Abstract
Cohesion establishment is central to sister chromatid tethering reactions and requires Ctf7/Eco1-dependent acetylation of the cohesin subunit Smc3. Ctf7/Eco1 is essential during S phase, and a number of replication proteins (RFC complexes, PCNA and the DNA helicase Chl1) all play individual roles in sister chromatid cohesion. While the mechanism of cohesion establishment is largely unknown, a popular model is that Ctf7/Eco1 acetylates cohesins encountered by and located in front of the fork. In turn, acetylation is posited both to allow fork passage past cohesin barriers and convert cohesins to a state competent to capture subsequent production of sister chromatids. Here, we report evidence that challenges this pre-replicative cohesion establishment model. Our genetic and biochemical studies link Ctf7/Eco1 to the Okazaki fragment flap endonuclease, Fen1. We further report genetic and biochemical interactions between Fen1 and the cohesion-associated DNA helicase, Chl1. These results raise a new model wherein cohesin deposition and establishment occur in concert with lagging strand-processing events and in the presence of both sister chromatids.
Keywords: CHL1, CTF7/ECO1, FEN1, cohesion, DNA replication, establishment, replication fork
Introduction
The temporal separation of DNA replication during S phase from chromosome segregation during mitosis requires cells to maintain the identity of duplicated chromosomes (sister chromatids) over time. Sister chromatid identity is maintained by molecular complexes known as cohesins that physically link the two sister chromatids together. Cohesin complexes consist of four main structural components that include Smc1, Smc3, Mcd1/Scc1 and Scc3 and maintain, in an as yet undefined manner, topological contacts with DNA molecules.1,2 In addition to a canonical role in tethering sister chromatids together, cohesins also play key roles in double stranded break repair,3,4 transcriptional regulation5 and heterochromatin assembly.
In all organisms studied, cohesin loading onto chromatin depends on a complex of two proteins, Scc2 and Scc4.2 Cohesin loading is not sequence-specific and localizes to poorly defined regions known as CAR (cohesin attachment regions).2 Currently, the timing of cohesin deposition relative to sister chromatid tethering reactions also remains unclear. For instance, budding yeast cohesins associate with DNA at the end of G1.6 This chromatin association, however, appears transient and highly dynamic. In contrast, cohesins loaded during S phase are quite stable and resistant to salt extraction.7 Cell cycle mapping studies further reveal that the essential function of Scc2 and Scc4 occurs during S-phase. Regardless of timing, cohesin loading is necessary but not sufficient to result in sister chromatid tethering. An additional activity, termed establishment, is required to convert chromatin-bound cohesins to a pairing competent state.8
The mechanism of cohesion establishment has recently come under intense scrutiny. These efforts revealed that the establishment factor Ctf7/Eco1 is an acetyltransferase that modifies the cohesin subunit Smc3 on a pair of highly conserved lysines.9-11 Similar to Scc2 and Scc4, Ctf7/Eco1 function is essential during S phase.12,13 Numerous studies reveal that Ctf7/Eco1 performs essential roles in cohesion establishment, chromosome condensation, DNA repair and transcription regulation.3,4,12,14 Despite its role in these fundamental processes, little is known regarding Ctf7/Eco1 function relative to the DNA replication fork. One popular model suggests that Ctf7/Eco1 acetylates cohesins that reside in front of the DNA replication fork. This pre-replicative cohesin acetylation is posited to both allow for fork progression and produce a cohesin state that promotes sister chromatid tethering.6,15,16 In contrast to this model, little evidence supports the notion that Ctf7/Eco1 binds pre-fork replication components, translocates with the replication fork or functionally alters cohesins prior to fork passage.17,18 Instead, a growing body of findings indicate that fork passage is required for establishment, and cohesins that associate with chromatin prior to fork passage are most likely not relevant to establishment.7,18,19
The above controversies highlight the importance of pinpointing the location and timing of cohesion establishment with respect to the DNA replication fork. To address this issue, we exploited the very well-characterized RAD27/FEN1 flap endonuclease (herein termed FEN1) that is critical for processing Okazaki fragments during DNA replication.20,21 Previous studies reveal that fen1 mutant cells exhibit cohesion defects,22 suggesting that cohesion establishment might be linked to Okazaki fragment processing events. Human Fen1 (hFen1) endonuclease also enhances the activity of hChlR1,23 a DNA helicase family previously identified as playing a role in cohesion establishment.24-26 In this study, we provide novel evidence that temporally link the activities of helicase, endonuclease and acetyltransferase and support a model in which cohesin deposition and establishment occur immediately behind the replication fork in a manner analogous to DNA chromatinization.
Results
Interrelated synthetic lethalities support coordination of cohesion establishment to lagging-strand processing.
