Abstract
Parental effects are ubiquitous in nature and in many organisms play a particularly critical role in the transfer of symbionts across generations; however, their influence and relative importance in the marine environment has rarely been considered. Coral reefs are biologically diverse and productive marine ecosystems, whose success is framed by symbiosis between reef-building corals and unicellular dinoflagellates in the genus Symbiodinium. Many corals produce aposymbiotic larvae that are infected by Symbiodinium from the environment (horizontal transmission), which allows for the acquisition of new endosymbionts (different from their parents) each generation. In the remaining species, Symbiodinium are transmitted directly from parent to offspring via eggs (vertical transmission), a mechanism that perpetuates the relationship between some or all of the Symbiodinium diversity found in the parent through multiple generations. Here we examine vertical transmission in the Hawaiian coral Montipora capitata by comparing the Symbiodinium ITS2 sequence assemblages in parent colonies and the eggs they produce. Parental effects on sequence assemblages in eggs are explored in the context of the coral genotype, colony morphology, and the environment of parent colonies. Our results indicate that ITS2 sequence assemblages in eggs are generally similar to their parents, and patterns in parental assemblages are different, and reflect environmental conditions, but not colony morphology or coral genotype. We conclude that eggs released by parent colonies during mass spawning events are seeded with different ITS2 sequence assemblages, which encompass phylogenetic variability that may have profound implications for the development, settlement and survival of coral offspring.
Introduction
Parental effects are fundamentally important in biological systems. They occur when the phenotype of the offspring is affected by the phenotype or environment of the parents [1], [2]. These effects consequently influence the life history [3], competitive ability [4], evolutionary trajectories, speciation rates [5] and population dynamics [6], [7] of future generations of individuals. Parental effects can also mediate host pathogen relationships and facilitate the perpetuation of symbiosis between generations by influencing the direct transmission of symbiotic microorganisms from parent to progeny [8]. Although parental effects have been extensively studied in plants, insects and terrestrial vertebrates, these effects have received much less attention in the marine environment [1], [9].
In the ocean, a variety of algal and cyanobacterial symbionts live in association with protists and animal hosts in habitats ranging from the coastal sediments to the deep-sea hydrothermal vents [10]. Coral reefs are ecosystems whose ecological success is framed by endosymbiotic associations between scleractinian corals and unicellular dinoflagellates in the genus Symbiodinium [11]. Symbiodinium photosynthesis contributes to ecosystem productivity by translocating newly fixed carbon to its coral hosts, powering respiration and enhancing the deposition of calcium carbonate skeletons thereby creating a complex habitat for the extraordinary biodiversity characteristic of coral reefs [12], [13].
Symbiodinium is a taxonomically diverse genus comprising nine major lineages called clades A through I [14], that each contain from 2 - >100 subclade types [15]–[17]. Some coral species form highly specific associations with one or two closely related Symbiodinium types from one clade (e.g. Poritids, [16], [18]), while others form relationships with multiple Symbiodinium types that span the taxonomic breadth of the genus Symbiodinium (e.g. Pocilloporids, [18]). The taxonomic composition of Symbiodinium assemblages found at different locations on the same coral colony, within a single coral species sampled from different reefs, depths or different times of the year may also vary [15], [19], [20], illustrating a high degree of spatio-temporal variation in these endosymbiotic associations [21]–[23]. The broad taxonomic divisions in the genus Symbiodinium reflect in functional diversity, with distinct clades having different physiological optima [24]–[27]. Importantly, these differences in performance influence physiological characteristics of the coral host, rendering them more or less susceptible to environmental disturbances and disease (e.g. [15], [19], [22], [28]).
Given the fundamental role that Symbiodinium plays in the basic biology of corals, the perpetuation of this symbiosis is pivotal to the persistence of corals through time. Most coral species release asymbiotic eggs that are fertilized in the water column and which must acquire Symbiodinium from environmental pools (horizontal transmission) as planula-larvae and/or newly metamorphosed primary polyps [29], [30]. In other species, however, Symbiodinium are passed directly from the parent to the developing eggs (vertical transmission). These eggs are released (in mass or non-mass spawning episodes depending on the species), fertilized in the water column, and then develop into symbiotic larvae. For the species studied to date, approximately 25% of coral species that spawn gametes transmit Symbiodinium vertically through their eggs (n∼44 species) and 90% of corals that brood larvae (n∼36 species), release larvae containing Symbiodinium [31]. For example, the reefs of the Hawaiian Archipelago are dominated by coral species (from the genera Montipora, Porites and Pocillopora) that transmit Symbiodinium vertically [32]. In general, vertically transmitting species exhibit higher specificity in their endosymbiotic unions than horizontal transmitters [33]–[35]. Symbiodinium assemblages in coral offspring can be comprised of a single Symbiodinium type or a range of types, and as in adults, the taxonomic composition of these assemblages affects the growth and physiological tolerance of the juvenile coral [24], [25], [36].
No studies to date have examined the Symbiodinium assemblages in the eggs of a coral that transmits Symbiodinium vertically. For coral species that host diverse Symbiodinium assemblages, the transmission of Symbiodinium directly from parent to the egg potentially provides an opportunity for the parent to select the type(s) of Symbiodinium transmitted to the egg, and thus influence the physiological range, survival and recruitment success of their offspring. Further, differences in the endosymbiotic assemblages among adult colonies that reflect environmental conditions, life-history stage, health state or morphology may result in eggs seeded with very different Symbiodinium assemblages. Such parental effects have never been examined in corals but likely have implications for larval behaviors (swimming and habitat selection), natural selection after settlement, the potential for acclimatization to different environments and ultimately the resilience of coral populations and reef communities.
The timing of spawning in many corals is temporally constrained and predictable, making the collection and comparison of assemblages of Symbiodinium in newly spawned eggs tractable. We used this approach to examine the Symbiodinium ITS2 sequence assemblages (sensu [37]) transmitted from parents to eggs in the coral Montipora capitata, a simultaneous hermaphrodite that releases egg-sperm bundles during the new moon from late spring through summer in Hawai’i [38]. This coral is one of the most broadly distributed, morphologically plastic and important reef building corals in the main Hawaiian Islands [39], [40]. This combination of traits makes this species an excellent model for examining Symbiodinium transmission in the context of environment and colony morphology and genotype. Specifically, this study tests the hypotheses that (i) Symbiodinium ITS2 sequence assemblages between parent and eggs differ, and that (ii) differences in the ITS2 sequence assemblages in the eggs reflect differences in environmental conditions or morphology of the coral adult.
Studying vertical transmission of Symbiodinium can greatly enhance our understanding of how parental effects influence host-symbiont combinations in corals worldwide, which may play an important role in the physiological performance and stress tolerance of next generations and the future of coral reefs.
Methods
Ethics Statement
This study was conducted under the research guidelines of the University of Hawaii Executive Policy E5.211 and corals and gametes collected under the State of Hawaii Special Activity Permit numbers 2007-02, 2008-02 issued to the Hawaii Institute of Marine Biology.
Study Sites and Sample Collections
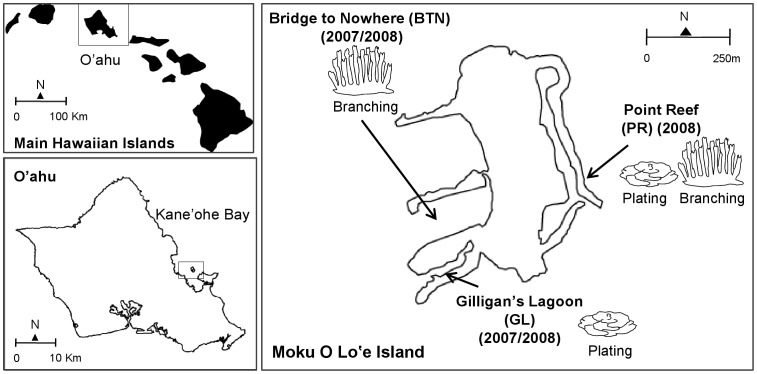
Parent colonies and gametes were sampled at three sites around Moku O Lo’e Island in Kane’ohe Bay, Hawai’i: Bridge to Nowhere (BTN), Gilligan’s Lagoon (GL), and Point Reef (PR) (Fig. 1) during the summers of 2007 and 2008. These sites are located at N 21° 25.893′ and W 157° 47.376′, N 21° 25.973′ and W 157° 47.392′, N 21° 25.988′ and W 157° 47.192′, respectively. Montipora capitata colonies exhibited primarily branching morphologies at BTN, plating at GL, and both branching or plating at PR (Fig. 2 a,b). The PR site was only sampled in 2008 and was included to examine whether Symbiodinium ITS2 sequence assemblages differed in corals sampled at a site where both branching and plating morphologies co-occur.
