Abstract
An obstacle in the treatment of human diseases such as cancer is the inability to selectively and effectively target historically undruggable targets such as transcription factors. Here, we employ a novel technology using artificial transcription factors (ATFs) to epigenetically target gene expression in cancer cells. We show that site-specific DNA methylation and long-term stable repression of the tumor suppressor Maspin and the oncogene SOX2 can be achieved in breast cancer cells via zinc-finger ATFs targeting DNA methyltransferase 3a (DNMT3a) to the promoters of these genes. Using this approach, we show Maspin and SOX2 downregulation is more significant as compared with transient knockdown, which is also accompanied by stable phenotypic reprogramming of the cancer cell. These findings indicate that multimodular Zinc Finger Proteins linked to epigenetic editing domains can be used as novel cell resources to selectively and heritably alter gene expression patterns to stably reprogram cell fate.
Keywords: Maspin, SOX2, artificial transcription factors (ATFs), breast cancer, cancer cell reprogramming, DNA Methylation, epigenetic reprogramming, epigenetics, Zinc Finger Proteins (ZFPs)
Introduction
Epigenetic processes, such as DNA methylation and histone post-translational modifications, ultimately define active or inactive states of chromatin that control gene expression.1 In general, DNA methylation is associated with repressive, inactive chromatin and is a key stable and heritable epigenetic modification.2 The majority of 5-methyl-cytosine methylation occurs at CpG dinucleotides that are modified up to 70–80% in a cell type-specific manner in human cells and are faithfully transmitted to daughter cells during cell division.3,4 DNA methylation is catalyzed by DNA methyltransferase enzymes DNMT1, DNMT3a, and DNMT3b. DNMT1 maintains existing DNA methylation patterns, whereas DNMT3a and DNMT3b are mainly involved in the de novo establishment of DNA methylation marks.5,6 Neoplastic transformation is associated with alterations in DNA methylation, including both global hypomethylation and gene-specific hypermethylation.7 Gains in DNA methylation in cancer cells typically reflect hypermethylation of CpG islands in gene promoter regions, which can lead to gene silencing. Methylation-dependent gene silencing is a normal mechanism for regulation of gene expression.8 However, in cancer cells methylation-dependent epigenetic gene silencing represents a mutation-independent mechanism of inactivation of tumor suppressor genes.9
Significant emphasis has been placed on developing novel therapeutic strategies to selectively target and change the inappropriate gene expression patterns in cells to re-direct cell fate in human disease such as cancer. However, it is recognized that any therapeutic approach must entail the ability to generate long-lasting transcriptome changes—something not readily achieved by siRNA- and/or shRNA-mediated alteration of gene expression, which is transient in nature.10,11 Artificial transcription factors (ATFs) provide an alternative strategy to siRNA and shRNA, in that a DNA-binding domain (DBD) can be fused directly with a transcriptional effector domain and targeted to selective promoters in cells to mediate gene expression changes.12 ATFs contain arrays of Cys2-His2 zinc finger (ZF) domains, which specifically interact with 3 bp (triplet of recognition) of DNA.13 Using the alphabet of recognition between the ZF domain and the DNA triplet, multimodular proteins have been engineered14-20 that recognize specific sequences of targeted promoters and regulate expression.12,21 6ZF proteins (ZFPs) that target 18 bps represent optimized, state-of-the-art designs that exhibit high specificity/affinity, and regulate single target genes with high selectivity.22-24
Given the advantages and potential of ATFs, we sought to develop a novel ATF-mediated approach to epigenetically regulate genes via direct targeting of DNA methylation through the catalytic domain of DNMT3a. For this study, we focused on two genes of high relevance in cancer, which control the onset of the tumorigenic state of cancer cells: the tumor suppressor Maspin and the oncogene SOX2. The levels of expression of these two genes are critically controlled by DNA methylation.25-27 Methylation of the SOX2 promoter irreversibly changes cell fate by promoting a switch from a proliferative state toward a differentiation state in multiple cell types, such as stem cells and cancer cells. In contrast, methylation of the Maspin promoter has been associated with onset of breast cancer tumorigenesis and metastasis. Herein, we show that the delivery of DNMT3a bearing ATFs that target the Maspin and SOX2 promoters causes stable and heritable downregulation of these genes in breast cancer cells via DNA methylation. These results open the door for ATFs to be used as a therapeutic strategy for the stable regulation of gene expression patterns to control cell fate and development.
Results
ATFs downregulate Maspin in breast cancer cells
Maspin is a tumor suppressor that is silenced by DNA methylation in metastatic breast cancer cells,25,26 contributing to metastatic progression and being associated with poor prognosis.28-31 On the other hand, SOX2 encodes a master regulator transcription factor (TF) and has been implicated as an oncogene in breast cancer.32,33 Although SOX2 is epigenetically silenced by DNA methylation in normal epithelial cells, its promoter is overexpressed in advanced stage breast cancers, where it is essential to maintain aberrant self-renewal and tumor progression.32,33 Therefore, both Maspin and SOX2 contribute to breast cancer initiation and development in opposite ways.
