Abstract
Most pathogenic mitochondrial DNA (mtDNA) mutations induce defects in mitochondrial oxidative phosphorylation (OXPHOS). However, phenotypic effects of these mutations show a large degree of variation depending on the tissue affected. These differences are difficult to reconcile with OXPHOS as the sole pathogenic factor suggesting that additional mechanisms contribute to lack of genotype and clinical phenotype correlationship. An increasing number of studies have identified a possible effect on the epigenetic landscape of the nuclear genome as a consequence of mitochondrial dysfunction. In particular, these studies demonstrate reversible or irreversible changes in genomic DNA methylation profiles of the nuclear genome. Here we review how mitochondria damage checkpoint (mitocheckpoint) induces epigenetic changes in the nucleus. Persistent pathogenic mutations in mtDNA may also lead to epigenetic changes causing genomic instability in the nuclear genome. We propose that “mitocheckpoint” mediated epigenetic and genetic changes may play key roles in phenotypic variation related to mitochondrial diseases or host of human diseases in which mitochondrial defect plays a primary role.
Keywords: DNA Methylation, epigenetics, epigenome, mitocheckpoint, mitochondria, OXPHOS
Introduction
Mitochondria are the major energetic sites of the cell, producing energy (ATP) through oxidative phosphorylation (OXPHOS). Mitochondrial OXPHOS is controlled by both mitochondrial DNA (mtDNA) and nuclear genomic DNA (gDNA). Mitochondria also perform a central role in many cellular processes including apoptosis, aging and oxidative metabolism.1,2 Mitochondrial function varies between different tissues. Tissues with high energy usage, including the brain, heart and muscle, contain a large number of mitochondria, allowing them to be more susceptible to a reduction in aerobic metabolism. Injury to the mitochondrial electron transport chain (ETC) or mutations of mtDNA leading to mitochondrial dysfunction have recently been suggested as an important factor in the pathogenesis of mitochondrial diseases, aging, cancer and a range of other human disorders.1-3 Most pathogenic mtDNA mutations induce defects in mitochondrial OXPHOS and thereby ATP synthesis. However, a large degree of phenotypic variation has been observed and associated with different mtDNA mutations. These are difficult to reconcile with OXPHOS defects as the sole pathogenic factor, implying that additional mechanisms must contribute to the phenotype, including epigenetic modifications. Here we review epigenetic and genetic factors associated with regulation of the mitochondrial and nuclear genomes with a particular emphasis on cancer.
Epigenetic modifications or mechanisms have been recognized as important factors regulating gene expression. Three broad categories of epigenetic modifications include genomic DNA methylation, post translational modification of histone tails within higher order chromatin and regulation of gene expression by non-coding RNAs (ncRNAs).4 Perturbed epigenetic mechanisms have been associated with a variety of human pathologies but their role in pathogenesis of mitochondrial diseases is unclear. The importance of the epigenome in the pathogenesis of mitochondrial diseases is likely to be as significant as any other human disease where epigenetic mechanisms are affected. Recent cancer studies identifying defects in the mitochondria have led to the discovery that mitochondrial dysfunction induces or is associated with epigenetic alteration within the nuclear genome, which could contribute to the perplexing complexity associated with mitochondrial diseases.5,6
The Mitochondrial Genome
The number of mitochondria within a particular cell varies from tissue to tissue. mtDNA represents less than 1% of total cellular DNA. However, its mitochondrial gene products are essential for normal cellular function. mtDNA within a single cell generally have identical sequences, which are described as homoplasmic. Heteroplasmy, or different mtDNA sequences within mitochondria of the same cell, can occur in response to somatic mutations.7 Since there could be over a thousand copies of mtDNA within a single cell, mitochondrial pathologies usually arise after a minimal threshold of heteroplasmy has been reached. That threshold is apparently different for different tissues and is dependent upon energetic and other mitochondrial functions.
The human mitochondrial genome is a 16.6 kb circular DNA encoding for 13 proteins in the ETC and the displacement loop (D-Loop), as well as two rRNAs (rRNAs) and 22 tRNAs (tRNAs) necessary for the translation of mitochondrial genes.1,8,9 The D-loop is a non-coding region within mtDNA (np 16024–516) contains cis-acting regulatory elements that are required for replication and transcription of mtDNA. All other mitochondrial proteins, including those involved in the replication, transcription and translation of mtDNA, are nuclear-encoded. Nuclear-encoded proteins are imported by specialized protein complexes on the inner and outer mitochondrial membrane.10 Mammalian mtDNA contains no introns and lacks histones.
