SUMMARY
Mitochondrial NLRX1 is a member of the family of nucleotide-binding domain and leucine-rich-repeat–containing proteins (NLRs) that mediate host innate immunity as intracellular surveillance sensors against common molecular patterns of invading pathogens. NLRX1 functions in antiviral immunity, but the molecular mechanism of its ligand-induced activation is largely unknown. The crystal structure of the C-terminal fragment (residues 629-975) of human NLRX1 (cNLRX1) at 2.65 Å resolution reveals that cNLRX1 consists of an N-terminal helical (LRRNT) domain, central leucine-rich repeat modules (LRRM) and a C-terminal three-helix bundle (LRRCT). cNLRX1 assembles into a compact hexameric architecture that is stabilized by inter-subunit and inter-domain interactions of LRRNT and LRRCT in the trimer and dimer components of the hexamer, respectively. Furthermore, we find that cNLRX1 interacts directly with RNA and supports a role for NLRX1 in recognition of intracellular viral RNA in antiviral immunity.
INTRODUCTION
Innate immunity provides the first line of host defense against invading microorganisms and subsequently stimulates adaptive immunity. Such an immediate response relies upon germline-encoded pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), nucleotide-binding domain and leucine-rich-repeat-containing proteins (NLRs), and RIG-1-like receptors (RLRs), which recognize pathogen-associated molecular patterns (PAMPs), such as bacterial lipopolysaccharide, flagellin, and viral RNA (Akira et al., 2006; Beutler et al., 2007; Medzhitov, 2007). NLRs function as intracellular surveillance sensors against microbial products and danger signals and, thereby, trigger host defense pathways via activation of the NF-kB pathway and inflammatory caspases (Inohara and Nunez, 2003). After discovery of the first NLR member, NOD1 (Inohara et al., 1999), the number of NLR family members has grown rapidly (34 in mice, 23 in humans) (Ting and Davis, 2005). Although NLR members share low primary sequence identities (~10 - 25 %) (Proell et al., 2008), most exhibit a characteristic multi-domain structure consisting of an N-terminal effector domain [ED; caspase activation and recruitment domain (CARD) or pyrin], a central nucleotide-binding and oligomerization domain (NOD; NACTH or NOD-NAD), and a C-terminal putative ligand-binding and regulatory leucine-rich repeat (LRR) domain (Harton et al., 2002). Recently, the NLR family has gained more attention, since polymorphisms or mutations in certain NLR genes, NOD1, NOD2, NLRP1, and NLRP3, and dysregulation of their products are associated with inflammatory disorders, such as Blau syndrome, Crohn's disease or early-onset sarcoidosis, or with susceptibility to inflammatory bowel diseases (Dieude et al., 2011; Lu et al., 2010; Molnar et al., 2007; Pontillo et al., 2011; Rosenstiel et al., 2007)
Amongst NLRs, NLRX1 (also known as NOD9) is unique in that it localizes to the mitochondria through an N-terminal mitochondrial targeting (MT) sequence (Arnoult et al., 2009; Moore et al., 2008; Tattoli et al., 2008). Like other NLR proteins, NLRX1 is a proinflammatory activator that stimulates the production of reactive oxygen species (ROS) via TNFα activation (Arnoult et al., 2009; Tattoli et al., 2008). Moreover, NLRX1 can also down-regulate inflammatory responses as a negative regulator of RIG-1 and TLRs through interaction with the mitochondrial antiviral signaling adaptor (MAVS) and TRAF6-IKK, respectively (Allen et al., 2011; Moore et al., 2008; Xia et al., 2011). Such findings have extended our knowledge of the function of NLRs from that of a simple immune activator to a more complex immune modulator. As another example, NLRC5 regulates both negatively in NF-kB and type-1 interferon responses (Cui et al., 2010) and positively in inflammasome activation (Davis et al., 2011). Thus, NLRs participate in diverse biological processes in innate immunity.
With such diverse biological outcomes, numerous efforts have been made to understand the molecular mechanism whereby NLR is activated. Collective analyses of NLR members and homologous proteins have lead to proposals that ligand-induced activation of NLR is accompanied by conformational changes from a closed, inactive, monomeric or dimeric form to an active, higher oligomeric complex through self- and hetero-oligomerization via NOD and ED domains, respectively (Faustin et al., 2007; Franchi et al., 2009; Riedl et al., 2005). Nevertheless, it has been debated whether NLRs recognize ligands directly or indirectly in the first step of the signaling process. In previous, mutational analyses and agonist screenings of NOD1 and NOD2, the cognate ligand recognition site was mapped onto the LRR domain (Girardin et al., 2005; Inohara et al., 2005; Moore et al., 2008; Tanabe et al., 2004). However, these data do not exclude an indirect sensing of PAMPs through an accessory molecule, as postulated that ligand recognition of NLRP3 was assisted by thioredoxin-interacting protein (TXNIP) (Zhou et al., 2010).