Figure 1. Location of study sites.
Parent colonies and gametes were sampled at three sites around Moku O Lo’e Island: Bridge to Nowhere (BTN), Gilligan’s Lagoon (GL), and Point Reef (PR). Moku O Lo’e Island is located in Kane’ohe Bay on the windward coast of the island of O’ahu, Hawai’i, USA. Montipora capitata colonies exhibited primarily branching morphologies at BTN, plating at GL, and both branching or plating morphologies at PR.
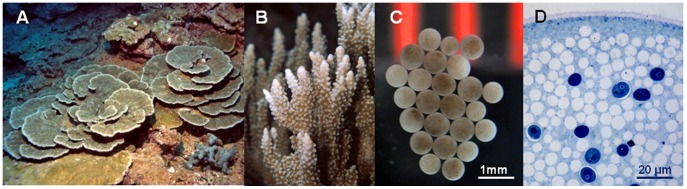
Figure 2. Montipora capitata colonies, their eggs and symbiotic Symbiodinium cells inside the eggs.
(a) plating and (b) branching morphologies, (c) eggs seeded with Symbiodinium cells acquired from parent colonies and (d) close up of Symbiodinium cells inside the egg, lighter circles are lipid droplets, darker circles are Symbiodinium cells.
Egg-sperm bundles were released (spawned) between 8∶45 and 9∶15 pm during the first quarter of the new moon in June, July and August of 2007 and 2008. Samples of parent colonies at depths of 1–2 m were collected 5 d before spawning nights by removing small tissue fragments 4 cm away from tips and edges of the colony. Egg-sperm bundles released by corals at the BTN and GL sites were collected in the field using cylindrical nets placed over the adult colonies [41]. Due to rough conditions at PR in 2008, ten fragments of adult colonies were moved from PR into tanks (∼2 weeks before spawning) exposed to ambient light levels and gametes were collected as in [42]. After collection, egg-sperm bundles were broken apart and eggs were rinsed using 0.2 µm filtered seawater to remove potential external contaminants. Eggs from a given parental colony (Fig. 2 c & d) were combined and isolated together. All coral fragments and eggs were immediately frozen after collection and stored at −80°C until processed.
To compare the thermal environments at the collection sites, temperature was measured at 10 min intervals for 1 yr in 2007 using StowAway Tidbits accurate to ±0.2°C (Onset Computer) at BTN and GL. Light and temperature levels were also measured at all sites during two 2-wk periods in 2008 (22 September – 1 October and 25 November – 5 December, 2008). Light measurements were taken at 10 min intervals using Odyssey Photosynthetic Irradiance Recording Systems (Odyssey). These measurements were made to compare the light levels among sites and the timing of these analyses constrained by the availability of instrumentation. Temperatures were also recorded at PR during the same two 2-wk periods in 2008 (22 September – 1 October and 25 November – 5 December, 2008) that light levels were recorded. Both, temperature and light measurements were performed at ∼1.5 m depth, at the coral collection sites.
Symbiodinium DNA Isolation, PCR, Cloning, Sequencing and Phylogenetic Analyses
Genomic DNAs from coral fragments and eggs were extracted using the guanidinium-based protocol described in [20]. The Symbiodinium ITS2 rDNA cistron was amplified (primers ‘ITS-DINO’ and ‘ITS2rev2’), cloned and sequenced following procedures detailed in [14]. DNA sequences were inspected and assembled using Sequencher v4.7 (Gene Codes Corporation, USA), identified via the Basic Local Alignment Search Tool (BLAST) in GenBank, and manually aligned with the BioEdit v5.0.9 sequence alignment software [43].
Sequences included in the downstream analyses met the following criteria: (1) they had either been published previously and the sequences retrieved and verified in multiple independent studies, or (2) were recovered in this study three or more times from clone libraries representing three or more independent coral samples. The remaining clone doubletons and singletons were excluded following [20]. Finally, ITS2 secondary structure folding was analyzed to identify potential pseudogenes as described in [37]. Two ITS2 alignments were then created for phylogenetic analyses, one for Symbiodinium clade C sequences and the other one for Symbiodinium clade D. Statistical parsimony haplotype networks were constructed using the software TCS version 1.21 [44] with a 95% connection limit and gaps were treated as a 5th state.
The Symbiodinium ITS2 sequence assemblages or ‘Symbiodinium ITS2 assemblages’ herein, refer to the total number and taxonomic nature of different ITS2 sequences recovered from each sample analyzed. These Symbiodinium ITS2 assemblages were used as a proxy for Symbiodinium communities and to explore how Symbiodinium sequences partition among individuals and life stages. Sequence assemblages were obtained from 26 adult colonies of Montipora capitata and the eggs produced by each of these colonies (n = 52). Six of these adult colonies and their eggs (Colony ID: 8, 10, 19, 23, 29 and 37) were analyzed over two reproductive seasons (summers 2007 and 2008) to examine interannual variability in the Symbiodinium ITS2 assemblages (n = 12).
Host DNA Isolation, PCR and Sequencing
A second subsample of the frozen adult corals was used to extract host DNA as described in [45]. The ATP synthetase subunit ß intron (atpsß) was amplified and sequenced as detailed in [37]. The resulting 300 bp sequences were edited and aligned using Sequencher v4.8 (Gene codes, Ann Arbor, MI). Gametic phases were determined computationally with Phase [46].
Statistical Analyses
Symbiodinium ITS2 sequence assemblages
Data from sites BTN and GL (2007 and 2008) and site PR (2008) were analyzed separately because colony morphologies did not co-occur at all sites. Environmental and temporal effects on sequence assemblages in both parent and eggs were analyzed using data from BTN and GL sites. Morphological effects were analyzed using data from PR where both morphologies co-occurred. BTN and GL data were analyzed according to a four-factor experimental design (site, life-stage, year and colony). PR data was analyzed using a three factor experimental design (morphology, life-stage and colony). Analyses of molecular variance (AMOVA, [47]) were used to test whether the composition of sequence assemblages differed between factors (environments, years, life stages and morphology) using the genetic distance between the sequences as described in [37]. Matrices of simple pairwise genetic distances were generated in Arlequin v3.11 [48], the square root of each distance was taken, and the matrices were imported to Permanova+ v1.0.2 software add on for Primer 6 [49]. Due to the inability to differentiate inter and intragenomic ITS2 variants and resulting issues associated with the interpretation of ITS2 ‘type diversity’ as a proxy of ‘species delimitations’ [37], [50], which preclude us from unequivocally estimating Symbiodinium ‘type diversity’, this study used the ITS2 marker for a comparative analysis of sequence assemblage variation among samples. The goal of our study was to examine patterns of Symbiodinium ITS2 sequence assemblages between parent and egg coral samples, not to distinguish whether or how these sequences relate to the number of Symbiodinium ‘species’ present within a given sample. Thus, although ITS2 sequences cannot represent the “true diversity” of Symbiodinium [37], the ITS2 sequence assemblage approach is useful in distinguishing differences in patterns of Symbiodinium diversity among samples. As demonstrated in Stat et al. [37], if there is a significant difference in the ITS2 sequence composition detected by the AMOVA, this implies that Symbiodinium ITS2 assemblages are partitioned, regardless of the actual number of Symbiodinium types represented.
AMOVA, using simple pairwise genetic distance among alleles, was used to test for differences in genetic composition of different life stages for each colony according to [47]. Φ ranges between 0 to 1, Φ = 0 indicates identical genetic composition between samples, and Φ = 1 indicates alternate fixation of alleles. The percent similarity index (PSI, [51]) was estimated between adults and eggs for each coral colony; zero values of PSI index correspond to no similarity in the ITS2 sequence assemblages between life stages.
Host genetics
AMOVAs performed in Arlequin v3.11 [48] were used to test whether host genetic variation was partitioned by morphology or collection site. Global exact tests of non-differentiation [52] were then performed (α = 0.05, Markov chain steps = 10,000) to verify the results from the AMOVA.
Environment
Means, standard deviations and ranges (minimum–maximum) were calculated for each site during the periods sampled. A Kruskal-Wallis test was used to evaluate the differences in temperature and light between sites.