In contrast to the popular notion that Ctf7/Eco1 acts on cohesins positioned in front of the DNA replication fork, Ctf7/Eco1 positioning relative to the fork remains unknown. Several studies point to factors that function behind the fork, such as the flap endonuclease Fen1, as critical for sister chromatid pairing and formally raise a model that establishment occurs as sister chromatids emerge from behind the fork.23,27 To test this alternate model, yeast cells harboring ctf7eco1-1:ADE were crossed with fen1::KANr mutant cells, and the resulting diploids were placed in sporulation medium. Notably, heterozygous ctf7eco1-1:ADE/CTF7 FEN1/fen1 cells exhibited extremely poor sporulation efficiency (< 2%). When diploid cells were first transformed with CEN-URA3-CTF7ECO1 plasmid, however, the resulting transformants sporulated with high efficiency (approximately 85%). Similar haplo-insuffiency in sporulation was previously observed in crosses involving ctf7/eco1 (Brands and Skibbens, unpublished observation), revealing that Ctf7/Eco1 performs an essential but dosage-dependent role during meiosis. Of the 112 spores obtained from sporulated transformed diploids, we recovered the expected number of wild-type cells and both fen1 and ctf7/eco1 single mutant cells (Table 1). In contrast, only seven double mutant fen1 ctf7/eco1 cells were recovered at 23°C, six of which harbored CTF7/ECO1 plasmid. Upon plating onto media supplemented with 5' FOA,28 all six plasmid-bearing double mutant strains were inviable, revealing a synthetic lethal interaction between ctf7/eco1 and fen1 mutations (Fig. 1). The single double mutant spore exhibited robust growth at all temperatures and, thus, likely results from meiotic gene conversion or incorporation of an extragenic suppressor (Fig. S1).
Table 1. Genetic interaction between fen1 and ctf7eco1–1 mutations.
|
Genotype |
Observed |
Expected |
|
fen1 |
25 |
28 |
|
ctf7/eco1–1 |
21 |
28 |
|
fen1, ctf7/eco1–1 |
7 (6 with CTF7/ECO1:URA) |
28 |
| Wild Type |
34 |
28 |
| Total | 87 | 112 |
Tetrad analyses of fen1::KANr cells crossed with ctf7/eco1–1: ADE cells.
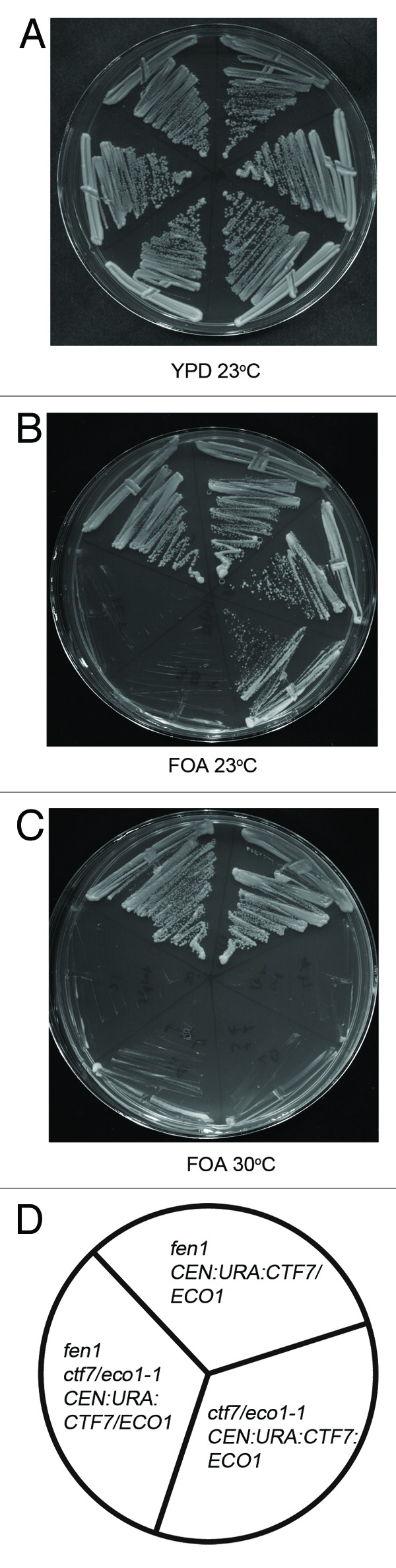
Figure 1.ctf7eco1–1 is synthetically lethal in combination with fen1. Yeast cells harboring ctf7eco1–1: ADE were crossed with fen1::KANr mutant cells and the resulting diploids were transformed with a CEN: URA3:CTF7ECO1 plasmid and sporulated. The resulting fen1 and ctf7eco1–1: ADE single mutants and fen1, ctf7eco1–1: ADE double mutants were plated on media with or without FOA (See also Table 1). Two independent isolates are shown for each strain. (A) Growth of fen1, ctf7eco1–1 single mutants and fen1::KAN ctf7eco1–1 CTF7: URA double mutants strains at 23°C on YPD. (B) Growth of fen1::KAN, ctf7eco1–1 single mutants and fen1 ctf7eco1–1: ADE CTF7: URA double mutants on FOA plates at 23°C. (C) Growth of fen1::KAN, ctf7eco1–1 single mutants and fen1 ctf7eco1–1: ADE CTF7: URA double mutants on FOA plates at 30°C (See also Fig. S1). (D) Schematic representation of fen1::KAN, ctf7eco1–1 single mutants and fen1 ctf7eco1–1: ADE CTF7: URA double mutant strains.