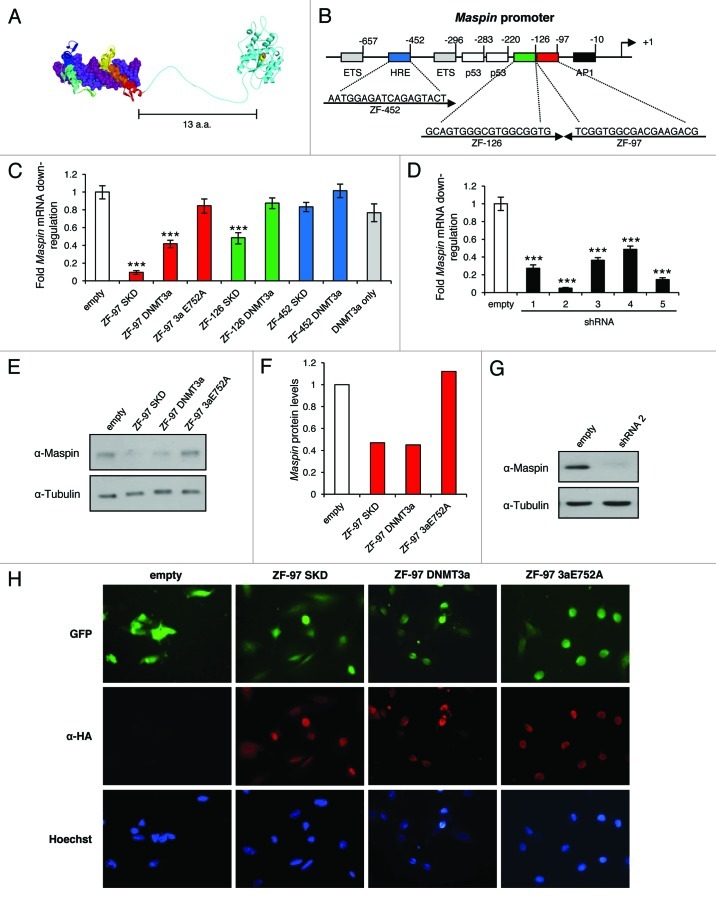
We first engineered ATFs containing ZFPs coupled with the KRAB (SKD) domain (used as a positive control) and the catalytic domain of DNMT3a to target and epigenetically repress the Maspin gene (Fig. 1A). We hypothesized that by directing the deposition of DNA methylation within the Maspin promoter we could trigger stable and heritable gene silencing, thereby increasing the oncogenic state of breast cancer cells. We have previously described the construction of three ZFPs designed to bind 18 bp sites in the Maspin proximal promoter34 (Fig. 1B). The ZFPs from ZF-97, ZF-126, and ZF-452 were C-terminally coupled to DNMT3a and a catalytic mutant that abolishes its enzymatic activity (DNMT3a E752A)35 with a flexible linker that enables DNMT3a to methylate target sequences proximal to the ATF binding site (upstream or downstream) (Fig. 1A). To determine if the designed ATFs were able to downregulate Maspin, we expressed the ATFs using the retroviral vector pMX-IRES-GFP in SUM159 breast cancer cells that express high levels of Maspin. Upon ATF transduction, ZF-97 SKD, ZF-97 DNMT3a and ZF-126 SKD, but not ZF-97 DNMT3a E752A or DNMT3a domain only, significantly downregulated Maspin expression relative to the empty vector control (Fig. 1C). DNMT3a appears to be a weaker repressor, but still significantly downregulates Maspin expression levels compared with empty vector control. ZF-126 DNMT3a, ZF-452 SKD, and ZF-452 DNMT3a exhibited similar levels to that of empty vector. Therefore, ZF-97 was selected for further subsequent studies given its ability to transcriptionally repress Maspin with the DNMT3a catalytically active domain. Transductions with five different shRNAs that target Maspin demonstrated significant transcriptional repression compared with the empty vector control (Fig. 1D). The Maspin shRNA 2 stable cell line was used in the subsequent studies as a positive control. Transduction of empty and ZF-97 DNMT3a E752A had no effect on Maspin protein levels; in contrast, ZF-97 SKD and ZF-97 DNMT3a cells exhibit significant alteration of Maspin protein levels, reflecting 50% reduction (Figs. 1E and F). The Maspin shRNA 2 almost completely abolished Maspin protein levels (Fig. 1G). Cells transduced with the empty vector, ZF-97 SKD, ZF-97 DNMT3a and ZF-97 DNMT3a E752A showed similar levels of infection based on GFP detection levels (Fig. 1H). Anti-HA was detected in all ZF-97 containing cells, but not in the empty vector (Fig. 1H). These results demonstrate that retrovirally delivered ZF-97 linked to silencing domains promoted Maspin downregulation in SUM159 cells similar (although weaker) to that of shRNA-directed silencing.
Figure 1. ATFs downregulate Maspin in SUM159 cells. (A) Structural model of 6 zinc finger domains linked by 13 amino acids to DNMT3a methyltransferase catalytic domain. (B) Schematic represents the ZF-452 (blue), ZF-126 (green), ZF-97 (red) sequences and their location from the transcription start site (arrow +1). (C) qRT-PCR results for Maspin expression in transduced cells. Red bars correspond to ZF-97 with SKD, catalytic domain of DNMT3a, and DNMT3a E752A. Green bars correspond to ZF-126 with SKD and DNMT3a domains. Blue bars correspond to ZF-452 with SKD and DNMT3a domains. Gray bar corresponds to the DNMT3a only and the white bar corresponds to empty vector control. Maspin expression level is depicted relative to that of the empty vector control. Error bars represent SEM. Statistical significance was analyzed using a t test (***p < 0.0001). (D) qRT-PCR results for Maspin expression. Black bars correspond to five different shRNA constructs that target Maspin and the white bar corresponds to empty vector control. Maspin expression level is depicted relative to that of the empty vector control. Error bars represent SEM. Statistical significance was analyzed using a t test (***p < 0.0001). (E) western blot analysis of Maspin protein levels in cells transduced with empty, ZF-97 SKD, ZF-97 DNMT3a, and ZF-97 3aE752A constructs. Tubulin is used as a loading control. (F) Quantification of Maspin protein levels relative to Tubulin. (G) western blot analysis of Maspin protein levels in cells after shRNA-mediated knockdown of Maspin. Tubulin is a loading control. (H) Detection of the retrovirus (GFP, green) and ZF-97 (α-HA, red) by immunofluorescence in cells. Nuclear staining was performed using Hoechst (blue). Images taken at 40x.