Results
Symbiodinium ITS2 Sequences in Montipora Capitata
A total of 659 sequences were recovered from the 64 samples (32 adults and their respective eggs), representing 7–13 Symbiodinium ITS2 sequences per sample (10±1.87, average ± SE; Table 1, Fig. 3, GenBank accessions JF683321-JF683339). Our initial screen of sequences resolved 29 different ITS2 sequences that have either been published before, or were retrieved from multiple samples here. 24 of these sequences belonged to Symbiodinium clades C, and 5 to clade D (GenBank accession numbers in Table S1). Nine of the sequences matched previously published sequences (C3, C17, C21, C21.6, C21.11, C31, C31.1, D1, and D1a). The remaining 20 sequences were novel and were assigned names indicating the clade, the number of the most closely related published sequence type, and a decimal and a number to distinguish them from published types and one another [20].
Table 1. Symbiodinium ITS2 sequences in adults and eggs of Montipora capitata, and ATP synthetase subunit β genotypes for the adult corals sampled at three sites on Moku O Lo’e Island, Kane’ohe Bay, Hawai’i.
| Site | Year | Colony ID | Symbiodinium ITS2 sequence/s ADULTS | S | Symbiodinium ITS2 sequences/s EGG | S | PSI | Φ | p | G |
| BTN | 2007 | 1 | C315, C31.22, C212, C171 | 1 | C32.28, C311 | 1 | 0.50 | 0.807 | <0.001* | U |
| 8 | D14, D1a4, D1a.12 | 1 | D1a2, C312, C31.41, C21.21,C21.101, C17.21, C31 | 2 | 0.22 | 0.734 | <0.001* | H | ||
| 10 | D1a4, D13, D1a.12 | 1 | C32.16, C312, C17.21 | 1 | 0.00 | 0.934 | <0.001* | F | ||
| 11 | D1a7, D11, D1a.11 | 1 | D1a5, D1a.13, D12 | 1 | 0.72 | −0.039 | 0.670 | G | ||
| 12 | D19, D1a4 | 1 | C314, C17.22, D1a1,C211, C31.11, C31.41, C32.11 | 2 | 0.10 | 0.875 | <0.001* | I | ||
| 18 | D13, D1a3, D1a.12, D1a.21 | 1 | C319, C211, C17.21 | 1 | 0.00 | 0.987 | <0.001* | M | ||
| 19 | C313, C213, D1a3, D11,D1a.11 | 2 | C315, C21.112, D1a1,C17.21 | 2 | 0.38 | 0.157 | 0.072 | N | ||
| 2008 | 8 | D15, D1a4 | 1 | C213, C31.12, D1a1,C21.31, C21.81, C21.51 | 2 | 0.11 | 0.856 | <0.001* | H | |
| 10 | D15, D1a5, D1a.11 | 1 | D14, D1a.13, D1a2, C21.111 | 2 | 0.69 | 0.006 | 0.339 | F | ||
| 19 | C212, C21.22, C21.11, C21.41,C21.71, C21.91, C21.111, C311 | 1 | C213, C21.51, C21.61, C21.81,C21.111, C17.11, C32.21, D1a1 | 2 | 0.30 | 0.007 | 0.387 | N | ||
| GL | 2007 | 23 | C17.25, C313, C171, C21.111 C3.11 | 1 | C17.23, C313,C17.11, C211, C32.21 | 1 | 0.65 | −0.045 | 0.912 | T |
| 29 | C318, C211 | 1 | C315, C31.11, C31.21,C3.11 | 1 | 0.63 | −0.055 | 0.527 | D | ||
| 30 | C214, C313,C17.22, C21.112 | 1 | C313, C17.23, C171, C21.31, C31.11, C31.31 | 1 | 0.45 | 0.095 | 0.104 | W | ||
| 35 | D1a6, D13, D1.31,D1a.11 | 1 | D1a5, D14, D1.31 | 1 | 0.86 | −0.074 | 0.811 | E | ||
| 36 | C315, C212,C17.11, C31.41, D11 | 2 | C317, C32.12,C31.11 | 1 | 0.50 | 0.004 | 0.414 | P | ||
| 37 | C17.26, C21.12,C211, C21.101,C31.21 | 1 | C318, C17.21, C31.11 | 1 | 0.10 | 0.500 | <0.001* | E | ||
| 39 | C316, C31.12,C17.11, C211, C31.31 | 1 | C316, C31.22, C31.11, C31.31 C31 | 1 | 0.73 | −0.028 | 0.718 | G | ||
| 2008 | 23 | C17.12, C312,C211, C21.41, C21.81 | 1 | C217, C171, C21.91, C31.21 | 1 | 0.14 | 0.044 | 0.241 | T | |
| 29 | C217, C312 | 1 | C219, C311 | 1 | 0.88 | −0.057 | 0.578 | D | ||
| 37 | C215, C21.103, C21.21, C21.111,C311 | 1 | C313,C212, C21.82, C21.31, C21.51, C21.111 | 1 | 0.38 | 0.060 | 0.121 | E | ||
| PR | 2008 | B1 | C214, C21.62, C21.102, C21.112, C31.41 | 1 | C315, C212, C21.111, C3.11, D11, D1a.21 | 2 | 0.27 | 0.119 | 0.009 | P |
| B2 | C214, C314, C21.112, C17.21 | 1 | C17.24, C212, C21.41, C21.61,C21.71, C21.111, C311 | 1 | 0.45 | −0.011 | 0.403 | D | ||
| B5 | C213, C313, C21.72,C21.11, C21.111, C17.21 | 1 | C214, C313, C21.21, C21.111,C17.21, D11 | 2 | 0.73 | −0.018 | 0.944 | B | ||
| B6 | C21.93, C312, C21.42, C17.11, C211, C21.101 | 1 | C315, C17.23, C21.51, C171,C32.21 | 1 | 0.20 | 0.126 | 0.039 | B | ||
| B7 | C214, C313, C17.11, C17.21, C21.61, C21.111 | 1 | C313, C31.12, C31.22, D1.32, C21.111,C32.21 | 2 | 0.36 | 0.111 | 0.025 | B | ||
| B8 | C315, C21.82, C211, C21.11, C21.111, C31.11 | 1 | C216, C17.22, C312,C21.51, C21.91,C21.101 | 1 | 0.24 | 0.099 | 0.108 | D | ||
| P1 | C314, C21.12, C21.22, C17.11, C211, C21.111 | 1 | C214, C21.62, C312, C17.22,C21.51 | 1 | 0.27 | −0.023 | 0.515 | J | ||
| P2 | C313, C213, C21.113, C17.21, C21.41 | 1 | C313, C213, C17.21, C21.61, D1a1 | 2 | 0.64 | −0.003 | 0.430 | L | ||
| P5 | C315, C31, C211, C21.71, C21.101, C21.111, C31.21 | 1 | D1a.15, D14, D1a3, D1a.21 | 1 | 0.00 | 0.973 | <0.001* | G | ||
| P6 | C313, C213, C21.32, C21.112, C21.21 | 1 | C215, C313, C21.41, C21.111, C31 | 1 | 0.64 | −0.055 | 0.839 | B | ||
| P7 | D1a.17, D1a3, C211, C21.81 | 2 | D1a4, D1a.11, D11, C211, C21.81 | 2 | 0.54 | −0.085 | 0.641 | N | ||
| P9 | C216, C21.51, C21.81, C311, C17.21 | 1 | C214, C21.112, C21.21, C21.31,C21.81, C311, D1a.11 | 2 | 0.55 | −0.002 | 0.514 | A |
Site abbreviations: BTN – Bridge to Nowhere, GL – Gilligan’s Lagoon, PR – Point Reef. Column Headings: S - number of clades/sample, PSI – Percent similarity index (between life stages), Φ - genetic differentiation, G – adult genotype. Numerals superscripts indicate the number of times a specific Symbiodinium ITS2 sequence was retrieved.
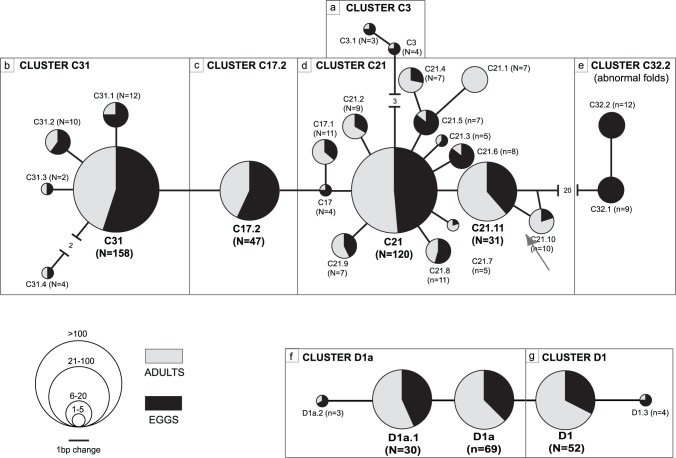
Figure 3. Symbiodinium sequence networks and folding clusters between parents and eggs.