Here, we present the crystal structure of the C-terminal fragment of human NLRX1 (cNLRX1) at 2.65 Å resolution. The cNLRX1 structure reveals a three-domain architecture consisting of an N-terminal helical domain, a central LRR motifs and a C-terminal three-helix bundle. Consistent with gel filtration and analytical ultracentrifugation analyses, cNLRX1 forms a hexamer through trimerization of dimers in the crystal via extensive inter-domain and inter-subunit interactions. Moreover, we show that a direct interaction of the C-terminal fragment of NLRX1 with RNA ligands plays a key role in NLRX1-mediated reactive oxygen species (ROS) activation. Finally, cNLRX1 displays a series of positively charged electrostatic patches on the surface that can correspond to a nucleic acid binding site on NLRX1.
RESULTS
Oligomerization of cNLRX1
The C-terminal fragment of human NLRX1 (residues 629-975, hereafter referred to as cNLRX1) (Figure 1A) was expressed in a baculovirus expression system and purified to homogeneity. Size exclusion chromatography analysis revealed that the purified recombinant cNLRX1 protein eluted in two major peaks corresponding to a monomer (apparent MW, ~39 kDa) and a hexamer (apparent MW, ~240 kDa) (Figure S1A). A minor peak was also observed between the two major peaks in a concentration-dependent manner and likely represents a dimer formation. In addition, the hexameric form dominates at higher concentrations and upon longer storage of the purified cNLRX1. Such monomer, dimer and hexamer forms were similarly found by analytical ultracentrifugation (AUC) analysis (Figure S1B). Thus, we conclude that cNLRX1 can adopt multiple forms from monomer to dimer to hexamer in solution.
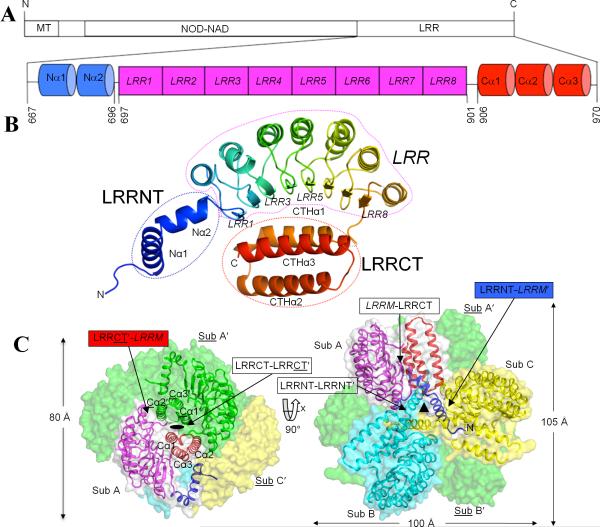
Figure 1. Overall structure of cNLRX1.
A. Schematic representation of NLRX1 (MT-NOD-NAD-LRR) and the cNLRX1 construct [LRRNT (Nα1 and Nα2; blue)-LRRM (LRR1-8; magenta)-LRRCT (Cα1-3; red)] that was used for structural and ligand binding studies. Residue numbers corresponding to the domain boundaries and the N- and C-terminal ends that are ordered in the crystal structure are indicated. B. A cNLRX1 monomer (subunit A) is rainbow-colored from blue (N-terminus) to red (C-terminus) in a ribbon diagram. Secondary structural elements of β-strands, α-helices and loops are indicated as arrows, helical ribbons and extended coil structures, respectively. C. cNLRX1 hexamer. The cNLRX1 hexamer is formed by trimerization of dimers through dihedral D32 symmetry. The crystallographic two-fold symmetry is highlighted by a side view of the cNLRX1 hexamer (left) and the 2-fold symmetry axis between subunit A and A′ is visualized as an oval. The 3-fold non-crystallographic symmetry is shown in the top view of the hexamer (right) and its axis is represented by a triangle at the trimer center. The cNLRX1 trimer is shown in ribbons [subunit A (LRRNT, blue; LRRM, magenta; LRRCT, red); subunit B, cyan; subunit C, yellow] and transparent surface representation (subunit A, gray; subunit B, cyan; subunit C, yellow), and its 2-fold symmetry-related partners (subunits A′, B′, C′) are in a green surface representation. Labels for the various domain interfaces are boxed with arrows. LRRM residues and descriptions are labeled in italics. A subunit (or residue) that is related by 2-fold symmetry to the other (reference) subunit (or residue) in a dimer is distinguished by underline and prime (′). These same colors symbols and fonts are used in the rest of figures.