ZF-97 DNMT3a represses Maspin by targeted DNA methylation
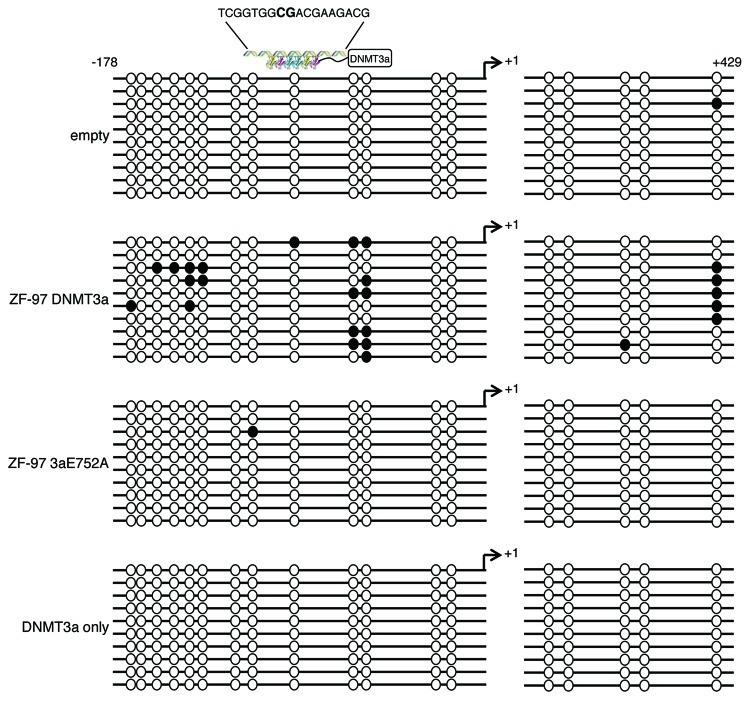
To verify that ZF-97 DNMT3a is catalytically active and that targeted DNA methylation was dependent on the specific binding of ZF-97, SUM159 cells were transduced with empty vector, ZF-97 DNMT3a, ZF-97 DNMT3a E752A, and DNMT3a only (untargeted domain). Genomic DNA of transduced cells was bisulfite converted and sequenced, and methylation patterns were examined within the Maspin promoter and exon 1 (Fig. 2). ZF-97 DNMT3a directly targeted DNA methylation to the Maspin promoter, with the highest density of methylated CpG dinucleotides immediately downstream of the ZF-97 binding site > 50% compared with controls (Fig. 2). Methylated CpG sites (50%) were found +429 downstream of the transcriptional start site (+1), which could reflect the folding and accessibility of these CpG sites within the higher order chromatin structure. Only background levels (< 1%) of CpG sites were methylated in cells containing the catalytically dead mutant (E752A) and untargeted DNMT3a domain (lacking ZF-97), indicating that a functional ZFP domain and an active DNMT3a enzyme domain are required for targeted methylation in vivo. Cells transduced with ZF-97 SKD did not exhibit targeted DNA methylation (Fig. S1). To determine if we could directionally target DNA methylation, we positioned the DNMT3a domain on the N-terminal region of the ZF-97 (originally cloned C-terminal). The DNMT3a ZF-97 significantly downregulated Maspin mRNA levels and similar levels of expression were seen in transduced cells (Figs. S2A and B). DNA methylation was targeted to the adjacent region surrounding the ZF-97 binding site, similar to that seen with the C-terminal DNMT3a domain (Fig. S2C). These results demonstrate the ability to selectively and efficiently target DNA methylation to the Maspin promoter, and that this methylation was not dependent on the orientation of the DNMT3a domain relative to ZF-97. This could be explained by the conformational freedom provided by our flexible linker separating the 6ZFs and the DNMT3a, which can mediate effective methylation in both orientations.
Figure 2. ZF-97 DNMT3a represses Maspin by targeted DNA methylation. Bisulfite sequence analysis for Maspin. Methylated CpGs are designated by closed circles, unmethylated CpGs are designated by open circles for cells transduced with empty vector, ZF-97 DNMT3a, ZF-97 3aE752A, and DNMT3a only (10 replicates each). Transcription start site indicated by arrow +1. ATF binding site depicted by 6 ZF proteins linked with a C-terminal DNMT3a domain with the sequence shown above. CG site within ZF-97 sequence shown in bold capital letters.
Downregulation of Maspin by targeted DNA methylation increases breast cancer cell colony formation
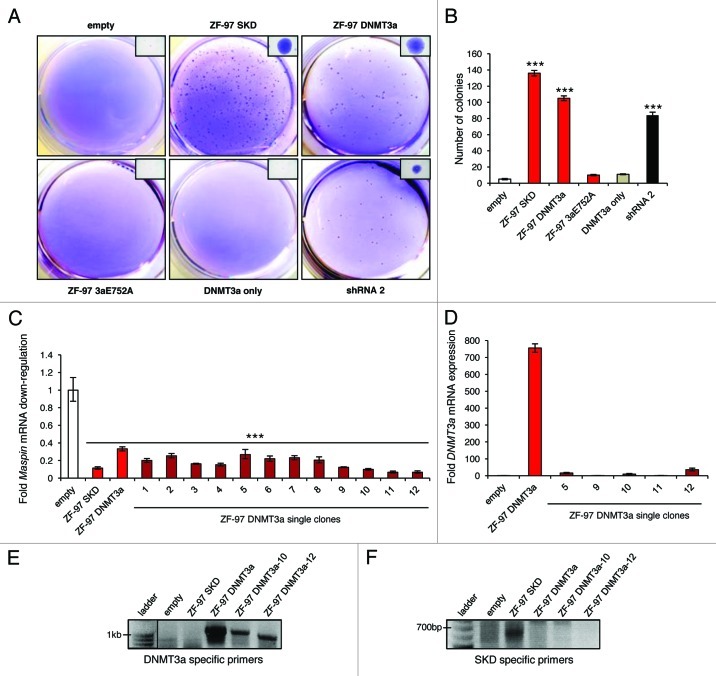
Maspin is silenced by DNA methylation in advanced breast cancers, contributing to metastatic progression. Therefore, we sought to examine if the downregulation of Maspin by ZF-97 DNMT3a targeted methylation led to anchorage-independent growth in soft agar similar to metastatic cells that lack Maspin expression. SUM159 is a poorly tumorigenic breast cancer cell line expressing high levels of Maspin. Transduced cells were seeded in soft agar and colony formation was examined. ZF-97 SKD, ZF-97 DNMT3a, and Maspin shRNA containing cells all induced colonies (> 100 colonies) relative to empty vector, DNMT3a mutant or DNMT3a only cells (< 10 colonies) (Figs. 3A and B). Select single colonies (ZF-97 DNMT3a clones 1–12) were recovered from the soft agar and expanded in culture medium. Importantly, qRT-PCR analysis of these clones revealed significant silencing of Maspin expression promoted by DNMT3a (Fig. 3C). Moreover, low levels of expression of the retrovirally integrated DNMT3a construct were observed via qRT-PCR compared with the parental ZF-97 DNMT3a cells (Fig. 3D). Genomic DNA from ZF-97 DNMT3a clone 10 and 12 were examined for the presence of the retrovirally integrated DNMT3a or SKD construct, using primers designed to overlap the retroviral vector with the DNMT3a or SKD domain, respectively. The integrated DNMT3a construct was detected in all of the constructs containing the DNMT3a domain, but not in cells containing the empty vector or SKD domains (Fig. 3E). The integrated SKD was only detected in the ZF-97 SKD containing cells (Fig. 3F). These results suggest that by 50 d post-infection, the ATF retrovirus exhibits diminished expression. Yet, the cells stably maintained Maspin downregulation, consistent with ATF-mediated epigenetic reprogramming of the Maspin promoter.