Symbiodinium ITS2 sequences (N = 659 sequences) identified from 64 Montipora capitata coral samples (see Table 1), showing the relationships among the 24 distinct ITS2 sequences retrieved in Symbiodinium clade C, and 5 in Symbiodinium clade D. The pie charts correspond to individual Symbiodinium ITS2 sequences, with the diameter of the pie charts proportional to the number of sequences retrieved corresponding to the circular inset scale (exact numbers given in brackets). Grey and/or black colors correspond to sequences obtained from adult coral colonies and coral eggs, respectively. Networks are subdivided into cluster groupings that each contains sequences with identical secondary structure folding. Details on secondary structures are shown in the Figure S1.
The 29 ITS2 sequences grouped into seven secondary structural folds, 5 representing clade C sequences, and 2 clade D (Fig. 3, Fig. 4, Fig. S1). Less abundant sequences generally exhibited identical folding structures to the most closely related dominant sequence. Two sequences, C32.1 and C32.2, exhibited secondary structures that diverged significantly from the fold of their closest relative C21.11, and each resulted in abnormal folding conformation of helix IIIb (Fig. 3, Fig. S1). Based on these structural abnormalities, these sequences did not meet all our criteria for inclusion in the downstream statistical analyses and were excluded.
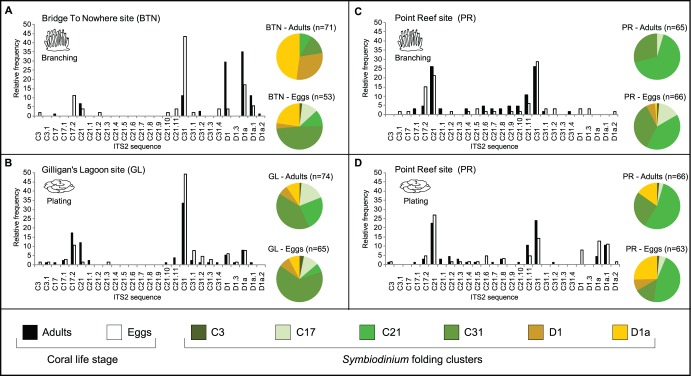
Figure 4. Abundance and distribution of Symbiodinium sequences and clusters between sites.
The frequency of Symbiodinium ITS2 sequences per site (adult and egg) is displayed as bar graphs. The pie charts represent the frequency of Symbiodinium based on six of the seven ITS2 secondary structures (folds a, b, c, d, f and g; see Figure S1); note that fold type e did not meet our criteria for inclusion and was omitted from the downstream analysis.
Patterns in Symbiodinium ITS2 Sequence Assemblages
Individual coral and egg samples harboured from two to seven co-occurring ITS2 sequences (Table 1). The ITS2 sequences C31, C21, D1a, D1, C17.2, and C21.11 were common in parents and eggs being recovered 159, 119, 69, 52, 44, and 31 times respectively. The less abundant sequences generally differed from a dominant sequence by one to three base pairs. All ITS2 sequences were identified at least once in both life stages except C21.1, which was only detected in adults (Table 1, Fig. 3, Fig. 4). Similarly, all ITS2 sequences were detected at least once in both morphologies, except C31.3, which was only found in adults and eggs of plating morphologies from GL site (Table 1, Fig. 4). Fourteen of the 29 ITS2 sequences were found at all three sites (Table 1), however ITS2 sequences were not distributed evenly among sites with some being more abundant at one or two sites than the others (Table 1, Fig. 4).
We examined how ITS2 assemblages partitioned among colonies, life stages, sites, and time (Table 1). Symbiodinium ITS2 assemblages differed between BTN and GL sites (p = 0.0199) and between the adult colonies and their eggs (p = 0.0202, Table 2). There was also a significant interaction among all factors investigated (p = 0.0001, Table 2a), suggesting that Symbiodinium ITS2 assemblages can differ at the colony level (and within life stages) depending on the year and site. Φ statistics were higher in colonies from BTN (p = 0.005) than both other sites, reflecting larger differences in the assemblages between life stages at this site.
Table 2. Results of AMOVA testing for differences in Symbiodinium ITS2 assemblages among sites, years and life stage (BTN, GL sites) (A) and between morphology and life stage (PR site) (B).
| Test | Source | df | MS | Φ | P (perm) | |
| A | Site, Years and Life stage | Si | 1 | 2041.9 | 0.315 | 0.0199 |
| Ye | 1 | 136.19 | −0.035 | 0.4783 | ||
| Li | 1 | 481.84 | 0.098 | 0.0202 | ||
| Co(Si) | 12 | 269.98 | 0.025 | 0.4674 | ||
| SixYe | 1 | 111.14 | −0.118 | 0.5283 | ||
| SixLi | 1 | 578.06 | 0.221 | 0.0108 | ||
| YexLi | 1 | 59.716 | 0.013 | 0.2624 | ||
| YexCo(Si) | 4 | 227.29 | 0.528 | 0.0331 | ||
| LixCo(Si) | 12 | 56.832 | 0.241 | 0.2677 | ||
| SixYexLi | 1 | 51.192 | 0.099 | 0.2581 | ||
| YexLixCo(Si) | 4 | 29.479 | 0.390 | 0.0001 | ||
| Res | 338 | 4.3134 | ||||
| Total | 377 | |||||
| B | Morphology andLife stage | Mo | 1 | 307.67 | 0.076 | 0.1944 |
| Li | 1 | 120.89 | 0.030 | 0.2185 | ||
| Co(Mo) | 10 | 147.9 | 0.274 | 0.1281 | ||
| MoxLi | 1 | 34.957 | −0.036 | 0.4386 | ||
| LixCo(Mo) | 10 | 58.791 | 0.453 | 0.0001 | ||
| Res | 236 | 5.9302 | ||||
| Total | 259 |
Significance was determined by permutation test (10,000 permutations) of the pseudo-F statistic. Significant values (p<0.05) are indicated with bold font. Factor abbreviations: Si - site, Li – life stage, Ye - year, Co - colony, Mo – morphology, Φ - genetic differentiation.
Pairwise comparisons of sites, grouping ITS2 sequences by life stage indicated a significant difference in the Symbiodinium ITS2 assemblages in adult corals from BTN and GL (p = 0.0093), but not in their eggs (p = 0.2566). Although adult corals from BTN had a higher abundance of clade D sequences (ITS2 sequences D1, D1a, D1a.1) than those from GL, ITS2 sequence C31 was the most abundant in eggs from both sites (Table 1, Fig. 4a,b). In addition, pairwise comparisons of life stage within sites indicated a significant difference in Symbiodinium ITS2 assemblages between adult corals and their eggs from the BTN (p = 0.0408), but not at GL (p = 0.4186), with BTN adults having a greater proportion of clade D sequences than their eggs (Table 2, Fig. 4a). Despite the fact that eggs did not differ in Symbiodinium sequence composition between the two sites, we found significant differences in assemblages in eggs released by different colonies within each site. This indicates that there are major differences in the Symbiodinium ITS2 assemblages transmitted to eggs that are simultaneously released by adjacent coral colonies during a mass spawning event. Pairwise comparisons of life stage by colony in 2007 revealed that adult and eggs had different ITS2 assemblages in four of the seven colonies sampled at BTN (#8, 10, 12, 18, p = 0.0005, p = 0.0057, p = 0.0003, p = 0.0001 respectively), and in one out of seven colonies at GL (#37, p = 0.0004). No significant differences in the Symbiodinium ITS2 assemblages between adults and eggs were found when comparing samples from the same colonies taken in 2007 and 2008 (p = 0.4783, Table 2a). Generally, differences in the ITS2 assemblages between life stages were observed in parental colonies harboring predominantly clade D; their eggs contained higher abundances of clade C (Table 1). Percent similarity indices (PSI) showed a large range of values within each site (Table 1), highlighting the variability in ITS2 assemblages between life stages for each colony. Montipora capitata colonies at the BTN were predominantly branching, while those at GL were plating. To test whether the site differences in assemblages between BTN and GL were due to morphology, we analyzed corals sampled in 2008 from a third site (PR) where the two morphologies co-occur. This analysis revealed no significant difference in assemblages between plating and branching morphologies at this site (p = 0.1944, Table 2b), nor between life stages (p = 0.2185, Table 2b). The only differences in assemblages were between life stages and morphology within individual colonies, p = 0.0001 (Table 2b), a pattern that is consistent with the interpretation that individual colonies at this site had different assemblages of Symbiodinium ITS2.