Chl1 (and the human homolog hChlR1) is a DNA helicase that promotes sister chromatid cohesion, and hChlR1 stimulates the flap endonuclease activity of hFen1.23-26 Prior evidence that ctf7/eco1 interacts genetically with chl1,24 coupled with the synthetic lethal ctf7/eco1 and fen1 interaction described above, predicts that fen1 might also interact genetically with chl1. To test this, chl1::HIS3 cells were crossed to fen1::KANr cells and the resulting diploids sporulated. High efficiency sporulation was observed. Tetrad analyses recovered the expected number of wild-type and both chl1 and fen1 single mutant strains (Table 2). In contrast, no chl1 fen1 double mutant spores were recovered (Table 2). This synthetic lethality extends findings obtained from a high-throughput assay that suggested that chl1 and fen1 interact genetically.29,30 The interrelated synthetic lethal network (ctf7/eco1-fen1; fen1-chl1; chl1-ctf7/eco1-current study and Skibbens),24 supports the model that cohesion establishment is temporally coupled to lagging strand processing.
Table 2. Genetic interaction between chl1 and fen1 mutations.
|
Genotype |
Observed |
Expected |
|
chl1 |
15 |
15 |
|
fen1 |
16 |
15 |
|
chl1, fen1 |
0 |
15 |
| Wild type |
15 |
15 |
| Total | 46 | 60 |
Tetrad analyses of chl1::HIS3 cells crossed with fen1::KANr cells.
Ctf7/Eco1 and Chl1 associate with lagging strand-processing factor Fen1.
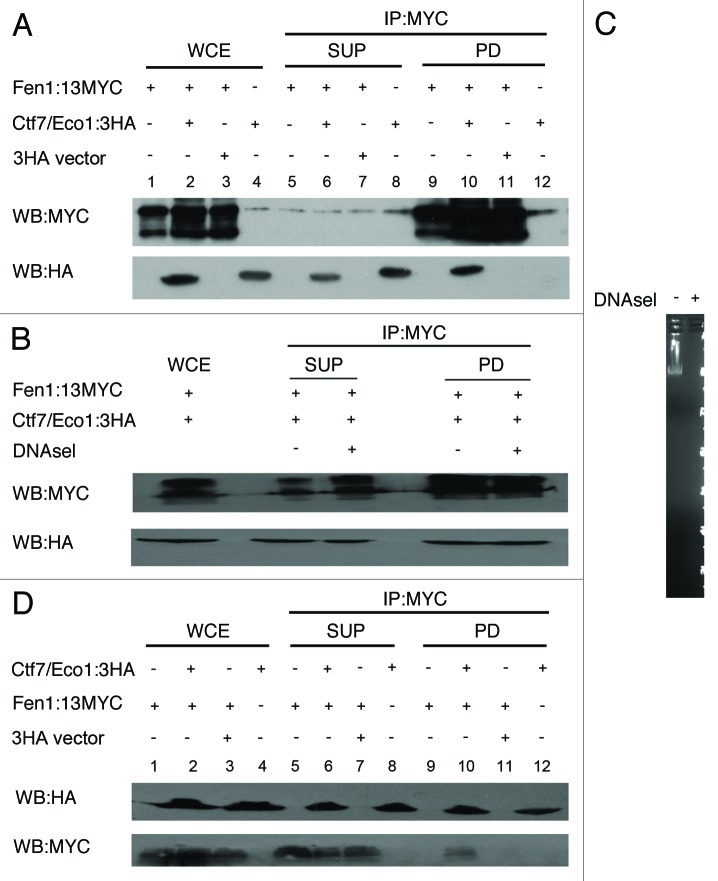
If the above lagging strand-coupled establishment model is correct, then each of these proteins may physically interact. To test this prediction, we transformed cells expressing Fen1-13Myc with either a construct directing elevated expression of Ctf7/Eco1–3HA or 3HA alone as a control. Logarithmically growing cultures of the resulting transformants were lysed, and cell extracts were incubated with anti-MYC beads. After several washes, bound protein complexes were eluted and assayed by western blot. As expected, Fen1-13MYC was efficiently immunoprecipitated through this procedure (Fig. 2A). Probing duplicate membranes with HA-directed antibodies revealed that Ctf7/Eco1–3HA co-immunoprecipitated with Fen1-13MYC but was not pulled down from lysates obtained from cells expressing untagged Fen1 (Fig. 2A). The role of Fen1 in DNA modification raised the possibility that the Fen1-Ctf7/Eco1 association might be mediated through DNA. To address this possibility, we repeated the co-immunoprecipitation but included DNaseI in the cell lysate. The results show that Ctf7-3HA continues to efficiently co-immunoprecipitate with Fen1-13MYC in the absence of DNA (Fig. 2B). Complete degradation of lambda DNA that was spiked into the co-immunoprecipitation reaction confirmed the efficacy of the DNaseI treatment (Fig. 2C).