Figure 3. Downregulation of Maspin by targeted DNA methylation increases breast cancer cell colony formation (A) Soft agar assay results from cells transduced with empty vector, ZF-97 SKD, ZF-DNMT3a, ZF-97 3aE752A, DNMT3a only, and shRNA 2. Representative images of the entire agar well stained with Crystal Violet are shown along with a closer image of a single colony (in upper right corner). (B) Quantification of colony number. The white bar corresponds to empty vector control, the red bars correspond to ZF-97, the gray bar corresponds to DNMT3a only, and the black bar corresponds to shRNA 2. Error bars represent SEM. Statistical significance was analyzed using a t test (***p < 0.0001). (C) qRT-PCR results for Maspin expression in ZF-97 DNMT3a single clones (dark red bars 1–12) picked from soft agar and grown in culture for > 50 generations. The white bar corresponds to empty vector control and the red bars correspond to ZF-97 SKD and ZF-97 DNMT3a (parental pool) included for technical reference. Maspin expression level is depicted relative to that of the empty vector control. Error bars represent SEM. Statistical significance was analyzed using a t test (***p < 0.0001). (D) qRT-PCR results for the DNMT3a construct expression in select ZF-97 DNMT3a single clones (dark red bars). DNMT3a expression level is depicted relative to that of the empty vector control and the parental ZF-97 DNMT3a is included for technical reference. (E) Agarose gels from genomic DNA extracted from cells transduced with the empty vector, ZF-97 SKD, ZF-97 DNMT3a, ZF-97 DNMT3a single clones 10 and 12. Line indicates a lane on the same gel but was separated from subsequent lanes. Amplicons demonstrate products obtained with DNMT3a specific primers and (F) SKD specific primers.
Maspin is stably and heritably repressed by DNA methylation
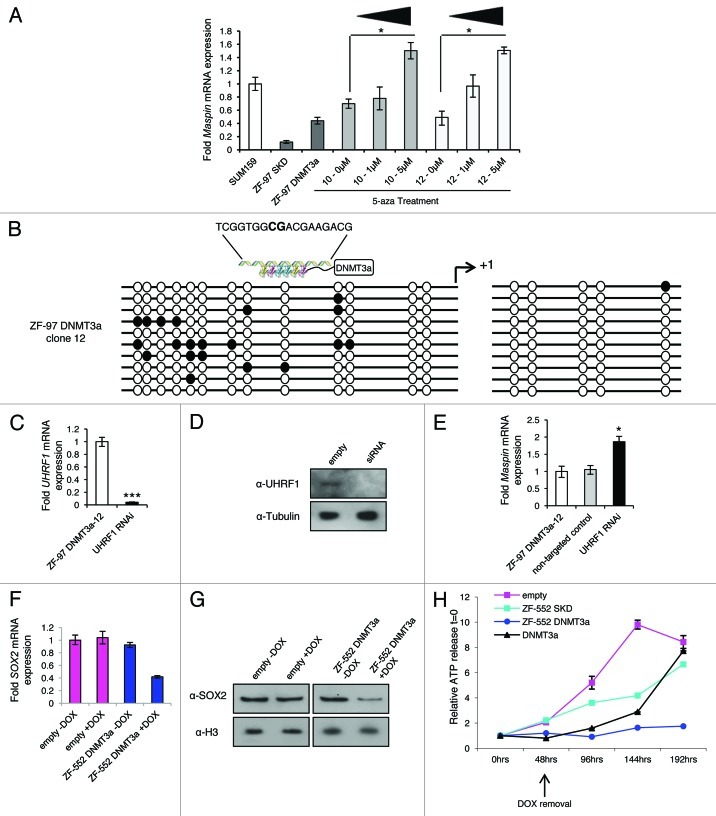
To address whether the initial methylation pattern of the targeted gene was inherited over multiple cell generations, we first treated ZF-97 DNMT3a clones 10 and 12 with 5-aza, an inhibitor of DNMTs, and Maspin gene expression was restored (100% of control) (Fig. 4A). The stable downregulation of Maspin seen in ZF-97 DNMT3a clone 12 was due to DNA methylation patterns similar to what was observed in the parental cells (ZF-97 DNMT3a)(Fig. 4B). These data suggest that DNA methylation patterns imposed by the ZF-97 DNMT3a construct were stably and heritably maintained over multiple cell generations.