The six ITS2 secondary structure folds were found in corals from all three sites. There were however, differences in the relative abundance of ITS2 folds at the three sites, which reflected the differences in the assemblages detailed above. For example, folding clusters D1 and D1a were most abundant at BTN site, whereas folding cluster C21 and cluster C31 were more abundant at the PR and GL sites, respectively (Fig. 4).
Host Phylogenetic Analysis
A 280 bp fragment of atpsß was amplified from 48 adult colonies (20 from BTN, 15 from GL, and 13 from PR). The alignment identified 7 polymorphic sites and 17 distinct atpsß alleles among the 47 individuals. Using the Akaike information criterion (AIC) with a likelihood approach in Modeltest v3.06 [53], the best fit model of sequence evolution was HKY with base frequencies A = 0.3171, C = 0.1400, G = 0.1333, T = 0.4097 and a transition/transversion (Ti/Tv) value of 1.4826.
Branching and plating colonies of Montipora capitata shared common alleles supporting the hypothesis that the two morphologies are the same species. AMOVA results indicate that the majority of variance could be explained at the among individual level for groupings based on both morphology and collection site (Pvar = 49.09%; p<0.001 and Pvar = 52.80% p<0.001 respectively, Table 3). Variance at the highest hierarchical level was low and non-significant in both tests (p>0.05). Global exact tests of overall non-differentiation are significant indicating no partitioning based on differences between morphologies (p<0.0001) or collection site (p<0.0001). These results suggest a lack of genetic structuring due to either morphology or collection site at the scale examined in this study (Table 3).
Table 3. Results of AMOVA showing how genetic variance is partitioned for M. capitata when grouped according to morphology (A) and collection site (B) for the diploid nuclear locus atpsβ.
| Test | Source of Variation | df | SS | Variance Components | % of variation | Statistic | Value | |
| A | Morphology | Among morphologies | 1 | 13.339 | 0.16381 | 10.86 | Φ AM | 0.109 |
| Among samples within morphologies | 2 | 8.576 | 0.12318 | 8.16 | Φ AS(AM) | 0.092* | ||
| Among individuals within samples | 44 | 86.908 | 0.75322 | 49.92 | Φ AI(AS(AM))) | 0.616*** | ||
| Within individuals | 48 | 22.500 | 0.46875 | 31.06 | Φ WI | 0.689*** | ||
| Total | 95 | 131.323 | 1.50895 | |||||
| B | Collection Site | Among sites | 1 | 3.197 | −0.12626 | −8.96 | Φ ASI | −0.090 |
| Among samples within sites | 2 | 18.718 | 0.31282 | 22.21 | Φ AS(WSI) | 0.204** | ||
| Among individuals within samples | 44 | 86.908 | 0.75322 | 53.48 | Φ AI(WS) | 0.616*** | ||
| Within individuals | 48 | 22.500 | 0.46875 | 33.280 | Φ WI | 0.667*** | ||
| Total | 95 | 131.323 | 1.40853 |
P<0.05.
P<0.005.
P<<0.001; statistical probabilities derived from 1023 permutations.
Environmental Characteristics of Sites
Temperature differed at the three sites in both late summer and late autumn (H = 972.5, df = 2, p<0.0001, H = 69.2, df = 2, p<0.0001 respectively, Table S2). Temperature was higher and more variable at the BTN site, with up to ∼3°C fluctuations observed over a single 24-hr period (data not shown). Light levels differed significantly among the three collection sites (H = 57.9, df = 2, p<0.0001, H = 59.7, df = 2, p<0.0001, summer and autumn respectively). The BTN site had the broadest range of light levels in both summer and autumn sampling times (Table S2), and exhibited a recorded summer maximum of 1540 µmol quanta/m2s. The Montipora capitata colonies at this site were predominantly branching morphologies. The GL site had the lowest light levels where M. capitata were predominantly plating in morphology. Medium light levels were observed at the PR site where plating and branching morphologies of M. capitata co-occurred (Table S2). Overall, PR and GL sites had around 45% and 23% the light levels of BTN.
Discussion
Parental effects in corals with vertical transmission of Symbiodinium have the potential to play a significant role in the phenotype of propagules, perpetuation of specific combinations of host-symbiont genotypes and ultimately the interaction of larvae with the environment. Vertical transmission increases the likelihood that offspring are seeded with Symbiodinium genotypes that are optimized to interact with the host. This strategy thus reduces the risk of forming unsuccessful symbiotic unions that might occur when acquiring Symbiodinium from the environment and that could reduce the growth and fitness of the coral [54]. This study is the first to explore Symbiodinium ITS2 assemblages vertically transmitted from parent to eggs in corals. Our results indicate that Symbiodinium ITS2 assemblages in the eggs of Montipora capitata are strongly influenced by the composition of the endosymbionts of the parent colony, and that the Symbiodinium ITS2 assemblages in the parent colonies differ and reflect characteristics of their physical environment.
A variety of Symbiodinium sequences were identified in the M. capitata adults and eggs, representing clades C and D, Symbiodinium lineages known to have different physiological characteristics and environmental thresholds. In this study, adults and eggs associated with clade D Symbiodinium were located in more challenging environments. For example, clade D Symbiodinium were found in branching colonies located in areas with high light and variable thermal regimes. This distribution is consistent with previous studies that document broader environmental thresholds for corals that associate with Symbiodinium clade D [22], [55]–[57].
Previous work on Symbiodinium diversity of M. capitata in O’ahu (Hawai’i) described a highly specific symbiosis between M. capitata brown morph and Symbiodinium ITS2 C31, and between the shallow orange morph and Symbiodinium ITS2 D1a [32]. In many cases the distinction between brown and orange morphs is ambiguous so we used shallow colonies that mostly resembled the “orange” morphotype. Our results revealed the presence of a much wider range of sequence types than previously reported in the “orange” morph, including the dominant types C3, C21, C31, D1 and/or D1a, and suggest that the presence and abundance of Symbiodinium ITS2 types are not specific to colony color or morphology. Despite the phenotypic plasticity of M. capitata, no host genetic differentiation was detected between sites or morphologies. This illustrates the important role that environment may play in structuring Symbiodinium ITS2 assemblages. For example, branching morphologies in a high light environment (BTN site) had higher abundances of ITS2 sequences D1 and D1a, whereas branching morphologies in a lower light environment (GL) had higher abundances of C31 and C21. Likewise, plate morphologies had C21 and C31 as dominant ITS2 sequences at PR and GL sites, respectively. Interestingly, plate morphologies at the PR site (medium light levels) showed higher abundances of ITS2 sequences D1 and D1a than plate morphologies at the GL site, which suggests that plate colonies may be experiencing a more challenging environment than branching colonies (which have more self-shading) at the PR site. M. capitata therefore appears to combine two strategies for acclimatizing to environmental change via differences in the composition of their Symbiodinium ITS2 assemblages and through its extraordinary morphological plasticity. Future experimental studies, using reciprocal transplants, should be conducted in order to fully validate the role played by the environment and the acclimatization strategies of the host.
The Symbiodinium ITS2 assemblages isolated from eggs were generally similar to their respective parent colony, encompassing anywhere from 2–7 Symbiodinium different sequences. It is unknown however, whether the nature of the Symbiodinium patterns in the eggs is controlled by the host or reflects competition within the Symbiodinium community and/or a race to occupy the less populated eggs [58]. To date, interactions between vertically transmitted symbionts remains underexplored and perhaps underestimated [59]. For example, it is unknown if a Symbiodinium type present in low abundance in an egg can proliferate and become dominant in the adult colony under the right environmental conditions, or if there is a threshold in abundance required for a Symbiodinium type to be viable in adult colonies.
The Symbiodinium ITS2 sequences in eggs from parents dominated by clade C were very similar in taxonomic composition to their parents. However, differences were detected in eggs originating from 4 of 7 parent colonies sampled at the BTN site, an environment where the corals exhibited branching morphologies. These 4 parent colonies all harbored clade D Symbiodinium; however, the eggs they produced all contained ITS2 assemblages with clade C and D. This result indicates that parent colony may preferentially transfer clade C Symbiodinium to their eggs rather than clade D. In coral species that are often dominated by Symbiodinium clade C versus D, clade D is often described as opportunistic [15], [60]. Although clade D has been shown to positively influence environmental thresholds in corals, there are known fitness tradeoffs, and corals hosting clade D do not grow as well as con-specifics that host clade C [24], [33], [55]. The idea that corals can detect these differences in physiology and preferentially select those that will provide the greatest benefit to their offspring is provocative and worthy of further investigation. Alternative explanations for these results include differences reflecting 1) environmental contamination of the eggs from free-living Symbiodinium cells, 2) sampling bias due to a single snapshot sampling of each adult coral colony investigated here, or 3) acquisition of Symbiodinium from the gastrovascular cavity environment rather than parental tissues.