Role of mtDNA in the Pathogenesis of Mitochondrial Dysfunction in Cancer
Inherited mutations in mtDNA as well as depletion of mtDNA content have been reported in a variety of diseases,11 including cancer.12 Mutations in mtDNA, involving somatic mutations as well depletion in mtDNA content are also associated with development of tumors. Interestingly, these somatic mutations within the D-loop region have been identified in a number of tumors.13,14 Since the D-Loop region contains regulatory elements involved in mtDNA replication, it could affect mtDNA copy number. Indeed tumor-specific changes in the mtDNA copy number have been reported in human cancers.12,15-20 Additionally, mtDNA dimers21 and mtDNA mutations in mitochondrial genes have been found in patients with leukemia, lung cancer, renal cell carcinoma, hepatocellular carcinoma, breast cancer and ovarian cancer.2,3,13 Interactions between proteins encoded by mitochondrial and nuclear genomes play critical roles in cellular function. Studies show that mitochondrial dysfunction induces changes in the expression of nuclear genes involved in cellular signaling, metabolism, growth and differentiation and apoptosis.22
Inherited mtDNA Mutations and Predisposition to Cancer
mtDNA is maternally inherited and genetic differences in mtDNA modulate the assembly of OXPHOS complexes and regulate its function.3,23,24 Recent studies have identified an increasing number of cancers associated with various mtDNA haplogroups within the human population. Tanaka’s group analyzed a population with 1,503 autopsied cases and identified genotypes for 149 polymorphisms within the coding region of the mitochondrial genome. Haplogroups were classified into 30 haplotypes. Subjects with the haplogroup M7b2 showed an increased risk for hematopoietic cancer. Results also indicated that haplogroup M7b2 is a risk factor for leukemia.25,26
Booker et al. determined an association of U haplogroup with prostate and renal cancers.27 Patients carrying the U haplogroup were found to have an increased risk of renal and prostate cancer. Bai et al.28 analyzed mtDNA polymorphisms in European-American females and reported that A10398G and T16519C polymorphisms increase breast cancer risk. In contrast, T3197C and G13708A polymorphisms were found to decrease breast cancer risk. Wang et al.29 evaluated polymorphisms in mtDNA associated with increased risk of pancreatic cancer in Caucasians, but found no significant associations with pancreatic cancer.
The 10398A allele localized in the NADH dehydrogenase-3 locus (ND3) of mtDNA has been associated with an increased risk of invasive breast cancer in African-American women.30,31 Similarly, the 10398A mutation is also associated with breast and esophageal cancer in Indian women, whereas 10398G has been shown to increase the risk of breast cancer in European-American women.28,32 Germ line mutations such as G10398A G9055GA, T16519C, G13708A, T3197C, and A10398G result in increased susceptibility to breast cancer in women.28,30 Using a cybrid based approach Kulawiec et al.12 analyzed the tumorigenic potential of the 10398A mutation (identified in African-American woman) and found that it induces complex I activity resulting in increased ROS production.12 The 10398A mutation also conferred resistance to apoptosis mediated by Akt activation. Additionally, the study demonstrated that the G10398A polymorphism leads to increased tumorigenesis and metastases in mice.12 A summary of mtDNA mutations associated with cancers is described in Table 1.
Table 1. Examples of certain mutations or polymorphisms in mtDNA associated with cancer susceptibility.
| mtDNA Mutations | Disease susceptibility | Reference |
|---|---|---|
| M7b2 Haplogroup |
Increased risk of hematopoietic cancer and leukemia |
25, 26 |
| U Haplogroup |
Prostate and renal cancers |
27 |
| A10398G and T16519C |
Increased breast cancer risk |
28 |
| T3197C and G13708A |
Decreased breast cancer risk |
|
| G10398A, G9055GA, T16519C, G13708A, T3197C, A10398G |
Increased susceptibility to breast cancer in women |
28, 30 |
| G10398A |
Increased risk of invasive breast cancer in African-American women |
30, 31 |
| G10398A |
Associated with breast and esophageal cancer in Indian women |
28, 32 |
| A10398G |
Increased risk of breast cancer in European-American women |
|
| G10398A |
Induces Complex I activity and increased ROS production |
12 |
| G10398A |
Resistance to apoptosis mediated by Akt activation |
|
| G10398A | Increased tumorigenesis and metastases in mice |
The Mammalian Nuclear and Mitochondrial Epigenomes
