Abstract
Smyd3 is a lysine methyltransferase implicated in chromatin and cancer regulation. Here we show that Smyd3 catalyzes histone H4 methylation at lysine 5 (H4K5me). This novel histone methylation mark is detected in diverse cell types and its formation is attenuated by depletion of Smyd3 protein. Further, Smyd3-driven cancer cell phenotypes require its enzymatic activity. Thus, Smyd3, via H4K5 methylation, provides a potential new link between chromatin dynamics and neoplastic disease.
Keywords: cancer, epigenetics, lysine, methylation, oncogene, oncology, Smyd3
Introduction
Methylation of histone proteins constitutes a principal chromatin-regulatory mechanism that influences fundamental DNA-templated processes such as gene transcription.1 Genetic abnormalities affecting the enzymes responsible for catalyzing the formation of histone methyl marks have been shown to play a significant role in the development of cancer and other diseases.1,2 For example, in multiple myeloma, a recurrent chromosomal translocation leads to overexpression of the lysine methyltransferase (KMT) MMSET (also named NSD2 and WHSC1), which dimethylates H3K36 to drive oncogenic programming.3 Neomorphic mutations of the KMT EZH2 have been found in patients suffering from diffuse large B-cell lymphoma and follicular lymphoma, implicating increased H3K27 trimethylation in the etiology of these cancers.4 These findings exemplify the potential clinical consequences of aberrant histone methylation, and suggest that increased understanding of histone methyltransferases will reveal important targets for oncology drug discovery.
Smyd3 (SET and MYND domain containing protein 3) is a KMT that is highly overexpressed in several cancers including liver, breast, and rectal carcinomas.5,6 Recent studies have begun to elucidate the oncogenic roles of Smyd3 including stimulation of proliferation, adhesion, and migration.7 While these studies highlight the involvement of Smyd3 in cancer and suggest an opportunity for chemotherapeutic intervention, a clear physiological function for the catalytic activity of Smyd3 remains obscure. On histones, Smyd3 has been reported to have conflicting activities, methylating either lysine 4 of histone H3 (H3K4) or lysine 20 of histone H4 (H4K20).5,8 We therefore took an unbiased substrate specificity screening strategy to identify the preferred histone residue methylated by Smyd3.
Results and Discussion
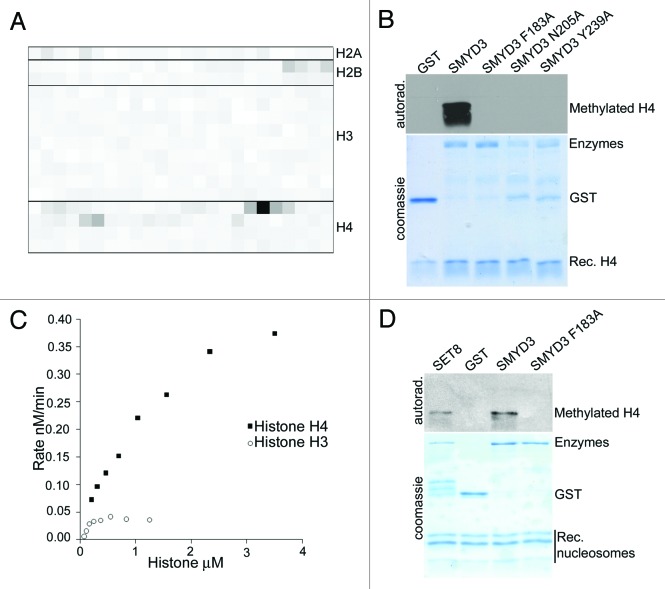
The methylation activity of recombinant full-length Smyd3 was screened against a library of 327 modified and unmodified histone peptides (see Table S1 for peptide content). The most highly methylated peptides were derived from histone H4 residues 1–21 (Fig. 1A, Table S1). None of the H3 peptides in the panel showed any activity as substrates including peptides that are unmethylated, mono- or di-methylated at H3K4 (Fig. 1A). Moreover, Smyd3 methylated recombinant histone, but three different Smyd3 catalytic mutants (Smyd3F183A, Smyd3N205A, and Smyd3Y239A) failed to do so (Fig. 1B and Fig. S1). Additionally, Smyd3 demonstrated 10-fold higher activity with recombinant histone H4 as compared to histone H3 (Fig. 1C). When reconstituted nucleosomes were used as substrates, Smyd3 methylated H4, like the positive control SET8/PR-Set7,9,10 but not the other core histones (Fig. 1D).