The first scenario (environmental contamination) is less likely since a recent study shows that sequence for free-living Symbiodinium in sediments and seawater (near our study site) do not overlap with any endosymbiotic sequences obtained in M. capitata or other corals at the same study site [61]. Although it is possible that eggs could have also been infected by Symbiodinium cells recently released by other spawning colonies, we think that contamination to the egg would be minimal, since eggs are released as an egg-sperm bundle (surrounded by bundle material) and are quickly transported to the surface [62]. Furthermore, it is unclear if spawn-released Symbiodinium have the capacity to infect the eggs in the water column since transfer of Symbiodinium from the parents to the eggs is mediated by follicle cells present in the adult [58].
The second scenario (sampling bias) is more likely and is based on the fact that the eggs examined were released from multiple polyps located across the colony; in contrast, adult samples were taken from a single location on the colony. M. capitata colonies are extremely plastic in their colony morphology, and this structural complexity creates microenvironments with very different light regimes, micro-spatial variations that could influence the distribution of Symbiodinium within colony [63]. Indeed, spatial patterning of Symbiodinium clades as a result of differences in irradiance has been reported in Montastraea sp. [21] and Acropora sp. [64], [65]. It is also noteworthy that M. capitata has tissues that penetrate deeply into a porous skeleton. M. capitata eggs develop deep into the skeleton [62] and as such, acquire Symbiodinium from adult tissues within the skeleton that represent different microenvironments to surface tissues [66]. These differences may drive micro-zonation of Symbiodinium within coral polyps that create different likelihoods of infection depending on the closeness of the symbionts to the egg. Thus, the Symbiodinium ITS assemblages in the egg could reflect a combination of both parental selection of Symbiodinium and/or a stochastic infection depending on the Symbiodinium diversity present near the fecund polyps. Although such micro-spatial patterns of Symbiodinium are not well understood, they may have important ramifications for the performance of these corals and are worthy of further investigation. For example, future analysis on vertical transmission should compare how Symbiodinium diversity in individual eggs relates to the diversity in the polyp tissue and in the case of perforate corals, in the tissue located within the skeleton, in and around locations where the eggs develop [62].
Finally, the third scenario to consider, is that eggs of M. capitata acquire Symbiodinium cells transiently present in the gastrovascular cavity that have recently been expelled/acquired but that are not endosymbiotic with the host colony. This scenario is plausible since eggs are infected by Symbiodinium cells ∼2 weeks before release [38], [67], when some of the eggs are located along the mesenterial filaments near the mouth (Padilla-Gamino, personal observation). If that were the case, eggs could exhibit differences in their capacities to acquire symbionts “horizontally” from the gastrovascular cavity, opportunities that would reflect their relative location with respect to the mouth. Having the capacity to obtain symbionts transiently available in the gastrovascular cavity could be an important strategy for the transmission of Symbiodinium diversity in “vertically” transmitting coral-algal symbioses and requires further exploration.
Our data show that M. capitata colonies simultaneously release eggs during spawning events that overall contain very different Symbiodinium sequence assemblages and could confer different physiological attributes to larvae and/or juvenile corals. For example, Little et al. [24] found that juvenile Acropora (same family as M. capitata) grow faster when infected with clade C than with clade D, regardless of whether clade C was the homologous or heterologous subclade type. Furthermore, Abrego et al. [25] showed that Acropora juveniles infected with Symbiodinium type C1 had enhanced physiological tolerance (measured by photosynthesis, respiration and fluorescence) over juveniles infected with clade D. Juveniles with Symbiodinium type C1 also had higher 14C photosynthate incorporation and increased carbon delivery to the host [26]. The production of a pool of eggs containing individuals with Symbiodinium assemblages that exhibit different physiological optima could potentially allow larvae to exploit a variety of habitats and survive a range of environmental conditions both in the water column and after settlement. As such, this characteristic may serve as an adaptive strategy to maximize reproductive success when the environments that offspring face, vary unpredictably. Settling in environments similar to the parent may be more advantageous for the offspring if the early-stage acclimatization capabilities are limited (i.e. inability to change Symbiodinium assemblages and/or acquire new Symbiodinium from the environment, “switching/shuffling” [15], inability to change host morphology).
This study showed for the first time the Symbiodinium sequence assemblages in coral eggs derived from a vertical transmission system. Our results demonstrate that eggs feature a much wider range of sequence types than previously considered and that environment may play a significant role in parental effects of a coral with vertical transmission. Thus, Symbiodinium diversity in the eggs can be a dynamic trait under parental influence. The diverse array of early-stage holobionts highlights the fundamental importance of multidimensional specificity and flexibility in vertical transmission, which have the potential to significantly influence the biology and ecology of host and the Symbiodinium [21], [68], the evolutionary processes (i.e. speciation rates) and the perpetuation and evolution of coral holobiont mutualisms [69]. Montipora capitata is a coral with high morphological plasticity that is able to host multiple Symbiodinium genotypes, and these genotypes differ in abundance depending on the environment and the colony. By releasing eggs with different Symbiodinium compositions, M. capitata populations maximize the chances of the early-stage holobionts to recruit and grow in microenvironments with very different environmental conditions and possibly reduce competition between the recruits. The diverse array of early-stage holobionts could enhance the resilience of future generations of M. capitata and may possibly increase the potential for adaptive responses to rapid environmental change [70].
Supporting Information
Symbiodinium ITS2 secondary structures. Distinct structure folds representing the 29 ITS2 sequences shown in Fig. 1 (schematized here on the upper left corner). Seven distinct fold clusters (a-g) were characterized based on criteria described in [37]. The seven secondary folding structures shown here correspond to the most dominant ITS2 sequence found in each cluster (i.e., C3, C31, C17.2, C21, C32.2, D1a, and D1). The location of mutations (insertions, deletions, or hemi-CBC changes) for each ITS2 sequence variant found in each cluster are indicated with a green arrow and corresponding variant number. Four sequence variants are not indicated here, because the observed mutations are found within the 5.8S rDNA (i.e., outside of ITS2 secondary structure). Furthermore, 1 out of 2 and 1 out of 3 observed mutations were also found within the 5.8S rDNA for sequences C21.4 and C21.1, respectively.
(TIF)
GenBank accession numbers for the Symbiodinium ITS2 sequences identified in the present study.
(DOCX)
Temperature (°C) and light (µmol quanta/m2s) data from the three study sites in Moku O Lo’e Island, Kaneohe Bay Hawai’i.