In addition to mtDNA mutations, recent studies have also provided valuable insight into epigenetic modifications associated with the regulation of the mitochondrial and nuclear genomes relative to mitochondrial dysfunction and disease.5,33,34 Epigenetic modifications are known to regulate gene expression either in a permanent or transient manner.35 These epigenetic mechanisms that alter the rate of gene expression without actually changing the physical sequence of nuclear encoded genomic DNA are categorized into three broad groups.36,37 Epigenetic modifications within the mammalian nuclear genome include DNA methylation (5-mC) or hydroxymethylation (5-hmC)38,39 at the carbon five position of cytosine residues juxtaposed to a guanine base (termed CpG dinucleotides), covalent histone modifications of N-terminal tails of the core nucleosome octamer (two H3, H4, H2A and H2B) and small/long non-coding RNA (ncRNA) regulation of gene expression.37,40-44 5-hmC has been associated with an increased presence in mammalian brain, particularly in Purkinjee cells. The function of this epigenetic modification and possible occurrence in other tissues is under intensive investigation,45,46 including roles the Tet1 family of proteins might have in addition to their catalytic and demethylating activities and association with Polycomb-mediated gene repression and H3K27me3 occupancy.46,47 In addition to mtDNA mutations and polymorphisms, epigenetic regulation of mtDNA by nuclear encoded proteins is unclear. Mammalian mitochondria have recently been identified to have mitochondrial DNA methyltransferase 1 (mtDNMT1) activity, 5-mC and 5-hmC.33 Shock et al.33 identified translocation of nuclear DNMT1 to the mitochondrial matrix is regulated by expression of a conserved mitochondria targeting sequence, upstream of the gene’s transcription start site within the nuclear encoded gene. Alterations in mtDNMT1 directly affected transcription from the light and heavy strands of mtDNA suggesting a correlation between 5-hmC and 5-mC mediated transcriptional regulation of mtDNA by a nuclear encoded gene. These findings provide new evidence implicating epigenetic regulation of the mitochondrial genome by nuclear encoded translocated mtDNMT1 relative to mitochondrial dysfunction.
Regulation of Genomic DNA Methylation and Chromatin Structure Associated with the Mitochondria
Hypermethylation of CpG dinucleotides present within CpG islands (CG-rich regions) at promoter regions of genes,48,49 CpG island shores (CG-rich regions near CpG islands)50 and non-coding regions within the human genome, including genes coding for microRNAs,51-53 are known to positively associate with their downregulation. In vertebrates, DNA methyltransferases (Dnmt1, Dnmt3A, Dnmt3B and Dnmt3L) catalyze the methylation of cytosine residues at their carbon five positions by the transfer of a methyl group (CH3) from S-adenosyl methionine (SAM-CH3 or AdoMet) to S-adenosyl-homocysteine (SAH), which acts as a donor.54-56 Mitochondrial dysfunction could cause alterations in redox (reduced oxygen) reactions and SAM-CH3 production in the cell, leading to perturbed methylation of nuclear encoded genomic DNA (Fig. 1). A host of factors are recruited to these regions including the methyl CpG binding proteins (e.g., MeCP2) and methyl binding domain proteins (MBDs) maintaining the repressive state. Hypermethylation of otherwise unmethylated regions or vice versa has been shown to arise de novo in certain types of cancers.57-60 In addition to the perturbed state of genomic DNA methylation, alterations at the level of chromatin structure associated with covalent modifications of histone tails has also been documented in cancer.61 The level of cross talk between DNA methylation and chromatin state are beginning to be identified. An understanding of factors, co-factors and enzymes generated both outside and inside the nucleus and within the mitochondria involved in the regulation of these complex processes remains elusive. These collectively include SAM,54 ATP (adenosine triphosphate) citrate lyase (ACL), Acetyl-CoA,62 flavin adenine dinucleotide (FAD)63 and lysine-specific demethylase 1 (LSD1).64,65 Understanding the influence these factors have on epigenetic states of nuclear genomic DNA and signaling pathways could help identify roles bioenergetics, biogenesis and the mitochondria might have in regulating epigenetic modifications within the nuclear genome.55
Figure 1. Schematic model showing a slowdown in redox reactions within the mitochondria and redox-dependent enzymes due to mitochondrial dysfunction. (A) Normal maintenance of hypermethylated CpGs within the nuclear genome associated with healthy mitochondria and synthesis of S-adenosyl-homocysteine (SAH) from S-adenosyl methionine (SAM-CH3). (B) Loss of DNA methylation in otherwise normally hypermethylated CpG dinucleotides within the nuclear genome due to reduced availability of SAM.