Figure 2. Fen1 and Ctf7/Eco1 physically associate in vivo. Cells expressing Fen1:13Myc and Ctf7/Eco1:3HA were mechanically lysed and clarified by centrifugation, the clarified whole cell extract was co-immunoprecipitated using anti-Myc beads (2A) and anti HA Beads (2D) and analyzed by immunoblotting for Fen1:13Myc and Ctf7/Eco1:3HA. Whole cell extracts (WCE, lanes 1–4), Supernatants (SUP, lanes 5–8) and pull down fractions (PD, 9–12) are shown. (A) Co-immunoprecipitation of Fen1:13Myc and Ctf7:3HA with anti-MYC beads. Cells expressing only 3HA tag (Lane 11) and cells expressing Ctf7:3HA but untagged Fen1 (Lane 12) were used to determine the specificity of the co-immunoprecipitation. (B) Clarified whole cell extracts of cells co-expressing Fen1:13Myc and Ctf7:/Eco1:3HA were treated with and without DNaseI before immunoprecipitation with anti-MYC beads. (C) 1 μg of λ DNA added in the clarified whole cell extract, with and without DNaseI treatment, run on a 1% agarose gel. (D) Reciprocal co-immunoprecipitation of cells expressing Fen1:13Myc and Ctf7:3HA using Anti-HA affinity beads. Cell expressing untagged Ctf7/Eco1 (Lane 9) and cells expressing 3HA tags alone (Lane 11) were used as a control to determine the specificity of the co-immunoprecipitation reaction.
Ctf7/Eco1 binding to Fen1 was confirmed using a reciprocal immunoprecipitation strategy. Briefly, logarithmically growing cells co-expressing Ctf7/Eco1–3HA and Fen-13MYC were lysed and cell extracts incubated with anti-HA affinity beads. After several washes, bound proteins were eluted and analyzed by western blot using HA-directed antibodies to reveal efficient Ctf7/Eco1 immunoprecipitation. Probing duplicate membranes with anti-MYC antibodies revealed Fen1-13MYC co-immunoprecipitated with Ctf7/Eco1–3HA (Fig. 2D). Importantly, Fen1-13MYC was not pulled down in lysates obtained from cells expressing untagged Ctf7/Eco1 or cells expressing only 3HA tags (Fig. 2D). Taken together, the reciprocal immunoprecipitation studies uncover a physical in vivo association between Fen1 and Ctf7/Eco1 that occurs independent of DNA.