(DOCX)
Acknowledgments
Thanks to all the wonderful volunteers that helped collect samples during the spawning events, especially the 2007 Edwin W. Pauley Summer Program students and the 2008 Super Spawning Team (S. Wagenhauser, C. Portocarrero, M. Sales, A. Cozo & R. Gabriel). Thanks to K. Stender for coral pictures and J.C. Ortiz, R. Kinzie, D. Karl, C. Smith and anonymous reviewers for their helpful comments. This is HIMB contribution number 1496, SOEST contribution number 8671 and 2007 Pauley Summer Program Contribution number 7.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: LPG was supported by the Mexican National Council for Science and Technology (CONACyT), the World Bank Coral Reef Targeted Research Project (www.gefcoral.org) and the Center for Microbial Oceanography Research and Education (C-MORE). The research was funded by the National Science Foundation (OCE-0752604 to RDG and OIA-0554657 administered by the University of Hawai'i), the Swiss National Science Foundation (PGBEA-115118 to XP), and the Edwin Pauley Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mousseau TA, Fox CW. The adaptive significance of maternal effects. . Trends in Ecology & Evolution. 1998;13:403–407. doi: 10.1016/s0169-5347(98)01472-4. Available:<Go to ISI>://000075997500009. [DOI] [PubMed] [Google Scholar]
- 2.Badyaev AV, Uller T. Parental effects in ecology and evolution: mechanisms, processes and implications. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:1169–1177. doi: 10.1098/rstb.2008.0302. Available:<Go to ISI>//000264661500012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donelson JM, Munday PL, McCormick MI. Parental effects on offspring life histories: when are they important? Biology Letters. 2009;5:262–265. doi: 10.1098/rsbl.2008.0642. Available:<Go to ISI>//000264371900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wulff RD. Seed size variation in Desmodium paniculatum. 2. Effects on seedling growth and physiological performance. Journal of Ecology. 1986;74:99–114. Available:<Go to ISI>//A1986A813800007. [Google Scholar]
- 5.Wade MJ. The evolutionary genetics of maternal effects. In: Mousseau TA, Fox CW, editors. Maternal effects as adaptations. Oxford, UK: Oxford University Press; 1998. pp. 5–21. [Google Scholar]
- 6.Ginzburg LR. Inertial growth: Population dynamics based on maternal effects. In: Mousseau TA, Fox CW, editors. Maternal Effects as Adaptations. Oxford, U.K.: Oxford University Press; 1998. pp. 42–53. [Google Scholar]
- 7.Donohue K. Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:1059–1074. doi: 10.1098/rstb.2008.0291. Available:<Go to ISI>//000264661500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proceedings of the Royal Society B-Biological Sciences. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. Available:<Go to ISI>//000235238100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marshall DJ, Allen RM, Crean AJ. The ecological and evolutionary importance of maternal effects in the sea. Oceanography and Marine Biology: An Annual Review. 2008;46:203–250. Available:<Go to ISI>//000256878700005. [Google Scholar]
- 10.Smith DC, Douglas AE. First. London: Edward Arnold; 1987. The Biology of Symbiosis. [Google Scholar]
- 11.Muscatine L, McCloskey LR, Marian RE. Estimating the Daily Contribution of Carbon from Zooxanthellae to Coral Animal Respiration. Limnology and Oceanography. 1981;26:601–611. Available http://www.jstor.org/stable/2836027. [Google Scholar]
- 12.Muscatine L, Falkowski PG, Porter JW, Dubinsky Z. Fate of Photosynthetic Fixed Carbon in Light-Adapted and Shade-Adapted Colonies of the Symbiotic Coral Stylophora pistillata. Proceedings of the Royal Society of London Series B-Biological Sciences. 1984;222:181–202. Available:<Go to ISI>//A1984TF60200005. [Google Scholar]
- 13.Allemand D, Ferrier-Pages C, Furla P, Houlbreque F, Puverel S. Biomineralisation in reef-building corals: from molecular mechanisms to environmental control. Comptes Rendus Palevol. 2004;3:453–467. Available:<Go to ISI>//000225802900003. [Google Scholar]
- 14.Pochon X, Gates RD. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Molecular Phylogenetics and Evolution. 2010;56:492–497. doi: 10.1016/j.ympev.2010.03.040. Available:<Go to ISI>//000278589500047. [DOI] [PubMed] [Google Scholar]
- 15.Baker AC. Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annual Review of Ecology Evolution and Systematics. 2003;34:661–689. Available:<Go to ISI>//000220102000024. [Google Scholar]
- 16.LaJeunesse TC. “Species” radiations of symbiotic Dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition (vol 22, pg 570, 2005). Molecular Biology and Evolution. 2005;22:1158. doi: 10.1093/molbev/msi042. Available:<Go to ISI>//000228139400061. [DOI] [PubMed] [Google Scholar]
- 17.Pochon X, Montoya-Burgos JI, Stadelmann B, Pawlowski J. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Molecular Phylogenetics and Evolution. 2006;38:20–30. doi: 10.1016/j.ympev.2005.04.028. Available:<Go to ISI>//000234461700003. [DOI] [PubMed] [Google Scholar]
- 18.Van Oppen MJH, Gates RD. Conservation genetics and the resilience of reef-building corals. Molecular Ecology. 2006;15:3863–3883. doi: 10.1111/j.1365-294X.2006.03026.x. Available:<Go to ISI>//000241388800001. [DOI] [PubMed] [Google Scholar]
- 19.Coffroth MA, Santos SR. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist. 2005;156:19–34. doi: 10.1016/j.protis.2005.02.004. Available:<Go to ISI>//000230105000012. [DOI] [PubMed] [Google Scholar]
- 20.Stat M, Pochon X, Cowie ROM, Gates RD. Specificity in communities of Symbiodinium in corals from Johnston Atoll. Marine Ecology-Progress Series. 2009;386:83–96. Available:<Go to ISI>//000268552500007. [Google Scholar]
- 21.Rowan R, Knowlton N. Intraspecific Diversity and Ecological Zonation in Coral Algal Symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2850–2853. doi: 10.1073/pnas.92.7.2850. Available:<Go to ISI>//A1995QP88900090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker AC, Starger CJ, McClanahan TR, Glynn PW. Corals’ adaptive response to climate change. Nature. 2004;430:741. doi: 10.1038/430741a. Available:<Go to ISI>//000223233600030. [DOI] [PubMed] [Google Scholar]
- 23.Iglesias-Prieto R, Beltran VH, LaJeunesse TC, Reyes-Bonilla H, Thome PE. Different algal symbionts explain the vertical distribution of dominant reef corals in the eastern Pacific. Proceedings of the Royal Society of London Series B-Biological Sciences. 2004;271:1757–1763. doi: 10.1098/rspb.2004.2757. Available:<Go to ISI>//000223338600015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little AF, van Oppen MJH, Willis BL. Flexibility in algal endosymbioses shapes growth in reef corals. Science. 2004;304:1492–1494. doi: 10.1126/science.1095733. Available:<Go to ISI>//000221795800041. [DOI] [PubMed] [Google Scholar]
- 25.Abrego D, Ulstrup KE, Willis BL, van Oppen MJH. Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. . Proceedings of the Royal Society B-Biological Sciences. 2008;275:2273–2282. doi: 10.1098/rspb.2008.0180. Available:<Go to ISI>//000258440500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantin NE, van Oppen MJH, Willis BL, Mieog JC, Negri AP. Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs. Available:<Go to ISI>//000265832100010 2009;28:405–414. [Google Scholar]
- 27.Rowan R. Coral bleaching - Thermal adaptation in reef coral symbionts. Nature . 2004;430:742. doi: 10.1038/430742a. Available:<Go to ISI>//000223233600031. [DOI] [PubMed] [Google Scholar]
- 28.Stat M, Morris E, Gates RD. Functional diversity in coral-dinoflagellate symbiosis. . Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9256–9261. doi: 10.1073/pnas.0801328105. Available:<Go to ISI>//000257645400024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Oppen M. In vitro establishment of symbiosis in Acropora millepora planulae. 000172560800002Coral Reefs. 2001;20:200. Available:<Go to ISI>://WOS000172560800002. [Google Scholar]
- 30.Hirose M, Yamamoto H, Nonaka M. Metamorphosis and acquisition of symbiotic algae in planula larvae and primary polyps of Acropora spp. . Coral Reefs. 2008;27:247–254. Available:<Go to ISI>//000255191100001. [Google Scholar]
- 31.Baird AH, Guest JR, Willis BL. Systematic and Biogeographical Patterns in the Reproductive Biology of Scleractinian Corals. Annual Review of Ecology Evolution and Systematics. 2009;40:551–571. Available:<Go to ISI>//000272455700026. [Google Scholar]
- 32.LaJeunesse TC, Thornhill DJ, Cox EF, Stanton FG, Fitt WK. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs. 2004;23:596–603. Available:<Go to ISI>//000226095100021. [Google Scholar]
- 33.Barneah O, Weis VM, Perez S, Benayahu Y. Diversity of dinoflagellate symbionts in Red Sea soft corals: mode of symbiont acquisition matters. Marine Ecology-Progress Series. 2004;275:89–95. Available:<Go to ISI>//000223426100009. [Google Scholar]
- 34.Thornhill DJ, Fitt WK, Schmidt GW. Highly stable symbioses among western Atlantic brooding corals. Coral Reefs. 2006;25:515–519. Available:<Go to ISI>//000242058400003. [Google Scholar]
- 35.Stat M, Loh WKW, Hoegh-Guldberg O, Carter DA. Symbiont acquisition strategy drives host-symbiont associations in the southern Great Barrier Reef. . Coral Reefs. 2008;27:763–772. Available:<Go to ISI>//000260616400006. [Google Scholar]
- 36.Abrego D, Van Oppen MJH, Willis BL. Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Molecular Ecology. 2009;18:3532–3543. doi: 10.1111/j.1365-294X.2009.04276.x. Available:<Go to ISI>//000268760100017. [DOI] [PubMed] [Google Scholar]
- 37.Stat M, Bird CE, Pochon X, Chasqui L, Chauka LJ. Variation in Symbiodinium ITS2 Sequence Assemblages among Coral Colonies. Plos ONE. 2011;6 doi: 10.1371/journal.pone.0015854. Available:<Go to ISI>//000286511200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter CL. Environmental cues controlling spawning in two hawaiian corals, Montipora verrucosa and M. dilatata Proceedings of the 6th International Coral Reef Symposium. 1988;2:727–732. Available:electronic. [Google Scholar]
- 39.Jokiel PH, Hildemann WH, Bigger CH. Clonal population structure of two sympatric species of the reef coral Montipora. Bulletin of Marine Science. 1983;33:181–187. [Google Scholar]
- 40.Jokiel PH, Brown EK. Global warming, regional trends and inshore environmental conditions influence coral bleaching in Hawaii. Global Change Biology. 2004;10:1627–1641. [Google Scholar]
- 41.Padilla-Gamino JL, Gates RD. Spawning dynamics in the Hawaiian reef building coral Montipora capitata. Marine Ecology Progess Series. 2012;449:145–160. [Google Scholar]
- 42.Cox EF. Continuation of sexual reproduction in Montipora capitata following bleaching. Coral Reefs. 2007;26:721–724. Available:<Go to ISI>//000248832400038. [Google Scholar]
- 43.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symposium Series. 1999;41:95–98. [Google Scholar]
- 44.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. Available:<Go to ISI>//000089998600020. [DOI] [PubMed] [Google Scholar]
- 45.Concepcion GT, Medina M, Toonen RJ. Noncoding mitochondrial loci for corals. . Molecular Ecology Notes. 2006;6:1208–1211. Available:<Go to ISI>//000242193300077. [Google Scholar]
- 46.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. American Journal of Human Genetics. 2003;73:1162–1169. doi: 10.1086/379378. Available:<Go to ISI>//000186493400017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Excoffier L, Smouse PE, Quattro JM. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes - Application to Human Mitochondrial-DNA Restriction Data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. Available:<Go to ISI>//A1992HW75900021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics. 2005;1:47–50. Available:<Go to ISI>//000207065900004. [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke KR, Warwick RM. Change in marine communities: an approach to statistical analysis and interpretation. 2001.