Co-factors Synthesized in the Mitochondria Regulate Chromatin Dynamics within the Nuclear Genome
The dynamic nature and plasticity associated with higher order chromatin structure renders a particular region of the nuclear genome either active or inactive in a tissue-specific manner. Catalysis of covalently modified histone tails at their N-terminals by histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), histone demethylases (HDMATs) or histone chaperones facilitates the dynamic rearrangement of particular loci rendering them loosely packed or highly compacted.66 These covalent modifications occurring primarily at histone tails include methylation, acetylation, ubiquitination and phosphorylation.67,68 For a review see refs.69-71 Deciphering the roles catalytic factors have in the active de/methylation or de/acetylation of histone tails could identify new co-factors associated with these highly dynamic metabolic reactions. Acetylation of histone tails primarily occurs on lysine residues; however, methylation occurs on both lysine (K) and arginine (R) residues.70 Monomethylation and trimethylation of histone H3 at lysine 4 (H3K4me1, H3K4me3) is associated with actively transcribed regions, whereas histone 3 trimethylated at lysine 27 (H3K27me3) is associated with transcriptionally silent regions within the compartmentalized nuclear genome.68,72
Classical metabolic reactions such as amine oxidation and hydroxylation have significant and active roles in the processes of histone demethylation, including the modification of histone 3 mono-methylated at lysine 4 (H3K4me1).73 These reactions involved in the dynamic alteration of histones allow for the active transcription or repression of regions within the nuclear genome.74 Acetylation and methylation of histone tails are dynamic processes regulated by histone de/acetyltransferases,74,75 methyltransferases or demethylases.76 Co-factors, including FAD (Flavin Adenine Dinucleotide),64 acetyl–CoA55,62 and α ketoglutarate (α-KG) are associated with the processes of active de/methylation or de/acetylation. Demethylation of covalently modified histones, including H3K4me2 and H3K4me1, is mediated by LSD1 (Lysine-Specific Demethylase 1) a component of the NuRD (Nucleosome Remodeling and Deacetylase) complex.65 The process of H3K4me2 demethylation catalyzed by LSD1 utilizes the classical amine oxidation reaction and FAD as a co-factor63 (Fig. 2A). The JmjC-domain proteins (JMJD), another family of histone demethylases, use α-KG and iron (Fe) as co-factors77 to demethylate the repressive H3K27me3/276,78(Fig. 2B). Both FAD and α-KG are known to be synthesized in mammalian mitochondria.63,79,80 Reduced levels of these co-factors due to mitochondrial impairment/dysfunction could have significant effects on regulation of the nuclear genome. This could lead to perturbed demethylation of histone lysine residues catalyzed either by LSD1 or JMJD. Tissue-dependent mitochondrial function established by the transcriptional status of nuclear encoded mitochondrial genes associated with differentially methylated regions (T-DMRs) could also serve to deduce important functional aspects of the mitochondria in different tissues and its perturbed disease states, particularly cancer.81,82
Figure 2. Illustrative model showing the catalysis of H3K4me2 (A) and H3K27me3 (B) demethylation mediated by LSD1 and the JMJD protein family, catalyzed using mitochondria synthesized co-factors FAD and α ketoglutarate, respectively.
Mitochondria to Nucleus Retrograde Response
Mitochondrial dysfunctions invoke mitochondria-to-nucleus retrograde responses in human cells.83 Perturbed DNA methylation profiles of certain loci within the human nuclear genome have recently been associated with depleted mitochondrial DNA. Smiraglia et al.5 developed and studied cells that were void of functional mitochondria (rho0 cells), enabling them to gain an insight into the possible role mitochondria might have in regulating or being associated with epigenetic alterations of the nuclear genome in a gene specific or genome-wide manner, particularly at the level of DNA methylation. Depletion of the mitochondrial genome in both chemically generated MCF12A rho0 or genetically modified 143B rho0 cell lines resulted in the aberrant methylation of promoter CpG islands (high CG-rich regions)84 that were previously unmethylated in the parental cell line,5 measured using restriction landmark genomic scanning (RLGS).85 The genetically altered 143B rho0 cell line showed a complete alteration in the methylation profile of the regions studied. One of the regions looked at further in this study was the aberrant promoter methylation of the BC030713 mRNA transcript that had been previously identified using RLGS.5,85 The study showed that the 5׀ UTR comprising a CpG island was partially hypo/unmethylated in 143B rho0 compared with the parental cell line that was completely hypermethylated at this region. The study concluded that this partial loss of genomic DNA methylation could be associated with the loss of mitochondrial DNA (mtDNA) and the mitochondria itself;5 however, it does not disqualify a perturbed genome-wide methylation profile. Repletion of wild-type mitochondria back into these 143B rho0 (mtDNA deficient cells) resulted in the partial re-establishment of some methylation profiles back to their original