Figure 1.
SMYD3 selectively methylates H4 in vitro. (A) Heat map summarizing results from a library of 327 histone peptides tested as Smyd3 methylation substrates. Peptides were derived from histones H2a, H2b, H3 and H4 and possessed various combinations of modifications (see Table S1). (B) Methylation assays using the indicated recombinant Smyd3 proteins on recombinant histone H4. Autoradiograph and Coomassie stained gels (loading control) are shown. (C) Quantitative evaluation of Smyd3 methylation using recombinant histones H3 and H4. (D) Methylation assays as in (b) using reconstituted nucleosomes. SET8 is known to methylate nucleosomal H4.9
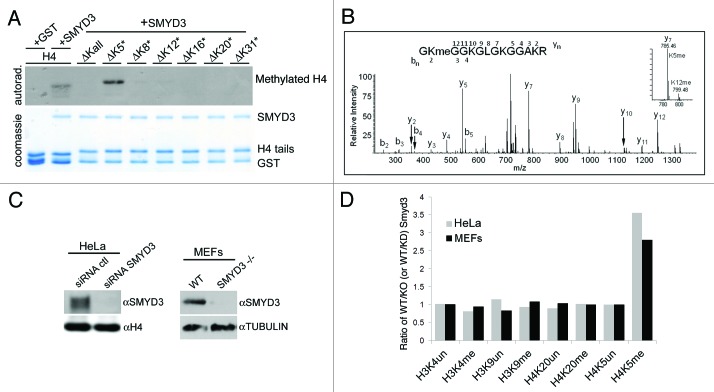
To determine which H4 lysine residue is modified by Smyd3, a recombinant library was generated in which only a single lysine residue on the H4 tail is present and available to undergo methylation. In this assay, H4K20 did not serve as a substrate, but methylation was observed primarily on K5, with very low activity also observed on K8 and K12 (Fig. 2A). Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) analysis of recombinant H4 methylated by Smyd3 identified H4K5me1 as the most abundant species, and also detected H4K5me2, H4K5me3 and H4K12me1 species, but no other methylation sites were detected including H4K20 (Fig. 2B). Thus, we conclude that in vitro Smyd3 primarily methylates histone H4 at lysine 5.
Figure 2.
Smyd3 methylates H4K5 in vitro and in vivo. (A) Methylation assay on H4 peptide residues 1–36, and H4 derived peptides with all lysines mutated to arginine (∆Kall) or single lysine maintained and all other lysines mutated to arginine as indicated (ΔK5* refers to a similar H4 peptide with all lysines mutated except K5). (B) LC-MS/MS analysis of Smyd3 methylated recombinant H4. (C) western blot analysis with the indicated antibodies of whole cell extracts (left panel) from HeLa cells expressing the indicated siRNAs and (right panel) wild-type and Smyd3−/− MEFs. (D) Quantitative mass spectrometry of the relative amounts of the indicated histone methylation marks in HeLa cells for Smyd3 positive/Smyd3 depleted cells (gray bars) and in wild type MEFs/ SMYD3−/− MEFs (black bars).
Smyd3 is detected in both the cytoplasm and nucleus (Fig. S2). Thus, while methylation of H4K5 in human cells has not been described to date, Smyd3 nuclear localization suggests that it could generate this mark in vivo. In this context, an unbiased MS/MS based approach was used to quantify the methylation states of all lysine residues present on histones H3 and H4 in two model cells systems, Smyd3 depleted HeLa cells and Smyd3 knockout mouse embryonic fibroblasts (MEFs) (Fig. 2C and D). H4K5me1 was detected in both cell types and the levels of this mark were significantly reduced upon Smyd3 knockdown or knockout (Fig. 2D and Table S2). Notably, in this analysis, no significant changes were observed in the methylation states of any of the other lysine residues including the previously implicated Smyd3 substrate sites of H3K4 and H4K20 (Table S2 and Fig. S3). Furthermore, global levels of H3K4 and H4K20 methylation in HeLa cells did not increase upon Smyd3 overexpression. (Fig. S4).