Overall structure
To gain insight into the atomic details of the LRR domain structure in NLR proteins, we determined the crystal structure of cNLRX1 by the multiple anomalous wavelength dispersion (MAD) method (Hendrickson, 1991) using a potassium tetranitro platinum (II) (K2(NO2)4Pt) derivative crystal. The crystal structure of cNLRX1 was refined to 2.65 Å (Table 1) and the final structure contains residues 667-970 out of the expressed protein residues 629-975 (Figure S2).
Table 1.
Crystallographic data and refinement statistics of cNLRXl structure.
| K2(NO2)4Pt derivative | |||
|---|---|---|---|
| Peak | Inflection | Remote | |
| Data collection | |||
| Space group | C2221 | ||
| Cell parameters (Å) | a = 99.01 | ||
| b = 123.47 | |||
| c = 145.85 | |||
| Resolution (Å) | 20.00 - 2.65 | ||
| Highest resolution (Å) | 2.79 - 2.65 | ||
| Wavelength (Å) | 1.0719 | 1.0716 | 0.8550 |
| No. observations | 194,003 | 193,637 | 219,316 |
| No. unique reflections | 25,742 | 25,749 | 26,342 |
| Rmerge(%)a | 6.0 (28.2)b | 3.1 (30.0)b | 7.0 (59.0)b |
| I/sigma | 6.9 (2.7)b | 7.4 (2.3)b | 6.8 (1.3)b |
| Completeness (%) | 97.9 (87.7)b | 97.9 (87.2)b | 100.0 (100.0)b |
| Multiplicity | 7.5 (4.7)b | 7.5 (4.7)b | 8.3 (8.7)b |
| Phasing | |||
| Resolution (Å) | 15.00 - 3.00 | ||
| Pt sites (SOLVE) | 4 | ||
| Overall figure of merit | 0.58 | ||
| Refinement | |||
| Resolution (Å) | 20.00 - 2.65 | ||
| No. reflections (total) | 24,448 | ||
| No. reflections (test) | 1,311 | ||
| Rcryst (%)c | 22.2 | ||
| Rfree (%)d | 26.5 | ||
| Average B-value (Å2) | 79.0 | ||
| No. protein atoms | 6,632 | ||
| No. water molecules | 8 | ||
| No. ligand atoms | 4 (Pt) | ||
| R.m.s.d bonds (Å) | 0.016 | ||
| R.m.s.d angles (°) | 1.56 | ||
| Ramachandrane (favored) | 97.0% | ||
| (outliers) | 0.4 % | ||
Rmerge= ΣhklΣi| Ii(hkl) - <I(hkl)> | / ΣhklΣi Ii(hkl).
Numbers in parenthesis were calculated from data from the highest resolution shell.
Rcryst = Σ| |Fobs|-|Fcalc| | / Σ|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5% of the total reflections chosen at random and omitted from refinement.
Calculated using MolProbity (http://molprobity.biochem.duke.edu).
The crystal structure reveals that the cNLRX1 monomer is composed of three distinct domains: an N-terminal helical domain (LRRNT, residues 667-696), central LRR modules (LRRM, residues 697-901) and a C-terminal three-helix bundle (LRRCT, residues 906-970) (Figures 1A, 1B and S2). The LRRNT domain contains two α-helices (Nα1, residues 672-686; Nα2, 688-696) that are oriented at 120° via a kink at Asn687 and Nα2 directly connects to the LRRM domain. The central LRR domain folds into a typical crescent shape and consists of 8 LRR motifs with their canonical, parallel β-strands disposed on the concave side. This concave face of the LRRM domain encircles one side of the LRRCT domain, which consists of three antiparallel α-helices (Cα1, residues 906-923; Cα2, 926-945; Cα3, 950-970) (Figure 1B).
As observed in solution, the cNLRX1 crystal structure assembles into a hexamer via 2-fold crystallographic symmetry (Figure 1C left) and 3-fold noncrystallographic symmetry (NCS) axes (Figures 1C right and S1B). In the crystal, the cNLRX1 dimer buries ~1550 Å2 of accessible surface area (ASA) (corresponding to 9.9% of the monomer surface) with the LRRCT domains at center of the dimer. In ASU, three cNLRX1 subunits (subunits A, B and C) come together as an NCS trimer in the shape of a three-petal pinwheel. The trimer is formed with three LRRNT domains positioned around the 3-fold NCS axis, which function as a pin holding the three protruding petals composed of the LRRM and LRRCT domains (Figure 1C right). Approximately 2054 Å2 of ASA is buried upon trimerization, which corresponds to 13.5% of the surface of each monomer. As a result, the LRRCT-mediated dimerization and LRRNT-centered trimerization assemble six cNLRX1 modules into a relatively compact hexameric structure (100 Å × 105 Å × 80 Å), which is more precisely a trimer of three dimers with approximate dihedral D32 symmetry.