parental state. Although one cannot rule out the effect other factors might have on maintaining the state of DNA methylation, this study provided an interesting insight into the possible role mitochondria might have in establishing or maintaining genomic (nuclear) DNA methylation. However, more evidence is needed to decipher the specific factors and co-factors affected by this loss in order to establish the direct or indirect relationship they might have in affecting the epigenetic profile at the level of genomic DNA methylation or chromatin structure. Cross-talk between the nucleus and the mitochondria could have significant implications in aging and cancer, where DNMT1 activity is known to be perturbed.86,87 Mitochondrial dysfunction has been associated with the occurrence of certain cancers5,88,89 and neurodegenerative disorders,80 including Alzheimer disease.90 Increasing evidence has shown there to be an interdependent relationship between the mitochondria and the nuclear genome, including DNA methylation in both the nucleus and the mitochondrial matrix.5,6,33,89 An understanding of the role these factors might have in regulating nuclear DNA encoded mitochondrial genes is of keen interest. Takasugi et al.91 looked at the possibility of epigenetic regulatory mechanisms affecting the transcriptional control of nuclear genomic DNA encoded mitochondrial proteins. They used a systematic approach to identify the presence of differentially methylated regions within nuclear encoded mitochondrial genes in a tissue-dependent manner in mouse tissue obtained from the cerebellum, liver and heart.91 The study identified the presence of differentially methylated regions in 636 out of 899 nuclear encoded mitochondrial genes, which are known to have functional roles related to the mitochondria, in a tissue-specific manner. The study established a direct correlation between the rate of transcriptional gene expression/repression relative to the methylation profile of tissue-dependent differentially methylated regions (T-DMRs) in the tissues that were studied, relative to the transcriptional start site of the gene.91
Mitochondrial Regulation of Genetic Changes in the Nuclear Genome
Studies suggest that retrograde cross-talk between mitochondria and nucleus plays an important role in tumorigenesis. Cybrid studies have revealed marked changes in the cellular proteome, including quantitative changes in the expression of several proteins in breast and ovarian tumors, which suggest that retrograde responsive genes may potentially function as tumor suppressors or oncogenes.19
We determined human rho0 cells to have a reduced rate of repair of oxidative DNA damage, and have increased frequencies of mutation within the nuclear genome.92,93 The mitochondria-mediated mutator phenotype detected in yeast rho0 cells can be suppressed by inactivating subunits of the error-prone DNA repair. Error-prone repair polymerases are involved in the bypass of several types of DNA lesions that have the potential to inhibit chromosome replication.93 Therefore, one interpretation of our findings is that mitochondrial dysfunction limits or decreases nuclear DNA repair, resulting in unrepaired DNA lesions, which are subsequently converted into mutations by error-prone repair. Another explanation for the mutator phenotype observed in rho0 cells is that mitochondrial dysfunction generates excessive DNA damage that is converted into mutations by error-prone repair polymerases. Increased error-prone bypass of DNA lesions has been suggested to be characteristic of imbalanced dNTP pools.94 Imbalance of these dNTP pools has been shown to induce base substitutions as well as frame-shift mutations.95 Furthermore, imbalance of the dNTP pools causes delay in the progression of the DNA replication fork and enhances fragile sites where chromosomes are susceptible to breakage. Consequently, imbalance of the dNTP pools can promote chromosome rearrangement, breakage and loss.96-98
DNA mutations may activate proto-oncogenes and/or inactivate tumor suppressor genes, leading to genomic instability, which plays an important role in development of human cancer. In this regard, it is noteworthy that POLG encoding mtDNA polymerase γ, (a nuclear-encoded gene involved in the maintenance of the mitochondrial genome), is involved in development of cancer. Mutations in polymerase γ, the major mtDNA polymerase, have been associated with breast cancer.2 Both in vitro and in vivo studies have shown that mutations in the exonuclease domain of polymerase γ lead to the accumulation of mitochondrial mutations, while mutations in the polymerase domain lead to the depletion of mtDNA.2,99 Interestingly the POLG promoter contains a CpG island and is regulated by changes in methylation.5
A Role for “Mitocheckpoint” in Pathogenesis of Human Diseases
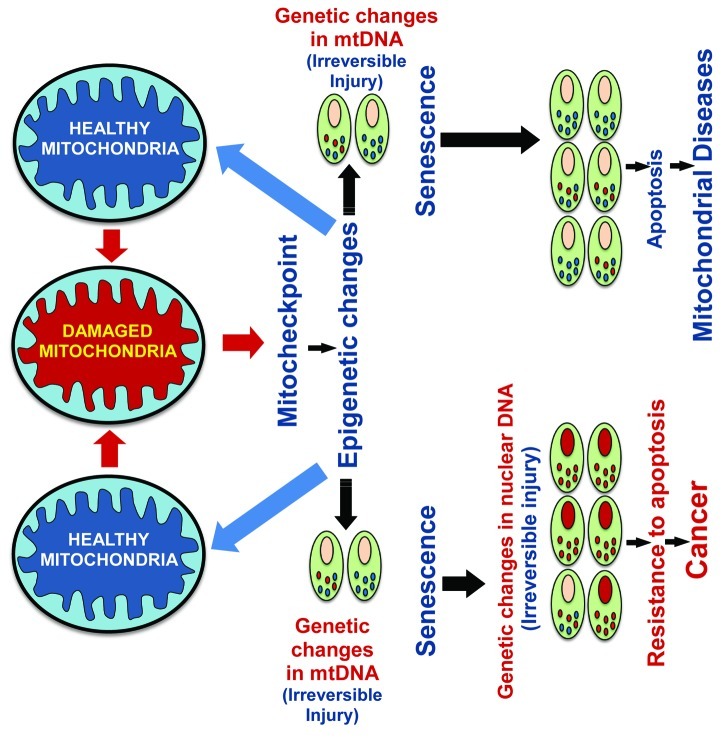
The functional state of mitochondria in human cells and the response to spontaneous or induced mtDNA or mitochondrial damage has been associated with a mitochondrial damage checkpoint (mitocheckpoint).3,12,83,100 The mitocheckpoint coordinates and maintains the proper balance between apoptotic and anti-apoptotic signals. Upon damage to mitochondria, mitocheckpoint is activated to help repair damage to mitochondria, restore normal mitochondrial function, and avoid induction of mitochondria-defective cells. This response may induce changes to the nuclear epigenome.5 Cross talk between the nucleus and mitochondria of individual cells may lead to a mitochondrial damage response as a result of incurred damage (Fig. 3). This could epigenetically alter mtDNA or gDNA expression.12,22,101
