nine passages can induce new, non-random patterns of DNA methylation,5 which may be critical for some other studies. DRG neurons have lost the ability to divide. The present study may therefore be most representative of postmitotic tissues or may highlight features characteristic of the nervous system.
Methods
Tissue samples
Dorsal root ganglia (DRG) from the L4 level were harvested from adult male Sprague Dawley rats, flash-frozen and immediately stored at -80°C. DRG used for HELP assay analysis (n = 5) and for RRBS studies (n = 5) were harvested from two groups of neurologically intact rats and compared with the control group of a previous study,33 which was also neurologically intact (i.e., the group without a nerve ligation), in order to match the condition under which the existing transcriptome data set was obtained. All animals were obtained from Harlan Laboratories, Inc. All procedures were approved by the Institutional Animal Care and Use Committee.
Reduced representation bisulfite sequencing (RRBS)
DNA extraction was performed using the spin columns (Qiagen) according to the manufacture instructions. One μg of genomic DNA from the L4 DRG of each of 5 adult male Sprague Dawley rats was digested with MspI. Restriction ends were blunted, 3′ adenylated, and ligated to pre-annealed forked Illumina adaptors containing 5′-methyl-cytosines. For each of the 5 samples, two library size ranges 150–175 bp and 175–225 bp, including adaptor length, were isolated from a 2% agarose gel. Subsequently, samples were treated with the EpiTect Bisulfite kit (Qiagen) extending the bisulfite conversion time beyond the manufacturer’s protocol to 14 h by adding 3 cycles of 95°Cx5 min, 60°Cx180 min., Purified samples were subjected to 18 cycles of PCR, and gel-purified. 50 bp sequences were obtained on an Illumina GA IIx genome analyzer.
Sequence read alignment and calculation of cytosine methylation levels
Sequence reads were mapped in the three-nucleotide space (A, G, T) to MspI fragments predicted from the forward and reverse strand of the rat reference genome (rn4) using the stand_alone_Eland_extended module. Sequence read alignment performed with an alternative aligner, Bowtie,38 yielded identical alignments for the large majority of reads and indistinguishable results in all downstream analyses. Cytosines were then scored in each read in a binary fashion as methylated or unmethylated according to the sequencing result in the respective position—C indicating methylation (base protected from bisulfite conversion) and T indicating lack of methylation (cytosine base converted to uracil by bisulfite reaction and subsequently amplified as thymidine by PCR). Read-level data was then aggregated in a MySQL database. A minimum of ten reads was required for inclusion of a cytosine in subsequent high-level analyses, which were performed using the statistical program package R. The average read depth for the CpG sites included in the analysis was 503-fold.
High-level methylation analysis of gene regions
Genes were grouped according to the criteria detailed in the figure legends and aligned according to their annotated TSS. A rectangular sliding window of 1000 bp width without further smoothing was moved across a region from -8000 to +8000bp relative to the TSS. Methylation levels of all CpG (covered by the RRBS experiment with at least ten reads) were grouped together to calculate the mean methylation level and percentile ranks of methylation levels at each nucleotide position. Statistical measures commonly used to describe differences between normally (Gaussian) distributed data such as the average are ineffective in capturing important differences in data sets such as of CpG methylation, where relatively few data points are near the average and most data points cluster at the extremes. (The distribution of methylation levels resembles a β-binomial distribution.) In such cases it is important to understand, which portion of the data is affected when a difference is observed. To clarify why the average (mean) may not be a good measure, we could picture a group of CpG methylation levels as people with different incomes in a country with great social disparity: Many are living in extreme poverty earning almost nothing (methylation = 0), some are very rich (methylation = 1.0) and the middle class is underrepresented (few methylation levels from 0.2 to 0.8). The “average” (income or methylation level) represents the ratio of the data points at the extremes and falls into a range, where very few individual data points lie. To capture the data in a more meaningful way we used percentile ranks. All genomic areas analyzed encompassed generally at least 10% of unmethylated CpGs, i.e., the 10th percentile rank is a flat line across all regions of most genes regardless of expression level or promoter CpG content and is therefore uninformative. Accordingly, the 20% trimmed mean was depicted in the figures to capture the middle section of the data set. The high percentile ranks were highly informative. Therefore, the 80th percentile is shown in the figures.