Figure 4.Maspin is stably and heritably repressed by DNA methylation and ZF-552 DNMT3a downregulates SOX2 in MCF7 cells resulting in reduced cell proliferation. (A) qRT-PCR results for Maspin expression in ZF-97 DNMT3a single colony cells treated with 1µM and 5µM of 5-aza. Error bars represent SEM. Statistical significance was analyzed using a t test (*p < 0.05). (B) Bisulfite sequence analysis for Maspin in ZF-97 DNMT3a clone 12. Methylated CpGs are designated by closed circles; unmethylated CpGs are designated by open circles (10 replicates each). Transcription start site indicated by arrow +1. ATF depicted by 6 ZF proteins linked with a DNMT3a domain with the sequence shown above. CG site within ZF-97 sequence shown in bold capital letters. (C) qRT-PCR results for UHRF1 expression after RNAi-mediated UHRF1 knockdown in ZF-97 DNMT3a clone 12 cells. Black bar correspond to RNAi and the white bar corresponds to ZF-97 DNMT3a clone 12 before knockdown. UHRF1 expression level is depicted relative to that of ZF-97 DNMT3a clone 12. Error bars represent SEM. Statistical significance was analyzed using a t test (***p < 0.0001). (D) western blot analysis of UHRF1 protein levels in cells after siRNA-mediated knockdown of UHRF1. Tubulin is used as a loading control. (E) qRT-PCR results for Maspin expression after RNAi-mediated UHRF1 knockdown in ZF-97 DNMT3a clone 12 cells. Black bar corresponds to RNAi, the gray bar corresponds to a non-targeted control, and the white bar corresponds to ZF-97 DNMT3a clone 12 before knockdown. Maspin expression level is depicted relative to that of ZF-97 DNMT3a clone 12. Error bars represent SEM. Statistical significance was analyzed using a t test (*p < 0.05). (F) qRT-PCR results for SOX2 expression in transduced cells. Pink bars correspond to empty vector (- or +DOX) and blue bars correspond to ZF-552 DNMT3a. (G) western blot analysis of SOX2 protein levels in cells either un-induced (-DOX) or induced (+DOX) with empty vector and ZF-552 DNMT3a. Histone H3 is used as a loading control. (H) A representative time course cell viability assay with cells induced with DOX at t = 0. At 48 h post-induction the DOX was removed from the cell medium. Cell viability is measured by fold increase in ATP release related to t = 0. Error bars represent SEM.
The protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is necessary for maintaining DNA methylation patterns during replication by recruiting DNMT1.36,37 To further verify the stable transmission of DNA methylation initiated by the ZF-97 DNMT3a ATF, we challenged ZF-97 DNMT3a clone 12 cells with siRNAs targeting the UHRF1 protein. We found that knockdown of UHRF1 (> 90%) (Figs. 4C and D) led to a significant re-expression of Maspin relative to that of the non-targeted control (Fig. 4E). This restored Maspin levels to those obtained after treatment with 5-aza (Fig. 4A). Collectively, these results demonstrate that the ZF-97 DNMT3a ATF directly targets CpG methylation adjacent to the ATF binding site. Over time the ATF expression is no longer detected and required for silencing, and these sites of methylation were read and transmitted by UHRF1/DNMT1 to stably convey the methylation pattern over multiple cell generations.
ZF-552 DNMT3a downregulates SOX2 in MCF7 cells resulting in reduced cell proliferation
Some breast cancers overexpress the SOX2 gene, and in these cancers SOX2 behaves like an oncogene.38 Therefore, the SOX2 gene was targeted to determine if directed, stable silencing by DNA methylation (similar to what was observed with Maspin) would repress an overexpressed oncogene found in many breast cancers and, in contrast to repression of Maspin, if this repression could result in inhibition of cancer cell growth. A ZFP (ZF-552) was utilized coupled to the DNMT3a domain designed to bind a unique 18 bp region in close proximity to the transcription start site within the SOX2 promoter (manuscript submitted). Upon ZF-552 DNMT3a induction (+DOX) in MCF7 breast cancer cells that overexpress SOX2, we observed a downregulation of SOX2 mRNA (≥ 60%) and protein levels (> 80%) compared with the empty vector and no DOX induction (Figs. 4F and G). A time-course cell viability assay was utilized to assess changes in cell proliferation after induction of the ATF (+DOX) and ATF shut-off (DOX removal). SOX2 repression was associated with arrest of cell proliferation following ATF induction. However, ZF-552 DNMT3a cells maintained robust cell arrest for at least 192 h after DOX removal (Fig. 4H). In contrast, cells containing the empty vector, ZF-552 SKD or untargeted DNMT3a were able to recover and resume proliferation (Fig. 4H). Our results indicate that the 6ZF-DNMT3a fusions nucleated a heritable transmission of DNA methylation over cell generations that were sufficient to promote stable cancer cell growth inhibition, even when the expression of the ZF agent was suppressed.
Discussion
We have generated novel molecular resources for ATF-mediated selective and heritable epigenetic repression of critical genes in cancer cells, showing for the first time that epigenetic targeting can stably reprogram cancer cells. Effector domains, such as the naturally occurring transcription factor KRAB domain (SKD), do not possess intrinsic enzymatic activity. Instead, these domains are used to repress gene expression by passively recruiting chromatin remodelers, histone-modifying enzymes, and other proteins.39 Here, we employed DNMT3a to repress the Maspin and SOX2 genes. In contrast to the action of the KRAB domain, which recruits enzymes to repress gene transcription,39 epigenetic effector domains such as DNMT3a are active enzymes that will directly repress promoter activity. Gene repression by DNA methylation occurs via recruitment of co-repressors to methylated CpG dinucleotides, such as mSin3 or Mi2-NuRD, and histone deacetylases, to form condensed, repressive chromatin leading to stable and heritable gene inactivation.40 The C-terminal catalytic domain of DNMT3a is active, and when fused with a ZFP, directs DNA methylation at the target sequence of reporter systems in vitro.35 While repression can be produced with the use of siRNAs, these methods are of limited utility.10,11 For example, several regions of mRNA are generally targeted by multiple siRNAs to ensure successful targeted repression, increasing chances of off-target effects. In contrast, ATFs allow the efficient downregulation of gene expression through targeting of a single sequence. Thus, the ability to stably and heritably alter gene expression states via epigenetic reprogramming in cancer cells could be utilized for new therapeutic approaches or to target genes of specific biological significance.