- 50.Correa AMS, Baker AC. Understanding diversity in coral-algal symbiosis: a cluster-based approach to interpreting fine-scale genetic variation in the genus Symbiodinium. Coral Reefs. 2009;28:81–93. Available:<Go to ISI>//000263071800011. [Google Scholar]
- 51.Wolda H. Similarity indices, sample size and diversity. Oecologia. 1981;50:296–302. doi: 10.1007/BF00344966. [DOI] [PubMed] [Google Scholar]
- 52.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. Available:<Go to ISI>//A1995TR69500027. [DOI] [PubMed] [Google Scholar]
- 53.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. Available:<Go to ISI>//000077489900010. [DOI] [PubMed] [Google Scholar]
- 54.Weis VM, Reynolds WS, deBoer MD, Krupp DA. Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbodinium spp and planula larvae of the scleractinian coral Fungia scutaria. Coral Reefs. 2001;20:301–308. Available:<Go to ISI>//000172560800017. [Google Scholar]
- 55.Berkelmans R, van Oppen MJH. The role of zooxanthellae in the thermal tolerance of corals: a “nugget of hope” for coral reefs in an era of climate change. Proceedings of the Royal Society B-Biological Sciences. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. Available:<Go to ISI>//000240400800007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garren M, Walsh SM, Caccone A, Knowlton N. Patterns of association between Symbiodinium and members of the Montastraea annularis species complex on spatial scales ranging from within colonies to between geographic regions. Coral Reefs. 2006;25:503–512. Available:<Go to ISI>//000242058400001. [Google Scholar]
- 57.Oliver TA, Palumbi SR. Distributions of stress-resistant coral symbionts match environmental patterns at local but not regional scales. Marine Ecology-Progress Series. 2009;378:93–103. Available:<Go to ISI>//000264955300009. [Google Scholar]
- 58.Hirose M, Kinzie RA, Hidaka M. Timing and process of entry of zooxanthellae into oocytes of hermatypic corals. Coral Reefs. 2001;20:273–280. Available:<Go to ISI>//000172560800013. [Google Scholar]
- 59.Vautrin E, Vavre F. Interactions between vertically transmitted symbionts: cooperation or conflict? Trends in Microbiology. 2009;17:95–99. doi: 10.1016/j.tim.2008.12.002. Available:<Go to ISI>//000264681800002. [DOI] [PubMed] [Google Scholar]
- 60.Stat M, Gates RD. Clade D Symbiodinium in Scleractinian Corals: A “Nugget” of Hope, a Selfish Opportunist, an Ominous Sign, or All of the Above? . Journal of Marine Biology. 2011. doi:10.1155/2011/730715.
- 61.Pochon X, Stat M, Takabayashi M, Chasqui L, Chauka LJ. Comparison of endosymbiotic and free-living Symbiodinium (Dinophyceae) diversity in a Hawaiian reef environment. Journal of Phycology. 2010;46:53–65. Available:<Go to ISI>//000273822800006. [Google Scholar]
- 62.Padilla-Gamino JL, Weatherby T, Waller RG, Gates RD. Formation and structural organization of the egg-sperm bundle of the scleractinian coral Montipora capitata. Coral Reefs. 2011;30:371–380. doi: 10.1007/s00338–010–0700–8. [Google Scholar]
- 63.Kaniewska P, Anthony KRN, Hoegh-Guldberg O. Variation in colony geometry modulates internal light levels in branching corals, Acropora humilis and Stylophora pistillata. Marine Biology. 2008;155:649–660. Available:<Go to ISI>//000260184400009. [Google Scholar]
- 64.Van Oppen MJH, Palstra FP, Piquet AMT, Miller DJ. Patterns of coral-dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host-symbiont selectivity. Proceedings of the Royal Society of London Series B-Biological Sciences. 2001;268:1759–1767. doi: 10.1098/rspb.2001.1733. Available:<Go to ISI>//000170834600002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulstrup KE, Van Oppen MJH. Geographic and habitat partitioning of genetically distinct zooxanthellae (Symbiodinium) in Acropora corals on the Great Barrier Reef. Molecular Ecology. 2003;12:3477–3484. doi: 10.1046/j.1365-294x.2003.01988.x. Available:<Go to ISI>//000186648400025. [DOI] [PubMed] [Google Scholar]
- 66.Santos SR, Toyoshima J, Kinzie RA. Spatial and temporal dynamics of symbiotic dinoflagellates (Symbiodinium: Dinophyta) in the perforate coral Montipora capitata. Galaxea. 2009;11:139–147. [Google Scholar]
- 67.Heyward AJ. Sexual reproduction in five species of the coral Montipora. Coral reef population biology. 1986. pp. 170–178. Available:electronic.
- 68.Herre EA, Knowlton N, Mueller UG, Rehner SA. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends in Ecology & Evolution. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. Available:<Go to ISI>//000079417200005. [DOI] [PubMed] [Google Scholar]
- 69.Thompson JN. Chicago, Illinois, USA: University of Chicago Press; 1994. The coevolutionary process. [Google Scholar]
- 70.Csaszar NBM, Ralph PJ, Frankham R, Berkelmans R, van Oppen MJH. Estimating the Potential for Adaptation of Corals to Climate Warming. Plos ONE. 2010;5 doi: 10.1371/journal.pone.0009751. Available:<Go to ISI>//000276456300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Symbiodinium ITS2 secondary structures. Distinct structure folds representing the 29 ITS2 sequences shown in Fig. 1 (schematized here on the upper left corner). Seven distinct fold clusters (a-g) were characterized based on criteria described in [37]. The seven secondary folding structures shown here correspond to the most dominant ITS2 sequence found in each cluster (i.e., C3, C31, C17.2, C21, C32.2, D1a, and D1). The location of mutations (insertions, deletions, or hemi-CBC changes) for each ITS2 sequence variant found in each cluster are indicated with a green arrow and corresponding variant number. Four sequence variants are not indicated here, because the observed mutations are found within the 5.8S rDNA (i.e., outside of ITS2 secondary structure). Furthermore, 1 out of 2 and 1 out of 3 observed mutations were also found within the 5.8S rDNA for sequences C21.4 and C21.1, respectively.
(TIF)
GenBank accession numbers for the Symbiodinium ITS2 sequences identified in the present study.
(DOCX)
Temperature (°C) and light (µmol quanta/m2s) data from the three study sites in Moku O Lo’e Island, Kaneohe Bay Hawai’i.
(DOCX)