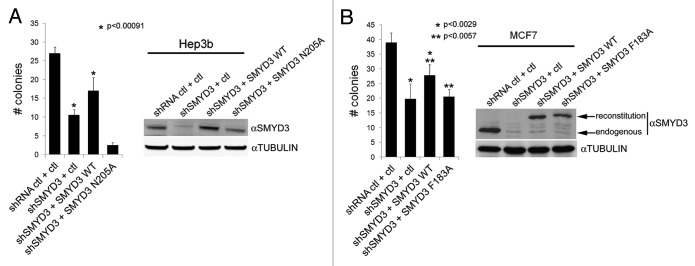
Consistent with previous reports, Smyd3 depletion attenuated proliferation of human carcinoma cell lines (Fig. S5).5 In addition, human breast carcinoma MCF7 cells and hepatoma Hep3B cells both lost the ability to form colonies in an anchorage-independent environment upon stable depletion of Smyd3 using shRNA directed to the 3′ UTR of Smyd3 (Figs. 3A and B). Colony formation was restored in Smyd3 depleted cells by complementation with wild-type Smyd3 (lacking the 3′ UTR and therefore RNAi-resistant) (Figs. 3A and B), whereas complementation with catalytically dead Smyd3 (Smyd3N205A (Fig. 3A) and Smyd3F183A (Fig. 3B) failed to reconstitute this activity. Moreover, global levels of H3K4me3 and H4K20me3 were unchanged upon Smyd3 knockdown in MCF7 cells (Fig. S6). Therefore, we conclude that while anchorage independent growth of MCF7 cells requires Smyd3 activity, maintenance of the global levels of H3K4me3 and H4K20me3 does not. Thus, Smyd3 is required for H4K5 methylation in cells and its enzymatic activity is important for maintaining transformed cell phenotypes associated with high Smyd3 expression.
Figure 3.
Smyd3 catalytic activity is required for anchorage-independent growth of cancer cells. (A-B) Complementation of Smyd3-depleted cells with wild-type Smyd3 but not catalytically dead Smyd3 restores anchorage-independent growth. (A) Left panel: Quantification of colony formation in methylcellulose after 10 d of Hep3b cells treated with either control shRNA or 3′UTR shSmyd3 reconstituted with GFP, Smyd3-WT or catalytically inactive Smyd3N205A. Right panel: western blot analysis of Hep3B whole cell extracts. (B) Left panel: Quantification of colony formation in soft agar after 14 d of MCF7 cells treated with either control shRNA or 3′UTR shSmyd3 reconstituted with Flag-control vector, Flag-Smyd3-WT, or catalytically inactive Flag-Smyd3F183A. Right panel: western blot analysis of MCF7 whole cell extracts. Bar graphs indicate the number of colonies per field. Error bars indicate the standard deviation (s.d.) from three independent experiments. The p values indicate the statistical significance as determined by t-test between the different conditions marked with * or **.
Here we report a novel site of histone modification, H4K5 methylation, which is catalyzed by the putative oncoprotein Smyd3. Future work aimed at understanding the molecular functions of H4K5 methylation in Smyd3-mediated oncogenic phenotypes should provide new insight into how chromatin methylation impacts human disease. Taken together, our results indicate that the likely physiologic chromatin target of Smyd3 is H4K5 methylation, and suggest that the catalytic methyltransferase activity of Smyd3 is an important target for anti-cancer drug discovery.
Materials and Methods
Materials and methods are detailed in the supplemental materials.
Supplementary Material
Acknowledgments
The authors are grateful to Song Tan for providing recombinant nucleosomes and Lydia Sanchez, Khyati Oza, Don Fisher and Linda Myers for technical support. This work was supported in part by grants to O.G. (NIH R01 GM079641 and GlaxoSmithKline), to B.A.G (DP2OD007447 and NSF CAREER Award), and J.S. and O.G. (CIRM RB1–01385). N.R. was funded by a Fondation pour la Recherche Medicale Postdoctoral Fellowship. O.G. is a recipient of an Ellison Senior Scholar in Aging Award.
Author Contributions
G.V., N.R., O.B., P.J.T., O.G. and R.G.K. conceived of and designed the experiments and wrote the manuscript. G.V., N.R., and O.B. contributed equally to the work and performed the majority of biochemical and cellular experiments. P.M., R.S. and B.L. helped generate reagents and perform experiments. Mass spectrometry analysis was performed by S.L., M.H., R.A., and B.A.G. Smyd3 knockout mice were generated and MEFs isolated by A.Z. and J.S.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/19506
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–20. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 6.Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–8. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–9. doi: 10.5483/BMBRep.2008.41.4.294. [DOI] [PubMed] [Google Scholar]
- 8.Foreman KW, Brown M, Park F, Emtage S, Harriss J, Das C, et al. Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS One. 2011;6:e22290. doi: 10.1371/journal.pone.0022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–99. doi: 10.1016/S0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 10.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–13. doi: 10.1016/S1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.