Abstract
Molecular dynamics ensures that proteins and other factors reach their site of action in a timely and efficient manner. This is essential to the formation of molecular complexes, as they require an ever-changing framework of specific interactions to facilitate a model of self-assembly. Therefore, the absence or reduced availability of any key component would significantly impair complex formation and disrupt all downstream molecular networks. Recently, we identified a regulatory mechanism that modulates protein mobility through the inducible expression of a novel family of long noncoding RNA. In response to diverse environmental stimuli, the nucleolar detention pathway (NoDP) captures and immobilizes essential cellular factors within the nucleolus away from their effector molecules. The vast array of putative NoDP targets, including DNA (cytosine-5)-methyltransferase 1 (DNMT1) and the delta catalytic subunit of DNA polymerase (POLD1), suggests that this may be a common and significant regulatory mechanism. Here, we discuss the implications of this new posttranslational strategy for regulating molecular networks.
Keywords: acidosis, heat shock, mobility, noncoding RNA, posttranslational regulation, protein dynamics, ribosomal intergenic spacer
The field of molecular dynamics was born nearly 40 years ago through the study of lateral mobility within a two-dimensional membrane. Early work focused on the analysis of cell surface particles and utilized fluorescent dyes to monitor mobility during recovery after photobleaching.1-5 Advances in time-lapse imaging technology and the cloning of green fluorescent protein6 has allowed scientists to pass through the barrier of the cell membrane, with minimal invasiveness and map the dynamic properties of intracellular particles. Historically, most have assumed that some level of mobility was necessary for molecules to carry out their cellular role, i.e., DNA polymerase must traverse along the genome to facilitate DNA replication (Fig. 1A). However, the question of how proteins and RNA are present in the right place at the right time is not well understood.
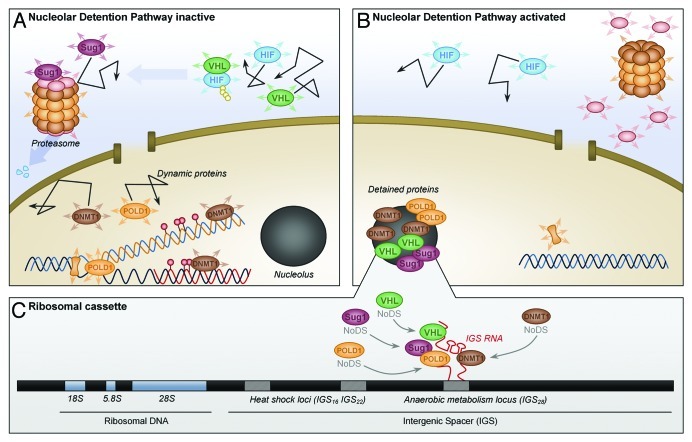
Figure 1. Regulation of molecular networks by the nucleolar detention pathway. (A) Under normal growth conditions cellular proteins are highly mobile and capable of executing essential cellular functions such as: ubiquitination (VHL), proteasomal degradation (SUG1), DNA replication (POLD1) and methylation (DNMT1); (B) Activation of the nucleolar detention pathway immobilizes proteins in the nucleolus away from their downstream effectors inhibiting basic cellular functions; (C) Capture and immobilization of NoDS-containing proteins in the nucleolus is mediated by inducible noncoding RNAs that originate from stimulus-specific loci within the ribosomal intergenic spacer.
In both the cytoplasm7-9 and nucleus,10-12 biologically active molecules diffuse throughout their cellular compartments in a random, rapid and energy-independent manner.11,13-16 Proper function of these factors requires the formation of complexes with other protein, RNA and/ or DNA molecules. This is believed to be accomplished through a stop-and-go scanning mechanism, whereby highly mobile particles randomly associate and dissociate from other molecules until transient, high-affinity and appropriate interactions can be found.13,14 Therefore, it appears that the highly chaotic and dynamic environment within the cell is, ironically, indispensible to generating order and maintaining proper cellular function.
Protein Dynamics as a Site of Posttranslational Regulation
This necessity for functional mobility presents the cell with an interesting opportunity to provide another layer of posttranslational control. To date the focus of posttranslational regulation has been on protein modifications through the addition of chemical/peptide groups or alterations in conformation and stability.17 Currently, hundreds of phosphatases, kinases, proteases and other modifying enzymes have been identified to fine-tune molecular networks by shifting the affinity of proteins toward one binding partner or another. However, many of these alterations are unable to affect global changes on multiple molecular networks in response to significant environmental stimuli. Altering protein mobility could provide a more systemic, rapid and reversible approach to regulating vast cellular pathways.