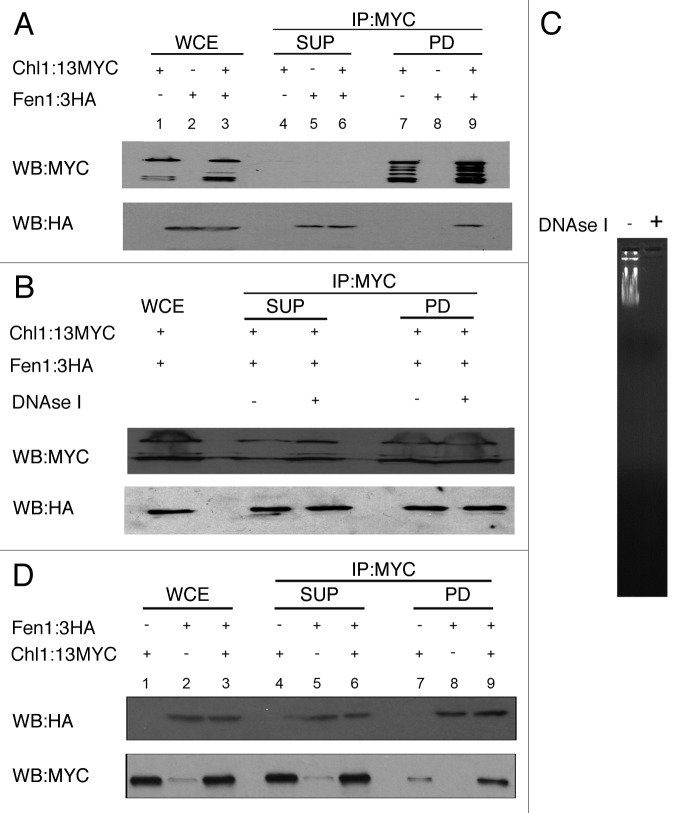
Fen1 flap endonuclease activity is stimulated by hChlR1,23 and both participate in cohesion.22,24-26 To test the possibility that Chl1 physically associates with Fen1, and thus provide positional information regarding Chl1 function relative to the DNA replication fork, cell lysates obtained from logarithmically growing cells co-expressing Chl1-13MYC and Fen1–3HA were incubated with anti-MYC beads. As before, bound proteins were eluted from washed beads and assayed by western blot. The results show efficient immunoprecipitation of Chl1-13MYC (Fig. 3A). Duplicate membranes probed for anti-HA antibodies reveals that Fen1–3HA co-immunoprecipitates with Chl1-13MYC but not with untagged Chl1 (Fig. 3A). We next tested whether Chl1 binding to Fen1 depended on DNA by including DNaseI in the cell lysate prior to pull down. The results show that Fen1–3HA efficiently co-immunoprecipitated with Chl1-13MYC even in the absence of DNA (Fig. 3B). Complete degradation of lambda DNA spiked into the co-immunoprecipitation reaction confirmed the efficacy of the DNaseI treatment (Fig. 3C).
Figure 3. Chl1:13Myc physically associate with Fen1:3HA in vivo. Cells expressing Chl1:13Myc and Fen1:3HA were mechanically lysed and clarified by centrifugation. The clarified whole cell extract was co-immunoprecipitated using anti-Myc beads (3A) and anti HA beads (3B) and analyzed by immunoblotting for Chl1:13Myc and Fen1:3HA. Whole cell extracts (WCE, lanes 1–3), Supernatants (SUP, lanes 4–6) and pull down fractions (PD, 7–9) are shown. (A) Co-immunoprecipitation of Chl1:13Myc and Fen1:3HA using anti-MYC beads. Cells expressing untagged Fen1 cell extracts were used to determine the specificity of the co-immunoprecipitation reaction (Lane 8). (B) Clarified whole cell extracts of cells co-expressing Chl1:13Myc and Fen1:3HA were treated with or without DNaseI treatment before co-immunoprecipitation with anti-MYC beads. (C) 1 μg of λ DNA added in the clarified whole cell extract with and without DNaseI treatment, run on a 1% agarose gel. (D) Reciprocal immunoprecipitation of cells co-expressing Chl1:13Myc and Fen1:3HA with Anti-HA beads. Cells expressing untagged Fen1 were used to determine the specificity of the co-immunoprecipitation reaction (Lane 7).
The interaction between Chl1 and Fen1 was confirmed using a reciprocal co-immunoprecipitation procedure. Briefly, cells co-expressing Chl1-13Myc and Fen1–3HA were lysed and incubated with anti-HA beads. Bound protein complexes were eluted from the washed beads and analyzed by western blotting. The results show that Fen1–3HA is efficiently immunoprecipitated (Fig. 3D), and that Chl1-13MYC co-immunoprecipitates specifically with Fen1–3HA such that only trace amounts pull down with untagged Fen1 (Fig. 3D).
Discussion
Two key features of current cohesion establishment models are that (1) cohesins are loaded in front of the DNA replication fork and (2) Ctf7/Eco1 acetylates pre-replicative cohesins to both allow for fork progression and engender sister chromatid tethering upon subsequent fork passage and synthesis of sister chromatids. One notion that appeared to confound this pre-replicative establishment model was that Ctf7/Eco1 is recruited to chromatin by PCNA.31 However, this interpretation is complicated in that Ctf7/Eco1 mutated within the PCNA binding PIP box appears to persist in binding chromatin at normal levels in yeast.18 Nor does this region appear necessary for ESCO2 (Ctf7/Eco1 homolog) chromatin-recruitment in human cells.32 Finally, mutation of PCNA produces only modest cohesion defects, inconsistent with a central role for PCNA in establishment. In fact, there is no convincing evidence to support the notion that Ctf7/Eco1 translocates with the DNA replication fork at all, despite claims to the contrary.17-19,33 Thus, the placement of Ctf7/Eco1 relative to the DNA replication fork had yet to be ascertained.