The tumor suppressor gene Maspin was utilized as a proof-of-principle because it allowed (a) determination of the ability to repress Maspin by DNA methylation near the ZF-binding site (b) examination of the resulting tumorigenic phenotype after downregulation and (c) characterization of stable and heritable methylation patterns over multiple cell generations. We focused our studies of targeted DNA methylation on the ZF-97 protein because this DNA-binding domain exhibited the strongest repressive potential when linked to both the SKD and DNMT3a domains. The higher repressive capabilities of ZF-97 DNMT3a relative to proteins ZF-126 DNMT3a and ZF-452 DNMT3a could be associated with several factors. (1) The location of the ATF binding site relative to the transcriptional start site. ZF-97 targets an 18 bp sequence located very close to the transcriptional start site; thereby the nucleation of DNA methylation by this protein could interfere with transcriptional activity more effectively than upstream methylation events. DNA methylation is expected to either directly interfere with the binding of RNA-polymerase or other proteins necessary for transcription or to indirectly engage the recruitment of methyl-binding proteins and co-repressors in the close proximity of the transcription start site. (2) The accessibility and chromatin structure of the 18 bp target could highly influence the regulatory outcome of the ATF. We have previously reported that the efficacy of regulation of a given ATF depends on the cell line examined, which could be associated with target site accessibility and with the endogenous promoter context (for example, chromatin folding, presence of specific TFs, co-activators or co-repressors).41 In breast cancer cell lines carrying a silenced Maspin gene, ZF-126 linked to a transcriptional activator (ATF-126) is in fact more potent than ATF-97 and ATF-452 in activating Maspin expression.34 In contrast, in cell lines carrying high Maspin levels, ZF-97 is more potent linked with both SKD and DNMT3a repressors, compared with ZF-126 and ZF-452. The relative affinities of ZF-97, ZF-126, and ZF-452 for their targets in vitro are very similar, in the lower nanomolar and even picomolar range.34 Thereby, the higher potency of ZF-97 linked to transcriptional repressors is possibly associated with endogenous factors, such as target site localization relative to the transcriptional start site and promoter context. It will be interesting to obtain more data sets of repression in other promoter contexts to evaluate the influence of the target site in DNMT3a-repression of endogenous genes. Nevertheless, our preliminary data suggest that more proximal ZFPs repress more effectively than distally targeted proteins.
Our data suggest that in the context of the Maspin promoter, the DNMT3a appears to be a weaker repressor than SKD, but still significantly downregulates Maspin expression levels compared with empty vector control. One potential reason by which SKD leads to a potent repression could be due to the recruitment of histone deacetylases,39,42 which might promote effective promoter condensation of the Maspin promoter. However, one potential problem of SKD-mediated repression is the maintenance of the histone deacetylation over time, which requires persistent expression of the ZFP. In contrast, the longevity of the repressive effect promoted by DNA methylation seems to be permanently maintained or reprogrammed, even in absence of expression of the ZF. In the future, the engineering of bi-functional ATFs carrying both DNA methylation and histone deacetylation domains could enhance both, the potency and longevity of the repression.
As shown in our Figure S1, the ZF-97 DNMT3a but not the ZF-97 SKD induced ~50% of methylation in the adjacent CpG sites. These methylation frequencies were not as high as those observed in metastatic cell lines lacking Maspin expression, such as MDA-MB-231 (average ~80% methylation26,43) and H157 (average ~99% methylation44). Consistent with the lower methylation frequencies induced by the ZF-97 DNMT3a, the repression of Maspin transcript in the SUM159 cells was not complete, but was significant and sufficient to induce colony formation. There could be multiple reasons as to why the ZF-97 DNMT3a did not reach methylation levels like it is observed in highly metastatic MDA-MB-231 cells. ZF-97 has a high degree of sequence selectivity, which could restrict the binding of the DNMT3a in the Maspin promoter. Future engineering could benefit from targeting multiple sites in a single promoter, for example by directing multiple ZF-DNMT3a fusions, which could enhance the frequency of promoter methylation. The delivery of a single domain such as DNMT3a is unlikely to entail the complex repressive effect observed in natural metastatic cell lines, where multiple epigenetic constituents interact resulting in effective silencing. The combinatorial delivery of multiple enzymatic functions, uncovering several epigenetic modulators, could more effectively and permanently silence gene expression.
There are four lines of evidence that support the suggestion that heritable DNA methylation patterns are established through the action of ZF-97 DNMT3a. First, single soft agar colonies that proliferated through ≥ 50 cell generations were examined and found to lack expression of the retrovirus (ZF-97 DNMT3a construct), yet Maspin repression was maintained. Transcriptional silencing of retroviral constructs has been previously described after long-term culture of infected cells.45,46 The observation that while the levels of ZF-DNMT3a expression diminished over time but the methylation pattern initially nucleated by the proteins were stably maintained, open the door for ATFs to be used as a therapeutic strategy for the stable regulation of gene expression patterns to control cell fate and development. Although the residual expression of ZF-97 DNMT3a could still methylate the Maspin promoter, the dramatic loss of DNMT3a expression in clone 12 was not accompanied by a decrease of DNA methylation. This result supports the notion that the endogenous proteins could recognize and transmit the marks initially incorporated by the engineered proteins. Second, selected cell clones were treated with 5-aza, an inhibitor of DNMTs, and Maspin expression was restored to wild-type SUM159 levels. Third, DNA methylation was observed in ZF-97 DNMT3a clone 12 demonstrating the maintenance after multiple cell divisions of the original methylation pattern imposed by the ZF-97 DNMT3a. In the replication fork during cell division UHRF1 recognizes hemi-methylated DNA and interacts with DNMTs to faithfully methylate the daughter strands properly. Fourth, when UHRF1 was knocked-down in ZF-97 DNMT3a clone 12 cells, the level of Maspin increased similar to that observed with 5-aza treatment. This demonstrates that UHRF1 maintains the methylation pattern observed in these cells. The ability to silence the oncogene SOX2 and to mitotically arrest cell growth by targeted DNA methylation, most likely in G0/G1, is an exciting finding given that our technology sustained permanent, indefinite silencing of Maspin. Whereas, the conical SKD repressor domain coupled with ZF-552 (described by Stolzenburg et al., submitted manuscript) returned to levels similar to that of empty vector, ZF-552 DNMT3a cells showed arrested growth for 8 d strongly suggesting that the silencing of SOX2 is persistent due to DNA methylation.