The study of regulated protein dynamics has been limited to a handful of molecules. Analysis of the lamin B receptor and the yeast protein Septin has shown that the same molecule can possess radically divergent kinetic properties, depending on its cellular localization18 or the stage of the cell cycle.19 Nucleostemin, a regulator of cancer/stem cell proliferation, further demonstrated that mobility can be affected by the GTP binding state of a molecule.20 The most dramatic display of altered protein dynamics was observed for the E3 ubiquitin ligases VHL and MDM2.21 Under normal physiological conditions, these highly dynamic molecules are diffused throughout the cytoplasm or nucleus, allowing them to locate their downstream effectors and target them for proteasomal degradation22,23 (Fig. 1A). However, in response to diverse stimuli, such as acidosis, heat shock and transcriptional stress, a novel class of inducible long noncoding RNA expressed from distinct loci within the ribosomal intergenic spacer (IGS RNA) has been shown to capture and immobilize these molecules within the nucleolus,21,24 away from their targets, rendering them functionally inert14 (Fig. 1B and C).
Subcellular Targeting vs. Subcellular Detention
Numerous factors have been shown to affect the subcellular distribution of proteins.7,25-31 Those studying these phenomena have used ambiguous terms such as targeting, recruitment and sequestration, to denote changes in the subcellular localization of molecules in response to environmental and cellular stimuli. While on the surface, the nucleolar detention pathway (NoDP) appears to emulate these other forms of subcellular redistribution, the term nucleolar “detention” has been specifically chosen to convey two fundamental distinctions unique to this form of localization.
First, live cell photobleaching analysis has demonstrated that proteins detained by the NoDP are both localized and statically immobilized within the nucleolus.21,24 In contrast, the more vague terms: targeting, recruitment and sequestration, generally overlook the concept of mobility and primarily focus on the assessment of steady-state localization by immunofluorescence microscopy of fixed cells. While the prevalent historical belief was that “targeted/recruited/sequestered” molecules were not dynamic and remained associated with their specific subcellular domains, recent studies have shown that most subcellular compartments are fluid structures composed of proteins that are rapidly entering and exiting the region.7,11,32 Second, the purpose of nucleolar detention appears to differ from other forms of subcellular trafficking. In many cases, molecules are targeted to a particular region in order to perform a specific cellular function. Conversely, nucleolar detention functions by removing important factors from their active sites, thereby disrupting molecular networks through the temporary imprisonment of key cellular factors within the nucleolus (Fig. 1).
Nucleolar Detention Signal as a Molecular Marker
The identification of proteins targeted to the nucleolus has historically been problematic, as most localization signals (NoLS) generally contain a seemingly random series of charged arginine and lysine residues.33 In contrast, the nucleolar detention signal (NoDS) is characterized by a position-independent consensus sequence, consisting of at least one arginine motif (RRI/L) and a minimum of two hydrophobic triplets LhL/v (where h represents a hydrophobic residue).34 While discreet, this motif is strongly predictive of NoDP activity. To date, localization and mobility studies have confirmed that 18 of 18 molecules tested are validated targets of nucleolar detention.21,24,34-36 In addition, analysis of the literature has found several putative NoDS-containing proteins that undergo stimuli-induced “sequestration” within the nucleolus, DAXX,37 SENP5,38 PML,39 TERT40 and TIP5,41 though their mobility has yet to be reported. Bioinformatic analysis using this motif has yielded numerous additional NoDP candidates, suggesting that the number of putative targets will increase substantially.
In conclusion, the potentially staggering array of NoDP targets hints at the systemic nature of RNA-mediated regulation of protein dynamics. These molecules have diverse functions in ubiquitination, proteasomal degradation, protein folding, DNA replication and methylation (Fig.≈1), indicating that the NoDP may control all aspects of cellular life.21,24,34,35 With the emergence of molecular dynamics, we recommend that photobleaching experiments become standard practice when studying the relocalization of molecules, especially within the nucleolus. Further examination of the nucleolar detentiome should highlight the significance of this novel form of posttranslational regulation and reveal other molecular networks under the control of the NoDP.