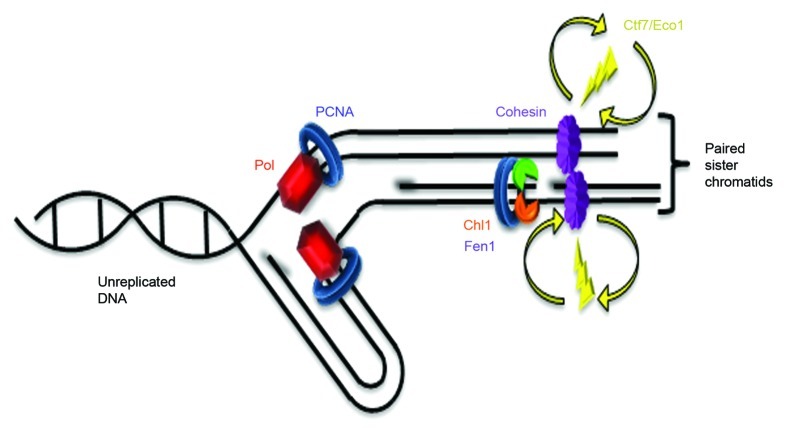
Here, we exploited the well-characterized lagging strand processing factor Fen1 to uncover novel Fen1-Ctf7/Eco1 binding and thus position Ctf7/Eco1 relative to the DNA replication fork. Findings from multiple genetic and biochemical studies indeed support a model in which cohesion establishment occurs immediately behind the fork and possibly coupled to lagging-strand processing.18,27,34 The genetic and physical interactions identified between Fen1, Ctf7/Eco1 and Chl1 (current study and Skibbens24) does not necessarily link Ctf7/Eco1 recruitment to Okazaki fragment maturation. For instance, neither Fen1 nor Chl1 are essential for cell viability20,21,35,36 and thus are unlikely as platforms critical for Ctf7/Eco1 recruitment. Currently, we cannot exclude the possibility that both cooperatively contribute to recruitment—a model consistent with the lethality of cells harboring deletions of both fen1 and chl1 (this study). Instead, we favor a model of cohesion establishment in which Ctf7/Eco1 transiently cycles in associating with cohesins loaded behind the DNA replication fork. We currently view these transient associations as occurring independent of replication fork factors, per se, but that establishment occurs in a post-fork chromatin context that exists in the vicinity of Okazaki fragment maturation as well as on leading-strand synthesis. This post-fork establishment model benefits from several accompanying features (Fig. 4). First, it posits that establishment occurs when sister chromatids are actually present, in contrast to current models that posit establishment requires acetylation of cohesins that reside in front of the fork and before sister chromatids are synthesized.13,16 Second, our model places Ctf7/Eco1 proximal to PCNA but does not require PCNA-dependent chromatin recruitment of Ctf7/Eco1. Nor do we envision Ctf7/Eco1 recruitment requiring Fen1 or Chl1. Instead, we favor a model that Ctf7/Eco1 transiently functions in the local environment that exists immediately behind the replication fork to convert cohesins to a pairing-competent state. Third, this model posits that the cohesins that participate in sister chromatid tethering reactions are most likely loaded behind the DNA replication fork. This is consistent with early reports in which the essential activity of Scc2 (and Ctf7/Eco1) was mapped to S phase.6,8 Thus, both cohesin deposition and Ctf7/Eco1-dependent cohesion establishment likely occur in this post-fork context—cohesin “association” prior to the replication fork being irrelevant to cohesion. This latter point is supported by the highly dynamic nature of cohesins during G1.7,33,37 Our current view of establishment parallels that of chromatinization, in which newly synthesized histone complexes are deposited onto nascent sister chromatid strands and subsequently posttranslationally modified to engender epigenetic states.38 Cohesin deposition/modifications that engender sister chromatin pairing and transcription regulation may be similarly temporally coupled. Toward this end, we note several chromatin-modifying complexes (INO80, RSC and SWI/SNF components) that promote efficient cohesion establishment.2 The physical link reported here between Chl1 and Fen1, coupled with recent findings that Chl1 exhibits 5'-3' unwindase capabilities, removes chromatin-bound proteins (possibly resolving chromatin structures such as G-quads) and is critical for cohesin deposition or stabilization, provides additional motivation in considering a chromatin-based post-fork model of cohesion establishment.39-41 Future research focused on testing cohesion establishment in the context of chromatinization is likely to provide significant insights applicable to multiple fields of inquiry and, given the role of cohesion pathway mutations in tumorigenesis and developmental maladies, may be of clinical interest.5,42
Figure 4. Cohesion establishment coupled to lagging strand processing. Replication fork (Pol = DNA polymerase coupled to PCNA) moves to the left: leading strand replication on the top and lagging strand replication on the bottom (RNA primers shadowed). Immediately behind the fork, PCNA associates with Fen1 (green) and Chl1 (orange). Ctf7/Eco1 (yellow) is not stably recruited to chromatin by any factor, but transiently interacts with chromatin to establish cohesion. Therefore, both cohesin deposition and subsequent cohesion establishment occur behind the replication fork. Cohesins (purple) depicted as unstructured to highlight the many models currently posited in the literature.1,44,45 MCM helicase, primase and RPA not shown (based on Burgers46).