Delivery of ZFPs into a cell in vivo has been a major limitation that has provided challenges for the clinical application of ZF technologies. Colleagues and we are designing novel state-of-the-art systemic delivery technologies for preclinical applications of ZFPs in mouse cancer models. We are utilizing nanoparticle technology to encapsulate ATF mRNA, which is less toxic and immunogenic than current delivery approaches. mRNA has a negligible chance of integration in the genome, therefore delivery of mRNA circumvents the problems associated with gene therapy. Overall our data demonstrate that it is possible to target “at will” DNA methylation to epigenetically and phenotypically reprogram cells. The ability to stably and heritably alter gene expression states via epigenetic reprogramming holds far reaching implications for treating human diseases, such as cancer.
Materials and Methods
ATF construction
The Maspin and SOX2 6ZFP construction conditions were previously defined.34 Maspin ZFP sequences are shown in Figure 1B and the SOX2 ZF-552 sequence is as follows 3′GGCCCCGCCCCCTTTCAT-5′ (manuscript submitted). The construction of the KRAB domain (SKD) was previously defined.21,42 The DNMT3a catalytic domain (598–908 amino acids) and flanking sequences were amplified from human fibroblast DNA and cloned in frame into pMX-IRES-GFP.47 Catalytically dead versions of DNMT3a were generated by PCR-mediated mutagenesis of the active site ENV motif, by changing ENV to ANV (E752A).35 The linker region between the ZFP and DNMT3a sequence is as follows: QASPKKKRKVGRA.
Cell culture, RNA and DNA preparation
Human breast cancer cell lines SUM159 and MCF7 were obtained from the Tissue Culture Facility of the UNC Lineberger Comprehensive Cancer Center (Chapel Hill, NC) and the 293TGagPol cell line was obtained from the American Type Culture Collection (ATCC). Cell lines were propagated in growth medium specified by the ATCC. Cell cultures were harvested for RNA and DNA preparation after retroviral or lentiviral transduction, siRNA transfection, and following demethylating treatment using the RNeasy Kit (Qiagen) or the Puregene DNA Purification Kit (Gentra Systems; Minneapolis, PA) respectively, according to the manufacturer’s protocol.
Retroviral and lentiviral shRNA infection
The retroviral vector pMX-IRES-GFP containing the Maspin ATFs and pRetroX-Tight-Pur containing the SOX2 ATF, and pRetroX-Tet-On-Advanced was first co-transfected with a plasmid (pMDG.1) expressing the vesicular stomatitis virus envelope protein into 293TGagPol cells to produce retroviral particles. Transfection was performed using Lipofectamine (Invitrogen) as recommended by the manufacturer. Recombinant lentiviruses were generated using a three-plasmid system as recommended by the manufacturer (Open Biosystems). The viral supernatant was used to infect the host cell lines (SUM159 and MCF7). pMX-IRES-GFP cells were collected 48 h after transduction for RNA, DNA, or protein preparation. pRetroX-Tight containing MCF7 cells were selected using 2 mg/ml geneticin (Gibco, Invitrogen) and 5 µg/ml puromycin (InvivoGen). Cells were induced with 100 µg/ml of doxycycline (DOX).
siRNA transfections
SUM159 ZF-97 DNMT3a-clone 12 breast cancer cells were transfected with Human UHRF1 (siGENOME MU-006977–01–0002) and Human UBB (siGENOME MU-013382–01–0002) siRNAs using DharmaFECT (Dharmacon) according to manufacturer’s protocol. siCONTROL Non-targeting siRNA no. 2 was used as a control. Cells were collected 72 h after transduction for RNA or protein preparation.
Quantitative real-time PCR
qRT-PCR was analyzed using the comparative ΔCt method (ABPrism software, Applied Biosystems) using GAPDH as an internal normalization control. Quantification is an average of at least 3 independent experiments and standard errors are indicated. Fold Maspin mRNA levels were expressed relative to control (cells transduced with an empty retroviral vector).
Western blotting and immunofluorescence
Transduced cells were lysed with RIPA Buffer (Sigma-Aldrich) for Maspin protein analysis and NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotechnology) was used for SOX2 protein analysis. Western blotting was performed using standard methods. Antibodies included: anti-Maspin (BD PharMingen) diluted 1:500; anti-Tubulin (Sigma-Aldrich) was diluted 1:5000; anti-UHRF1 (Abcam) diluted 1:1000; anti-SOX2 (Cell Signaling Technology) diluted 1:1000; and anti-H3 (Active Motif) diluted 1:10,000. For Immunofluorescence staining, transduced cells were grown on glass coverslips and stained according to standard methods. anti-HA (Covance) antibody was diluted 1:000 and detected with Alexafluor-594 (Invitrogen) goat anti-mouse diluted 1:500.