Figure 3. Schematic model depicting restoration of nuclear or mitochondrial gene expression associated with functional mitocheckpoint control. Induction of apoptosis or cancer due to damaged mitochondria resulting in a failure of mitocheckpoint control.
If mitochondria are severely damaged, such an event will trigger apoptosis (Fig. 3). If damage to mitochondria is persistent and defective mitochondria accumulate in the cell, it would lead to instability of the nuclear genome.12,19,93,98 Accumulated nuclear genome instability may help cells acquire new functions such as resistance to apoptosis,12,101 migration, and invasive characteristics which, in turn, could induce tumorigenesis12,102 (Fig. 3).
Recent studies suggest that cellular senescence provides an important barrier to tumorigenesis. These studies conducted in various cell types provide in vivo evidence that senescence is a defining feature in premalignant tumors.103-105 Cellular senescence limits the capacity to replicate, thus preventing the proliferation of cells. Senescence bypass appears to be an important step in the development of cancer.106 Our study suggests that an OXPHOS defect in a variety of cell types leads to cellular senescence101 and senescence bypass induced due to instability of the nuclear genome.12,98 We propose that OXPHOS-induced cellular senescence may act as an additional checkpoint mechanism that can function as a tumor suppressor (Fig. 3).
Future Perspectives
Finally, we suggest that damage to mitochondria could activate mitocheckpoint, which allows the restoration of mitochondrial function. When mitochondrial damage is severe or persistent, mitocheckpoint can fail and lead to cellular senescence (aging) or development of tumors (tumorigenesis) due to the accumulation of mutations or epigenetic modifications in mitochondrial or nuclear genomes. The mitocheckpoint may share characteristics of a signal-transduction pathway and may contain components such as damage/dysfunction sensors, mediators, signal-transducers and effectors. Sensor protein(s) may recognize damage to the mitochondria and mediators could signal the presence of damaged mitochondria and initiate biochemical cascade(s). Transducers are likely to be protein kinases that can relay and amplify the signal. Effectors may include transcription factors,12 involved in regulation of cell-cycle, DNA repair and apoptosis. Identifying the epigenetic profile of tissue-dependent differentially methylated regions of genes and their possible perturbed state in mitochondrial diseases,107 neurological disorders79,80,90 and cancers6 could further our understanding of the pathogenesis of complex human diseases due to defective mitochondria.19
Acknowledgments
This work was supported in part by a National Institutes of Health RO1 CA121904 and 113655 (K.K.S.). S.M. is a Marie Curie Early Stage Research (ESR) fellow funded by the Marie Curie Research Training Network, “Chromatin Plasticity” (EU FP6). The Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation, Denmark.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/19547
References
- 1.Singh KK. Mitochondrial DNA mutations in aging, disease and cancer. Springer, New York, NY, 1998. [Google Scholar]
- 2.Singh KK, Costello C. Mitochondria and Human Cancer. Springer, New York, NY, 2008. [Google Scholar]
- 3.Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbabio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol Ther. 2008;7:1182–90. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA, et al. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun. 2007;364:656–61. doi: 10.1016/j.bbrc.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee AME, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–74. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi NO, Bianchi MS, Richard SM. Mitochondrial genome instability in human cancers. Mutat Res. 2001;488:9–23. doi: 10.1016/S1383-5742(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 9.Taanman J. The mitochondrial genome: structure, transcription, translation and replication. Biochimica et Biophysica Acta (BBA)- Bioenergetics. 1999;1410:103–23. doi: 10.1016/S0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 10.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–6. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 11.Alberio S, Mineri R, Tiranti V, Zeviani M. Depletion of mtDNA: syndromes and genes. Mitochondrion. 2007;7:6–12. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A Variant in African-American with breast cancer provides resistance to apoptosis and promotes metastasis in mice. T Hum Genet. 2009;54:647–54. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–62. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvanayagam P, Rajaraman S. Detection of mitochondrial genome depletion by a novel cDNA in renal cell carcinoma. Lab Invest. 1996;74:592–9. [PubMed] [Google Scholar]
- 16.Lee MS, Yuet-Wa JC, Kong SK, Yu B, Eng-Choon VO, Nai-Ching HW, et al. Effects of polyphyllin D, a steroidal saponin in Paris polyphylla, in growth inhibition of human breast cancer cells and in xenograft. Cancer Biol Ther. 2005;4:1248–54. doi: 10.4161/cbt.4.11.2136. [DOI] [PubMed] [Google Scholar]