Experimental Methods
Media and yeast strains.
Saccharomyces cerevisiae strains, plasmids, growth and sporulation media are as described previously in reference 24, and as listed in Table 3. To construct fen1::KANr cells, PCR fragments were generated using primers 5'-CGA TGA AAA GCG TTG ACA GCA TAC ATT GGA AAG AAA TAG CGG ATC CCC GGG TTA ATT AA-3' and 5'-CAA GGT GAA GGA CCA AAA GAA GAA AGT GAA AAA AGA ACC CCC GAA TTC GAG CTC GTT TAA AC-3' and pFA6a-kanMX6.43 The resulting PCR product was transformed into YBS 1039 (Table 3). FEN1::KANr was confirmed by PCR using primers 5'-GGT GAC TTT CGT TAA TGG GGA-3' and 5'-GCA AAC GAA TTA CAG CCA GTG-3'.
Table 3. All strains are of S288C background except those marked with * are W303 background strains.
|
Strain YBS1019 YBS1020 *YBS1039 *YMM360 *YSR005 *YSR004 YSR010 YBS1129 YBS040 YSR051 YSR052 YSR054 YSR071 |
Genotype MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 [4] MATα ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 [4] MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 [4] MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 ctf7eco1–1::ADE2 [4] MATα ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 fen1::KANr MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 chl1::HIS3 MATα ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 FEN1:3HA:TRP1 MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 CHL1:13Myc:URA3 [6] MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 FEN1:3HA:TRP1 CHL1:13Myc:URA3 MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 FEN1:13MYC:TRP1 MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 FEN1:13MYC:TRP1 (YBS 1070, CTF7:3HA:LEU2) MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 FEN1:13MYC:TRP1 (YBS 1074, 3HA:LEU2) MATa ade2–101 his3∆200 leu2 lys2–801 trp1∆63 ura3–52 (YBS 1070, CTF7:3HA:LEU2) |
To construct FEN1–3HA:TRP cells, PCR fragments were generated using primers 5'-AGA GCA CAA GAA AAT AAA AAA TTG AAC AAA AAT AAG AAT AAA GTC ACA AAG GGA AGA AGA CGG ATC CCC GGG TTA ATT AA-3' and 5'-CAA GGT GAA GGA CCA AAA GAA GAA AGT GAA AAA AGA ACC CCC GAA TTC GAG CTC GTT TAA AC-3' on pFA6a-3HA-TRP1 43 and transformed into YBS 1020 (Table 3). FEN1:3HA: TRP was confirmed by western blotting and PCR analysis.
To construct chl1::HIS3 cells, PCR fragments were generated using the primers 5'-GTA GAA AAC CAG GCT AAA AAC AGT CAC ACT AGT CCA AAA AAC GGA TCC CCG GGT TAA TTA A-3' and 5'-ATA TAG TAG TAA TCA CAG TAT ACA GGT AAA CGT ATT CCT TGA ATT CGA GCT CGT TTA AAA C-3' on p-FA5a-His3MX6 43 and transformed into YBS 1019 (Table 3). CHL1::HIS3 was confirmed using primers 5'-TGC CTG GCT GAC TTC TTA GAC-3' and 5'-CGT GAG CAA ACA ACG GGT AAT-3'.
Co-immunoprecipitations and western blot analyses.