Significance testing
Differences in the methylation levels were assessed for gene regions. At each nucleotide position within the region the 20% trimmed mean was determined for each gene group resulting in a value pair (one value each for the repressed and highly active gene group). For instance in a 2000 nucleotide wide region up to 2000 such value pairs could be formed, while only those nucleotide positions were included for which data was available in both gene groups. A paired, two-sided t-test was then performed.
Gene expression levels
Gene activity was determined from publicly available raw read data of our recent DRG transcriptome study,33 which had used RNA-seq on poly-A purified RNA from the rat DRG. Gene expression levels were quantified as reads aligned to specific genes normalized for the total number of reads aligned in each sample. Repressed genes were defined as those lacking reads. Active genes were divided into five groups of equal size according to expression levels, whereby the top group was referred to as highly active genes.
HELP assay
The HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) assay was performed as described in the original report32 employing a published rat-specific Nimblegen oligonucleotide array design.39 In brief, genomic DNA from each DRG (n = 5) was divided into two aliquots, which were subjected to restriction digestion with MspI or HpaII. These enzymes are isoschizomers recognizing the same sequence motif, 5′-C’CGG-3′, while only HpaII is inhibited by methylation of the CpG in the center of the motif and MspI is always active independently of the cytosine methylation state. The pattern of restriction fragments generated with MspI and HpaII will therefore be different reflecting the methylation state of 5′-C’CGG-3′ sites throughout the genome. To detect the differences, restriction fragments were amplified by ligation mediated PCR following published protocols32 and subjected to competitive hybridization on a custom rat oligonucleotide array.39 Hybridization was performed at the Nimblegen contract research laboratory, which was accessed through the genomics core facility contract services at Albert Einstein College of Medicine, where the HELP assay was originally developed.39 Program scripts from a published HELP assay bioinformatics pipeline available through bioconductor40 were utilized for processing HELP data. The threshold for fragment detection was set at 2.5x background fluorescence for HpaII and MspI hybridizations. Fragments with lower fluorescence in the MspI samples were non-informative. Informative fragments were included in the validation analysis for the above RRBS assay if the CpG sites flanking the HELP assay fragment were also assayed by RRBS. CpG sites were scored as methylated in the HELP assay if the respective fragment was detected only in the MspI sample and as unmethylated if it was detected in both the MspI and the HpaII sample.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank Dr. Maria Figueroa for early advice in performing the HELP assay and for helpful critiques during the manuscript preparation. Support was provided by the National Institute of Neurological Disorders and Stroke (NINDS) of the NIH (R01NS063022 and R21NS062271) (A.S.B.) and by the Schulze Family Foundation (A.S.B.).
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/19565
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- 3.Elango N, Yi SV. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 2008;25:1602–8. doi: 10.1093/molbev/msn110. [DOI] [PubMed] [Google Scholar]
- 4.Landolin JM, Johnson DS, Trinklein ND, Aldred SF, Medina C, Shulha H, et al. Sequence features that drive human promoter function and tissue specificity. Genome Res. 2010;20:890–8. doi: 10.1101/gr.100370.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–7. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–29. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–8. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 11.Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982;79:3418–22. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busslinger M, Hurst J, Flavell RA. DNA methylation and the regulation of globin gene expression. Cell. 1983;34:197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 13.Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–9. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 14.Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, et al. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–70. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelizzola M, Ecker JR. The DNA methylome. FEBS Lett. 2011;585:1994–2000. doi: 10.1016/j.febslet.2010.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010;11:554–60. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–84. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han H, Cortez CC, Yang X, Nichols PW, Jones PA, Liang G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20:4299–310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Ladd-Acosta C, Wen B, Wu ZJ, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, Kalyana-Sundaram S, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–41. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh S, Yates AJ, Frühwald MC, Miecznikowski JC, Plass C, Smiraglia D. Tissue specific DNA methylation of CpG islands in normal human adult somatic tissues distinguishes neural from non-neural tissues. Epigenetics. 2010;5:527–38. doi: 10.4161/epi.5.6.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–96. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Fan GP. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 25.Szyf M. The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics. 2011;6:971–8. doi: 10.4161/epi.6.8.16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–29. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 28.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 29.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–60. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–31. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5:e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–55. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer P, Banck MS, Amberg R, Wang C, Petznick G, Luo S, et al. mRNA-seq with agnostic splice site discovery for nervous system transcriptomics tested in chronic pain. Genome Res. 2010;20:847–60. doi: 10.1101/gr.101204.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 35.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong CB, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107:2926–31. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem. 2010;285:15111–8. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, et al. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24:1161–7. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.