- 17.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90:2390–6. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438:981–7. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- 19.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, Matsui S-I. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–6. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–38. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 21.Clayton DA, Vinograd J. Circular dimer and catenate forms of mitochondrial DNA in human leukaemic leucocytes. Nature. 1967;216:652–7. doi: 10.1038/216652a0. [DOI] [PubMed] [Google Scholar]
- 22.Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pello R, Martín MA, Carelli V, Nijtmans LG, Achilli A, Pala M, et al. Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum Mol Genet. 2008;17:4001–11. doi: 10.1093/hmg/ddn303. [DOI] [PubMed] [Google Scholar]
- 24.Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, et al. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma M, Kumar D. Application of mitochondrial genome information in cancer epidemiology. Clin Chim Acta. 2007;383:41–50. doi: 10.1016/j.cca.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Verma M, Naviaux RK, Tanaka M, Kumar D, Franceschi C, Singh KK. Meeting report: mitochondrial DNA and cancer epidemiology. Cancer Res. 2007;67:437–9. doi: 10.1158/0008-5472.CAN-06-4119. [DOI] [PubMed] [Google Scholar]
- 27.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, et al. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol. 2006;175:468–72, discussion 472-3. doi: 10.1016/S0022-5347(05)00163-1. [DOI] [PubMed] [Google Scholar]
- 28.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–94. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, et al. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1455–9. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 30.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–33. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 31.Hall IJ, Moorman PG, Millikan RC, Newman B. Comparative analysis of breast cancer risk factors among African-American women and White women. Am J Epidemiol. 2005;161:40–51. doi: 10.1093/aje/kwh331. [DOI] [PubMed] [Google Scholar]
- 32.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–55. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proceedings of the National Academy of Sciences 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–8. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergman Y, Cedar H. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nat Rev Immunol. 2004;4:753–61. doi: 10.1038/nri1458. [DOI] [PubMed] [Google Scholar]
- 37.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 38.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Wang X, He K, Ma Y, Su N, He H, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–76. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 45.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–64. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–35. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ezzeldin HH, Lee AM, Mattison LK, Diasio RB. Methylation of the DPYD promoter: an alternative mechanism for dihydropyrimidine dehydrogenase deficiency in cancer patients. Clin Cancer Res. 2005;11:8699–705. doi: 10.1158/1078-0432.CCR-05-1520. [DOI] [PubMed] [Google Scholar]
- 50.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 53.Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31:864–70. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, et al. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–80. [PubMed] [Google Scholar]
- 55.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet. 2005;14(Spec No 1):R139–47. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 57.Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, et al. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br J Cancer. 2005;92:1574–80. doi: 10.1038/sj.bjc.6602478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–5. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 59.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 60.Feinberg AP. Genomic imprinting and gene activation in cancer. Nat Genet. 1993;4:110–3. doi: 10.1038/ng0693-110. [DOI] [PubMed] [Google Scholar]
- 61.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 62.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barile M, Brizio C, Valenti D, De Virgilio C, Passarella S. The riboflavin/FAD cycle in rat liver mitochondria. Eur J Biochem. 2000;267:4888–900. doi: 10.1046/j.1432-1327.2000.01552.x. [DOI] [PubMed] [Google Scholar]
- 64.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 66.Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 68.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–91. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–73. doi: 10.1016/S0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 71.Minocherhomji S, Madon PF, Parikh FR. Epigenetic regulatory mechanisms associated with infertility. Obstet Gynecol Int. 2010;2010:2010. doi: 10.1155/2010/198709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–6. doi: 10.1016/S0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 74.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 75.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 76.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 77.Koehntop KD, Emerson JP, Que L., Jr. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 78.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 79.Berkich DA, Xu Y, LaNoue KF, Gruetter R, Hutson SM. Evaluation of brain mitochondrial glutamate and alpha-ketoglutarate transport under physiologic conditions. J Neurosci Res. 2005;79:106–13. doi: 10.1002/jnr.20325. [DOI] [PubMed] [Google Scholar]