Bisulfite conversion, cloning and sequencing
Bisulfite modification of genomic DNA was performed using a procedure adapted from Grunau et al.,48 as described previously.49 Cloning and sequencing of PCR products were described previously.50
Anchorage-independent growth and cell viability assays
Anchorage-independent growth assays were performed using standard methods and evaluated visually for the presence of colonies that were greater than 2 mm in diameter. Cell viability was assessed by CellTiter Glo (Promega) according to the manufacturer’s protocol.
Demethylating treatment with 5-aza-2’-deoxycytidine
5-aza-2’-deoxycytidine (5-aza) (Sigma-Aldrich; St Louis, MO) was employed as a demethylating agent. ZF-97 DNMT3a clones 10 and 12 were propagated in freshly made growth medium containing 1 µM or 5 µM 5-aza for 3 d, with re-feeding of fresh media containing the drug every day. Cells were collected after treatment for RNA preparation.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. Lee M. Graves and William B. Coleman at the University of North Carolina School of Medicine for critical reading and scientific discussions of this manuscript. This work was supported by Postdoctoral Fellowship 116066-PF-08–166–01-GMC from the American Cancer Society awarded to AGR and from National Cancer Institute/National Institutes of Health grants 1R01CA125273, 3R01CA125273–03S1 and Department of Defense (DoD) awards W81XWH-10–1-0265 (to PB) and W81XWH-10–1-0266 (to BDS).
Glossary
Abbreviations:
- ATFs
artificial transcription factors
- DNMT3a
DNA methyltransferase 3a
- DBD
DNA-binding domain
- ZF
zinc finger
- ZFPs
6ZF proteins
- TF
transcription factor
- SKD
KRAB domain
- UHRF1
ubiquitin-like, containing PHD and RING finger domains 1
- PBS
phosphate-buffered saline
- 5-aza
5-aza-2’-deoxycytidine
- BSA
Bovine Serum Albumin
- NLS
nuclear localization signal
- HA
terminal hemagglutinin
- DOX
doxycycline
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/19507
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–69. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 6.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–17. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 8.Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–83. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 10.Campbell TN, Choy FY. RNA interference: past, present and future. Curr Issues Mol Biol. 2005;7:1–6. [PubMed] [Google Scholar]
- 11.Kim VN. RNA interference in functional genomics and medicine. J Korean Med Sci. 2003;18:309–18. doi: 10.3346/jkms.2003.18.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blancafort P, Segal DJ, Barbas CF., 3rd Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361–71. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 13.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 14.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–3. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 15.Choo Y, Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci U S A. 1994;91:11168–72. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamieson AC, Kim SH, Wells JA. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–95. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 17.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci U S A. 1999;96:2758–63. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreier B, Segal DJ, Barbas CF., 3rd Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 19.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–78. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 20.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, et al. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–97. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 21.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–33. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–48. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 23.Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci U S A. 2003;100:11997–2002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol. 2001;19:656–60. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domann FE, Rice JC, Hendrix MJ, Futscher BW. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer. 2000;85:805–10. doi: 10.1002/(SICI)1097-0215(20000315)85:6<805::AID-IJC12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–9. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 27.Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, et al. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. J Neurosci Res. 2008;86:1680–93. doi: 10.1002/jnr.21635. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M, Shi Y, Magit D, Furth PA, Sager R. Reduced mammary tumor progression in WAP-TAg/WAP-maspin bitransgenic mice. Oncogene. 2000;19:6053–8. doi: 10.1038/sj.onc.1204006. [DOI] [PubMed] [Google Scholar]
- 29.Shi HY, Liang R, Templeton NS, Zhang M. Inhibition of breast tumor progression by systemic delivery of the maspin gene in a syngeneic tumor model. Mol Ther. 2002;5:755–61. doi: 10.1006/mthe.2002.0602. [DOI] [PubMed] [Google Scholar]
- 30.Cher ML, Biliran HR, Jr., Bhagat S, Meng Y, Che M, Lockett J, et al. Maspin expression inhibits osteolysis, tumor growth, and angiogenesis in a model of prostate cancer bone metastasis. Proc Natl Acad Sci U S A. 2003;100:7847–52. doi: 10.1073/pnas.1331360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe M, Nasu Y, Kashiwakura Y, Kusumi N, Tamayose K, Nagai A, et al. Adeno-associated virus 2-mediated intratumoral prostate cancer gene therapy: long-term maspin expression efficiently suppresses tumor growth. Hum Gene Ther. 2005;16:699–710. doi: 10.1089/hum.2005.16.699. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–78. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltran A, Parikh S, Liu Y, Cuevas BD, Johnson GL, Futscher BW, et al. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene. 2007;26:2791–8. doi: 10.1038/sj.onc.1210072. [DOI] [PubMed] [Google Scholar]
- 35.Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–12. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 37.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 38.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Müller F, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 41.Blancafort P, Beltran AS. Rational design, selection and specificity of artificial transcription factors (ATFs): the influence of chromatin in target gene regulation. Comb Chem High Throughput Screen. 2008;11:146–58. doi: 10.2174/138620708783744453. [DOI] [PubMed] [Google Scholar]
- 42.Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., 3rd Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci U S A. 1994;91:4509–13. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltran AS, Sun X, Lizardi PM, Blancafort P. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol Cancer Ther. 2008;7:1080–90. doi: 10.1158/1535-7163.MCT-07-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltran AS, Blancafort P. Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs) Epigenetics. 2011;6:224–35. doi: 10.4161/epi.6.2.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–9. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Constantinescu SN, Sun Y, Bogan JS, Hirsch D, Weinberg RA, et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem. 2000;280:20–8. doi: 10.1006/abio.2000.4478. [DOI] [PubMed] [Google Scholar]
- 48.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–5. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivenbark AG, Jones WD, Risher JD, Coleman WB. DNA methylation-dependent epigenetic regulation of gene expression in MCF-7 breast cancer cells. Epigenetics. 2006;1:32–44. doi: 10.4161/epi.1.1.2358. [DOI] [PubMed] [Google Scholar]
- 50.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.