Acknowledgments
We thank John Copeland for his critical reading of the manuscript. This work was supported by grants from the Canadian Institute of Health Research (CIHR) to S.L.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20140
References
- 1.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–69. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elson EL, Schlessinger J, Koppel DE, Axelrod D, Webb WW. Measurement of lateral transport on cell surfaces. Prog Clin Biol Res. 1976;9:137–47. [PubMed] [Google Scholar]
- 3.Koppel DE, Axelrod D, Schlessinger J, Elson EL, Webb WW. Dynamics of fluorescence marker concentration as a probe of mobility. Biophys J. 1976;16:1315–29. doi: 10.1016/S0006-3495(76)85776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlessinger J, Koppel DE, Axelrod D, Jacobson K, Webb WW, Elson EL. Lateral transport on cell membranes: mobility of concanavalin A receptors on myoblasts. Proc Natl Acad Sci U S A. 1976;73:2409–13. doi: 10.1073/pnas.73.7.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaz WL, Jacobson K, Wu ES, Derzko Z. Lateral mobility of an amphipathic apolipoprotein, ApoC-III, bound to phosphatidylcholine bilayers with and without cholesterol. Proc Natl Acad Sci U S A. 1979;76:5645–9. doi: 10.1073/pnas.76.11.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–33. doi: 10.1016/0378-1119(92)90691-H. [DOI] [PubMed] [Google Scholar]
- 7.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vikstrom KL, Lim SS, Goldman RD, Borisy GG. Steady state dynamics of intermediate filament networks. J Cell Biol. 1992;118:121–9. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 10.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–6. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 11.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–9. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 12.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–5. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–7. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 15.Politz JC, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14:4805–12. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9:285–91. doi: 10.1016/S0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 17.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–72. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 18.Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–57. doi: 10.1016/S1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 20.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–84. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. 2005;170:733–44. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 24.Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–57. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Fayolle C, Pourchet J, Cohen A, Pedeux R, Puisieux A, de Fromentel CC, et al. UVB-induced G2 arrest of human melanocytes involves Cdc2 sequestration by Gadd45a in nuclear speckles. Cell Cycle. 2006;5:1859–64. doi: 10.4161/cc.5.16.3119. [DOI] [PubMed] [Google Scholar]
- 26.Stark LA, Dunlop MG. Nucleolar sequestration of RelA (p65) regulates NF-kappaB-driven transcription and apoptosis. Mol Cell Biol. 2005;25:5985–6004. doi: 10.1128/MCB.25.14.5985-6004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch WJ, Feramisco JR. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J Biol Chem. 1984;259:4501–13. [PubMed] [Google Scholar]
- 28.Audas TE, Li Y, Liang G, Lu R. A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response. Mol Cell Biol. 2008;28:3952–66. doi: 10.1128/MCB.01439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, Yoshida N, Murakami N, Kawata K, Ishizaki H, Tanaka-Okamoto M, et al. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol Biol Cell. 2007;18:1701–9. doi: 10.1091/mbc.E06-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss C, Goldberg M. Recruitment of proteins to DNA double-strand breaks: MDC1 directly recruits RAP80. Cell Cycle. 2011;10:2850–7. doi: 10.4161/cc.10.17.17341. [DOI] [PubMed] [Google Scholar]
- 31.Peddibhotla S, Wei Z, Papineni R, Lam MH, Rosen JM, Zhang P. The DNA damage effector Chk1 kinase regulates Cdc14B nucleolar shuttling during cell cycle progression. Cell Cycle. 2011;10:671–9. doi: 10.4161/cc.10.4.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 33.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–8. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mekhail K, Rivero-Lopez L, Al-Masri A, Brandon C, Khacho M, Lee S. Identification of a common subnuclear localization signal. Mol Biol Cell. 2007;18:3966–77. doi: 10.1091/mbc.E07-03-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004;6:642–7. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- 36.Mekhail K, Rivero-Lopez L, Khacho M, Lee S. Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle. 2006;5:2401–13. doi: 10.4161/cc.5.20.3387. [DOI] [PubMed] [Google Scholar]
- 37.Ivanchuk SM, Mondal S, Rutka JT. p14ARF interacts with DAXX: effects on HDM2 and p53. Cell Cycle. 2008;7:1836–50. doi: 10.4161/cc.7.12.6025. [DOI] [PubMed] [Google Scholar]
- 38.Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–98. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattsson K, Pokrovskaja K, Kiss C, Klein G, Szekely L. Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc Natl Acad Sci U S A. 2001;98:1012–7. doi: 10.1073/pnas.031566998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong JM, Kusdra L, Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol. 2002;4:731–6. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 41.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–61. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]