Structure overlay of three cNLRX1 monomers (A, B, and C) shows that the LRRNT and LRRM domains are almost identical between subunits with an r.m.s.d. of 0.1 Å, whereas the LRRCT domains display greater structure displacement (Figure S1D). We consider that those observed structure differences are a result of crystal packing (Figure S1E) and do not affect the description and interpretation of the NLRX1 structure. Hereafter, the cNLRX1 structure of subunit A (cNLRX1A) is mostly described unless otherwise specified.
LRR domain
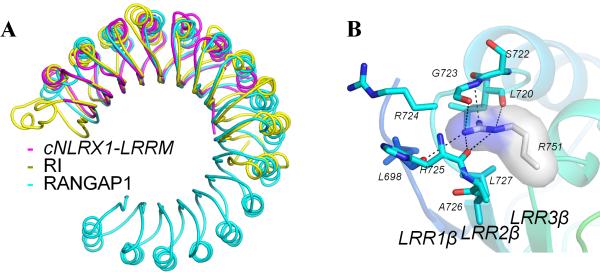
A DALI database search reveals that the overall fold of cNLRX1 LRRM domain resembles those of other LRR containing proteins (Kobe, 2001) (Figure 2A). The closest structural homologues of cNLRX1 LRRM are from segments of ribonuclease inhibitor (RI) (Kobe and Deisenhofer, 1996) and RAN GTPase activating protein 1 (RANGAP1) (Hillig et al., 1999). Although cNLRX1 LRRM can be superimposed on RI and RANGAP1, cNLRX1 LRRM nevertheless deviates substantially from RI and RANGAP1 both in primary sequence (sequence identity, 23% and 19%, respectively) and in tertiary structure (r.m.s.d.'s, of 2.5 Å for 195 Cα atoms and 2.4 Å for 188 Cα atoms, respectively).
Figure 2. Structure of cNLRX1 LRRM domain.
A. Comparison of the cNLRX1 LRRM structure with other LRR structures. Porcine RI (PDB ID code 1bnh; yellow) and RANGAP1 (PDB ID code 1yrg; cyan) were superposed onto cNLRX1 LRRM (magenta) using Cα atoms. B. Close-up view of the cNLRX1 LRR2 interactions. The aberrantly short LRR2 introduces a groove in the loop connecting LRR2 and LRR3 (LRR2-3 loop). Arg751 is shown in a stick model with surface representation (carbon, gray; nitrogen, blue) and its interacting residues are represented by stick models (carbon, cyan; nitrogen, blue; oxygen, red). H-bonds are depicted in black dashed lines.
Each motif of LRR1-7 consists of a single β-strand on the concave side, an α-helix on the convex side, and two connecting loops, forming a typical β-strand-ascending loop-α-helix-descending loop structure, similar to RI LRRs (Kobe and Deisenhofer, 1993). However, the last LRR motif, LRR8, does not contain an α-helix and proceeds directly to LRRCT domain. LRR2 is also aberrantly short with only 25 residues, unlike the other LRR motifs (LRR1 and LRR3-7) that are either 28 or 29 residues long (Figure S2). In other LRR structures, the presence of an irregular length LRR motif in consecutive LRRs often causes a distortion in LRR curvature, or a formation of a protruding loop or a groove on the surface of LRR structure. In the cNLRX1 LRR structure, the short LRR2 introduces a groove at Arg751 (position 1 of LRR3), which is a connecting residue between the LRR2 α-helix and the LRR3 β-strand. The Arg751 side chain points toward LRR1 and is deeply inserted below Ser722 and Gly723 of the protruding LRR1-2 loop (Figure 2B). The Arg751 guanidinium group is extensively coordinated through H-bonds or polar contacts to five carbonyl oxygen atoms from the main chain of the surrounding Leu698, Leu720, Ser722, Gly723 and His725 residues. In addition, Leu727 provides a hydrophobic stacking interaction below Arg751 to sandwich Arg751 between the LRR2-3 loop and Leu727.
cNLRX1 LRR modules
Primary sequence and tertiary structure comparisons of individual cNLRX1 LRR motifs confirm that they contain the expected conserved residues of the LRR consensus motif (xL2xxL5xL7xxN10xL12xxxxL17xxL20xxxL24) as seen in other LRR structures (Kobe and Kajava, 2001) (Figure S2). Positions 2, 5, 7, 12, 17, 20 and 24 are occupied by apolar residues, such as leucine, isoleucine, valine and phenylalanine, which construct the hydrophobic core of the LRR structure. Position 10 selectively places the buried polar residues (asparagine or threonine) in a location that stabilizes the ascending loop between a β-strand and an α-helix via H-bonds with neighboring main-chain atoms of the loop.