- 80.Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF. Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim Biophys Acta. 2010;1802:122–34. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sisková Z, Mahad DJ, Pudney C, Campbell G, Cadogan M, Asuni A, et al. Morphological and functional abnormalities in mitochondria associated with synaptic degeneration in prion disease. Am J Pathol. 2010;177:1411–21. doi: 10.2353/ajpath.2010.091037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–40. doi: 10.1016/S0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 83.Singh KK. Mitochondria damage checkpoint in apoptosis and genome stability. FEMS Yeast Res. 2004;5:127–32. doi: 10.1016/j.femsyr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 84.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 85.Smiraglia DJ, Kazhiyur-Mannar R, Oakes CC, Wu YZ, Liang P, Ansari T, et al. Restriction landmark genomic scanning (RLGS) spot identification by second generation virtual RLGS in multiple genomes with multiple enzyme combinations. BMC Genomics. 2007;8:446. doi: 10.1186/1471-2164-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu L, van Groen T, Kadish I, Li Y, Wang D, James S, et al. Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clinical Epigenetics 2011:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rhee I, Bachman KE, Park BH, Jair K-W, Yen R-WC, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 88.Dakubo G. Functional Importance of Mitochondrial Genetic Alterations in Cancer. Mitochondrial Genetics and Cancer 2010:213-36. [Google Scholar]
- 89.Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA, et al. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun. 2007;364:656–61. doi: 10.1016/j.bbrc.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iijima-Ando K, Hearn SA, Shenton C, Gatt A, Zhao L, Iijima K. Mitochondrial mislocalization underlies Abeta42-induced neuronal dysfunction in a Drosophila model of Alzheimer’s disease. PLoS One. 2009;4:e8310. doi: 10.1371/journal.pone.0008310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takasugi M, Yagi S, Hirabayashi K, Shiota K. DNA methylation status of nuclear-encoded mitochondrial genes underlies the tissue-dependent mitochondrial functions. BMC Genomics. 2010;11:481. doi: 10.1186/1471-2164-11-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Delsite RL, Rasmussen LJ, Rasmussen AK, Kalen A, Goswami PC, Singh KK. Mitochondrial impairment is accompanied by impaired oxidative DNA repair in the nucleus. Mutagenesis. 2003;18:497–503. doi: 10.1093/mutage/geg027. [DOI] [PubMed] [Google Scholar]
- 93.Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–17. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kunz BA, Kohalmi SE. Modulation of mutagenesis by deoxyribonucleotide levels. Annu Rev Genet. 1991;25:339–59. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 95.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proc Natl Acad Sci U S A. 1990;87:4946–50. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jacky PB, Beek B, Sutherland GR. Fragile sites in chromosomes: possible model for the study of spontaneous chromosome breakage. Science. 1983;220:69–70. doi: 10.1126/science.6828880. [DOI] [PubMed] [Google Scholar]
- 97.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–74. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 98.Desler C, Munch-Petersen B, Stevnsner T, Matsui S, Kulawiec M, Singh KK, et al. Mitochondria as determinant of nucleotide pools and chromosomal stability. Mutat Res. 2007;625:112–24. doi: 10.1016/j.mrfmmm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 99.Stumpf JD, Copeland WC. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh KK. Mitochondria damage checkpoint, aging, and cancer. Ann N Y Acad Sci. 2006;1067:182–90. doi: 10.1196/annals.1354.022. [DOI] [PubMed] [Google Scholar]
- 101.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, et al. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279:7512–20. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- 102.Kulawiec M, Arnouk H, Desouki MM, Kazim L, Still I, Singh KK. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer. Cancer Biol Ther. 2006;5:967–75. doi: 10.4161/cbt.5.8.2880. [DOI] [PubMed] [Google Scholar]
- 103.Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436:636–7. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- 104.Collado M, Serrano M. The senescent side of tumor suppression. Cell Cycle. 2005;4:1722–4. doi: 10.4161/cc.4.12.2260. [DOI] [PubMed] [Google Scholar]
- 105.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 106.Dimri GP. What has senescence got to do with cancer? Cancer Cell. 2005;7:505–12. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Naviaux RK. Mitochondrial control of epigenetics. Cancer Biol Ther. 2008;7:1191–3. doi: 10.4161/cbt.7.8.6741. [DOI] [PubMed] [Google Scholar]