109 cells in YPD medium were harvested, washed with sterile water and resuspended in 500 ml, IPH150 buffer [150 mM NaCl, 50 mM TRIS pH 8, 5 mM EDTA, 0.5% IGEPAL-CA 630 (Sigma), 1 mM DTT, 10 mM Sodium Butyrate, Roche protease inhibitor cocktail]. 0.5 mm glass beads were added and the cells were frozen immediately in liquid nitrogen and stored at -80°C. The frozen cells were then thawed on ice and mechanically lysed by bead beating (Biospec mini bead beater). The soluble fraction was separated from the insoluble fraction by centrifugation at 10,500 rpm (TOMY TX-160, TMA 24 rotor). To test for DNA based interactions, IPH150 was supplemented to 10 mM MnCl2 and 500 ng of DNaseI (Roche) per ml and incubated for 1 h at 4°C. The clarified cell extract was then incubated with EZ view anti-C-MYC affinity gel (Sigma) or Anti-HA affinity matrix (Roche) overnight at 4°C. The beads were harvested by centrifugation and washed extensively with IPH50 buffer [50 mM NaCl, 50 mM TRIS pH 8, 5 mM EDTA, 0.5% IGEPAL-CA 630 (Sigma), 1 mM DTT, 10 mM Sodium Butyrate, Roche protease inhibitor cocktail] and bound proteins solubilized with 2x Lamelli buffer (Sigma). Proteins were then separated by SDS PAGE and analyzed by immunoblotting. Immunodetections were performed using anti-MYC 9E10 (1:1,000) (Santa Cruz) and anti-HA (1:500) (Santa Cruz) in combination with goat anti mouse HRP (1:10,000) (Bio-Rad) and ECL plus (GE healthcare) for visualization.
Supplementary Material
Acknowledgments
The authors thank Skibbens and Cassimeris lab members for comments throughout the course of these experiments. RVS is supported by awards from the National Institute of General Medical Sciences (2R15GM083269–02) and a Faculty Innovation Grant from Lehigh University. Any opinions, findings, and conclusions or recommendations expressed in this study are those of the authors and do not necessarily reflect the views of the National Institutes of General Medical Sciences or Lehigh University.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20547
References
- 1.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–29. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 2.Skibbens RV. Mechanisms of sister chromatid pairing. Int Rev Cell Mol Biol. 2008;269:283–339. doi: 10.1016/S1937-6448(08)01005-8. [DOI] [PubMed] [Google Scholar]
- 3.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–8. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 4.Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–5. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 5.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–54. doi: 10.1016/S1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 7.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–8. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 8.Skibbens RV. Establishment of sister chromatid cohesion. Curr Biol. 2009;19:R1126–32. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–9. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 10.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–51. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–19. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–33. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mönnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–4. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherwood R, Takahashi TS, Jallepalli PV. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 2010;24:2723–31. doi: 10.1101/gad.1976710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–99. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Skibbens RV. Sticking a fork in cohesin--it’s not done yet! Trends Genet. 2011;27:499–506. doi: 10.1016/j.tig.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, et al. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2008;27:111–21. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gary R, Park MS, Nolan JP, Cornelius HL, Kozyreva OG, Tran HT, et al. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol Cell Biol. 1999;19:5373–82. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommers CH, Miller EJ, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–6. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 22.Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, et al. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell. 2004;15:1724–35. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, et al. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem. 2008;283:20925–36. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skibbens RV. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 2004;166:33–42. doi: 10.1534/genetics.166.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–45. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, et al. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci. 2004;117:3547–59. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- 27.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol. 2005;25:5445–55. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–75. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 29.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, et al. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst) 2005;4:459–68. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 31.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–32. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–18. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol. 2010;30:4940–51. doi: 10.1128/MCB.00642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skibbens RV. Holding your own: establishing sister chromatid cohesion. Genome Res. 2000;10:1664–71. doi: 10.1101/gr.153600. [DOI] [PubMed] [Google Scholar]
- 35.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–71. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–58. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNairn AJ, Gerton JL. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res. 2009;17:155–63. doi: 10.1007/s10577-008-9007-9. [DOI] [PubMed] [Google Scholar]
- 38.Jasencakova Z, Groth A. Restoring chromatin after replication: how new and old histone marks come together. Semin Cell Dev Biol. 2010;21:231–7. doi: 10.1016/j.semcdb.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Brosh RM., Jr. Put on your thinking cap: G-quadruplexes, helicases, and telomeres. Aging (Albany NY) 2011;3:332–5. doi: 10.18632/aging.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Brosh RM., Jr. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks039. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM., Jr. Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287:1007–21. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum Mutat. 2010;31:623–30. doi: 10.1002/humu.21252. [DOI] [PubMed] [Google Scholar]
- 43.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth K. How might cohesin hold sister chromatids together? Philos Trans R Soc Lond B Biol Sci. 2005;360:483–96. doi: 10.1098/rstb.2004.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surcel A, Koshland D, Ma H, Simpson RT. Cohesin interaction with centromeric minichromosomes shows a multi-complex rod-shaped structure. PLoS One. 2008;3:e2453. doi: 10.1371/journal.pone.0002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.