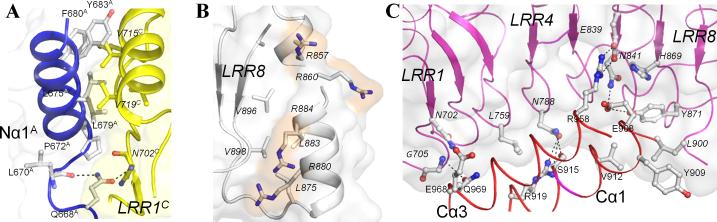
In cNLRX1, the N-terminal end of the LRRM hydrophobic core is protected by the adjacent LRRNT domain (e.g., LRRMC by LRRNTA, as described below, i.e. LRRNT-LRRM contact) from exposure to solvent (Figure 3A). The amphipathic Nα1 helix places its apolar residues, Pro672, Leu675, Leu676, Leu679, Phe680 and Tyr683, near LRR1 and buries ~560 Å2 of ASA. Furthermore, a more distal LRRNT residue, Gln668A intercalates between the N-terminal region of subunit A (residues 669A-672A) and the LRR1-β strand of subunit C (LRR1-βC) and its side-chain nitrogen and oxygen atoms form H-bonds with the main-chain oxygen of Leu670A and the amide nitrogen of Asn702C, respectively. At the C-terminal end of cNLRX1 LRRM, four arginine residues (Arg857, Arg860 of LRR6 and Arg880, Arg884 of LRR7) are found on the surface, resulting in a burial of ~470 Å2 of ASA (Figure 3B). While the non-polar carbon atoms of the arginines make van der Waals interactions with the hydrophobic residues of LRR6 and LRR7, their polar guanidinium groups are disposed on the cNLRX1 surface. In addition, the C-terminal end of LRRM domain (LRR8) is further stabilized by the 2-fold symmetry related LRRCT domain in the cNLRX1 dimer (see below).
Figure 3. cNLRX1 LRRM contacts.
A. Inter-subunit LRRNT-LRRM′ interactions. Nα1A (blue cartoon and gray sticks) provides an N-cap to cover the hydrophobic residues from LRR1C (yellow structure). B. A series of arginine residues displayed on the C-terminal NLRX1 LRRM surface. The NLRX1 LRRM is shown in ribbon and surface representation. Four arginine residues (857, 860, 880 and 884) on the surface of the C-terminal LRR are depicted in ball-and-stick models (carbon, gray; nitrogen, blue) and highlighted in orange. Their neighboring hydrophobic LRR residues are also shown in a stick model. C. Intra-subunit LRRCT-LRRM contact. Two α-helices of the LRRCT domain, Cα1 and Cα3, diagonally buttress the concave face of the LRRM domain. Side-chain and main-chain atoms of residues involved in the LRRCT-LRRM contact are depicted in a ball and stick representation. Hydrogen bonds are shown in black dashed lines. LRRM domain is shown in surface representation.
Numerous inter-domain contacts in the cNLRX1 structure
Three domains, LRRNT, LRRM and LRRCT, mutually interact with each other, generating various inter-domain interfaces to stabilize the monomer structure and enable formation of multiple oligomers, such as dimer, trimer and hexamer. First, an inter-domain contact between the LRRM and LRRCT domains (LRRM-LRRCT) occurs in the cNLRX1 monomer, where the LRRCT domain diagonally buttresses the concave face of the LRRM structure, burying ~630 Å2 of ASA of the LRRM domain (Figures 1B and 3C). The LRRM-LRRCT interactions include 7 hydrogen bonds, one salt bridge and 25 van der Waals contacts composed from residues between Cα1-Cα3 and the LRRM β-strands (Figure 3C).
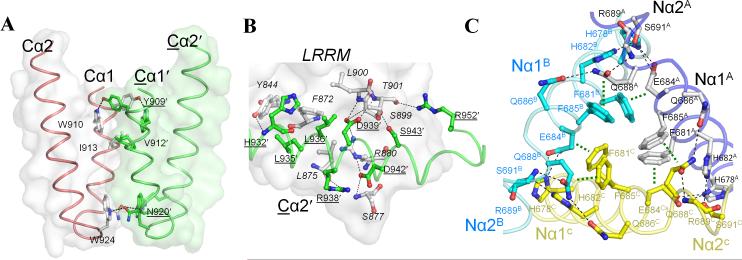
Besides the intra-subunit interactions with the LRRM, the LRRCT domain forms a parallel homodimeric interaction with its 2-fold symmetry-related partner (i.e. LRRCTA-LRRCTA′, LRRCTB-LRRCTC′, and LRRCTC-LRRCTB′) (Figure 1C left). The 2-fold assembly introduces two discrete symmetry contact areas of LRRCT-LRRCT′ (Figure 4A) and LRRCT′-LRRM (Figure 4B). The LRRCT-LRRCT′ contact results in a burial of ~530 Å2 of ASA through symmetrical interactions between Cα1 and Cα1' (Tyr909, Trp910, Val912, Ile913, Asn920 and Trp924) (Figure 4A). At the LRRCT′-LRRM contact, Cα2' partially covers the C-terminal end of cNLRX1 LRR7 and LRR8 with 10 H-bonds, 42 van der Waals contacts and ~510 Å2 of ASA buried (Figure 4B). In particular, H-bonds between Cα2' (Asp939', Ser943' and Arg952') and main chain of LRR8 (Ser899, Leu900 and Thr901) appear to stabilize the LRR8 β-strand and loop. Thus, the LRRCT domain plays a key role in cNLRX1 dimerization and provides structural support for the cNLRX1 LRR modules through intra-subunit and inter-subunit interactions.
Figure 4. Inter-subunit contacts for cNLRX1 oligomerization.
A. In the cNLRX1 dimer, the LRRCT domain packs parallel to the 2-fold symmetry-related LRRCT (LRRCT-LRRCT′ interaction) and the view shown is perpendicular to the 2-fold symmetry axis. Residues in Cα1-Cα1′, which symmetrically interact with each other, are shown in a ball-and-stick representation on their respective gray and green surfaces. B. LRRCT′-LRRM interactions in the cNLRX1 dimer. The C-terminal region of LRRM domain makes contacts with Cα2′ residues and Cα3′ Arg952 from its dimerization partner. LRRM residues are shown in gray with surface representation and Cα2′ residues and Arg952′ in green with cartoon display. Interface residues are highlighted in a ball-and-stick model. Hydrogen bonds are displayed in dashed lines. C. LRRNT-LRRNT′ interactions in the cNLRX1 trimer. Inter-subunit LRRNT-LRRNT′ interactions are formed at Nα1 and N-terminal Nα2 around the NCS three-fold axis. Subunit affiliation (A, B and C) is designated by a superscript with a different color scheme (subunit A, black; B, blue; C, yellow). Hydrogen bonds and stacking interactions are shown in black and green dashed lines, respectively. A network of hydrogen bonds is formed between the C-terminal region of Nα1 and the beginning of Nα2, including pairs of Glu686 and Gln688 residues that connect from one subunit to the other. For clarity, only main-chain atoms are drawn for Arg689.
The cNLRX1 forms a trimer in the ASU of the crystal by three-fold non-crystallographic symmetry (Figure S1C). Three Nα1 helices from subunits A, B, and C are arranged in a triangle (Nα1-triangle) at the center of the cNLRX1 trimer. The Nα1-triangle is surrounded by Nα2 and LRR1, generating LRRNT-LRRNT′ and LRRNT-LRRM′ contacts among three subunits (Figures 1C right and 4C). Around the three-fold axis, three equivalent sets of two phenylalanine residues, Phe681 and Phe685 from Nα1, form a six aromatic ring stack in a double-layered circle with an average ring-to-ring distance of ~ 4 Å. At the intercept of the Nα1-triangle, these phenylalanine residues make inter-subunit π···Cβ-Cγ and π···Cγ-Cδ stacking interactions such that, for example, Phe681A and Phe685A interact with the aliphatic moieties of Glu684C and Gln688C, whose side-chains H-bond with His678A, and Gln686A and Arg689C, respectively. His678A and His682A, nearby the phenylalanines (Figure 4C), provide additional inter-subunit interactions with Nα1C and Nα2C, through van der Waals interactions with Leu670C, Ala692C and Leu695C, and H-bonds with Gln688C and Ser691C. Such LRRNT-LRRNT′ interactions are buttressed by other NCS-related inter-domain contacts between Nα1A and the proximal N-terminal surface of LRRC (LRRNT-LRRM′) (Figures 1C right). Therefore, we conclude that the LRRNT domains function to stabilize the trimerization of three cNLRX1 dimers and the resulting hexamer buries of 20,210 Å2 of ASA.
To address the role of cNLRX1 oligomerization in NLRX1 function, we introduced dimerization- or trimerization-disrupting mutations by replacing interface residues with aspartate to introduce an unfavorable negative charge into their corresponding interfaces, which would thus disfavor oligomerization. When mutant cNLRX1 constructs were expressed in a baculovirus expression system, as for the wildtype cNLRX1, each mutant protein was found to be severely aggregated and could not be biophysically characterized. These mutations were then introduced into the full-length NLRX1 construct for cellular ROS study. Compared to the wildtype NLRX1, the mutant constructs substantially decreased poly I:C-stimulated ROS production in HeLa cells (Figure S3). These results suggest that NLRX1 dimerization and trimerization are both likely conserved in the full-length protein and should be maintained to provide NLRX1 stability and cellular function.
Specific RNA interaction of cNLRX1
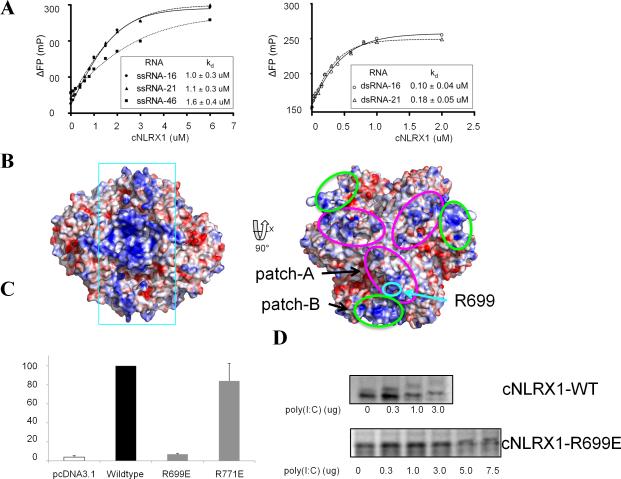
Taking advantage of known NLRX1 function in antiviral responses against viral RNA (Tattoli et al., 2008), we tested whether the RNA ligand binding site is located in the C-terminal region of NLRX1. We performed native polyacrylamide gel electrophoresis (native PAGE) analysis (Figure S4). The presence of poly I:C and single-stranded RNA (ssRNA) resulted in a mobility change of the purified cNLRX1 protein, whereas none of the double-stranded (ds) and ssDNAs shifted the position of the cNLRX1 bands. In order to assess binding affinity between the purified cNLRX1 and various RNA ligands, we monitored polarization shifts of 5′-fluorescein-labeled ssRNAs and dsRNAs in 16- and 21-mers, as well as a 46-mer F-ssRNA (ssRNA-46), upon cNLRX1 binding. All ssRNA and dsRNA ligands bound to cNLRX1 and their binding affinities were estimated to be 1~1.6 μM of Kd for ssRNAs and 0.1~0.2 μM of Kd for dsRNA ligands, indicating a 5 to 10 fold increase in binding affinity with dsRNA ligands (Figure 5A). These observations suggest that NLRX1 directly recognizes viral RNA for antiviral innate immunity and that this interaction occurs through the C-terminal region of NLRX1.
Figure 5. cNLRX1-RNA interaction.
A. Fluorescent polarization (FP) analysis using fluorescein-labeled ssRNA and dsRNA to determine binding affinities of cNLRX1 for RNA ligands. The y-axis indicates ΔFP, for which the FP value is normalized using that without cNLRX1. B. Electrostatic potential representation of the cNLRX1 structure. Electrostatic surface potentials of the cNLRX1 hexamer are shown in the same orientation as Figure 1C and contoured from red (-10kT/e) to blue (+10kT/e). A continuous positive surface is highlighted in cyan rectangle (Left). Two positive patches, patch-A and -B are depicted in magenta and green ellipses, respectively (Right) and expand to become a continuous basic surface via 2-fold crystal symmetry. Arg699 is highlighted in turquoise circle. C. The critical role of patch-A residue Arg699 in the cellular ROS response. Poly I:C-stimulated ROS production of NLRX1 mutants (R699E or R771E) in HeLa cells was normalized to that of full-length wildtype NLRX1. Arg771 is located on the LRRM convex surface away from the proposed RNA binding sites (patches-A and -B) and its mutant, R771E, was included as a control. As expected, NLRX1 R771E exhibited ROS production comparable to full-length, wildtype NLRX1. In contrast, Arg699 belongs to RNA binding patch-A (Figure 5B right) and its mutation to glutamate completely disrupted ROS production. D. Reduced RNA binding of cNLRX1 R699E compared to wildtype cNLRX1 (cNLRX1 WT). cNLRX1 WT and its mutant cNLRX1 R699E proteins were purified and their RNA binding were monitored by native PAGE. The cNLRX1 R699E binding to RNA was decreased by at least ~15-20 fold compared to that of cNLRX1 WT.
Potential RNA binding site on cNLRX1
Using our cNLRX1 crystal structure, we further investigated whether any discrete positive patch or residue was positioned on the cNLRX1 surface that could accommodate negatively charged RNA molecules. The surfaces of LRRM ascending lateral side and LRRCT residues on the same plane are highly negatively charged, thus implying that those residues are probably excluded from direct recognition of RNA ligands. On the other hand, a series of continuous positive patches on the cNLRX1 surface could be responsible for RNA binding (rectangle in Figure 5B left). A large continuous patch (patch-A, magenta ellipse in Figure 5B right) is created by residues from the LRR1-3 descending lateral side (Arg699, Arg724, His725, Arg753 and K754) and Arg689 of the LRRNT. In addition, three positive LRRCT residues (Arg944, Lys953 and Arg964) and C-terminal end of the LRRM domain (Arg857, Arg860, and Arg895) form another patch (patch-B, green in Figure 5B right). The patch-B expands its surface with its symmetry-related equivalent residues and the resulting combined patch-B-B′ is continuous with patch-A-A′ in the cNLRX1 hexamer (Figure 5B left). To support our prediction, a point mutation of Arg699 to a glutamate (cNLRX1 R699E) on patch-A ablated cellular ROS stimulation in HeLa cells (Figure 5C) and substantially disrupted poly I:C interaction in native PAGE (Figure 5D). Indeed, Arg699 is presumed to be present on the surface of full-length NLRX1 using a homology model based on the Apaf-1 structure (PDB ID code 1z6t, Riedl et al., 2005) (Figure S5). Therefore, we conclude that the positive charged arginine at residue 699 (patch-A) on the cNLRX1 is a putative RNA binding site, although other residues on positive patch-A or -B could be involved in NLRX1-RNA ligand interaction.
DISCUSSION
Here, we have presented the crystal structure of the C-terminal fragment of human NLRX1 protein (cNLRX1) and provided evidence to support specific interaction of cNLRX1 with an RNA ligand. The cNLRX1 exhibits a three-domain architecture of an N-terminal α-helical domain (LRRNT), central LRR modules (LRRM), and C-terminal three-helix bundle (LRRCT). Extensive inter-domain and inter-subunit interactions are observed and result in the trimerization of three dimers that leads to formation of a hexamer in the crystal. The cNLRX1 hexamer is also found in solution by size exclusion chromatography and analytical ultracentrifugation analyses.
Our cNLRX1 structure includes LRRNT that consists of two α-helices (Nα1 and Nα2). While the LRRNT helices are intertwined with each other, their associated interactions occur exclusively between subunits (inter-subunit interactions) and thereby deliver stabilizing elements to the NLRX1 hexamer. Simultaneously, the amphipathic Nα1 helix of LRRNT protects the hydrophobic core of the LRRM domain of the adjacent subunit in the trimer (e.g. LRRNTA for LRR1C) from exposure to solvent as an N-terminal capping motif of LRR structure. Thus, NLRX1 is unique in that N-terminal α-helices provide inter-subunit stabilization of the N-terminal LRRM as well as mediate subsequent higher oligomerization. However, the well-characterized TLR family, another innate immune receptor with LRRs displays a disulfide-linked β-hairpin that flanks the N-terminal LRRs to protect the LRR hydrophobic core, but the β-hairpin is not involved in TLR-oligomerization (Bella et al., 2008; Botos et al., 2011). An additional NLRX1-specific feature is that Gln668A at the N-terminus participates in an H-bond network of the β-sheet structure to a neighboring LRR motif by interacting with Asn702C-amide nitrogen of LRR1βC. The critical role of the Gln668 in NLRX1 protein folding was underscored by the extreme instability of a cNLRX1 variant (residues 669-975) that does not contain Gln668. Although we obtained crystals of this truncated protein, x-ray diffraction data were observed only to ~6 Å resolution. Taken together, LRRNT is indispensible for LRR structural integrity through mutually dependent LRRNT-LRRNT′ and LRRNT-LRRM′ interactions, resulting in NLRX1 oligomerization.
In general, NLRs are described as having a three-domain organization of ED-NOD-LRR domains (Ting and Davis, 2005). Alternatively, a four-domain architecture of ED-NOD-NAD-LRR can be designated, where NOD is subdivided into a nucleotide binding NOD (a region with Walker-A and -B motifs in a length of ~100 amino acids residues) and a NOD-associated domain (NAD, 200~300 residues) (Albrecht and Takken, 2006; Proell et al., 2008). In the conventional three-domain description, the NAD was neglected due to very low sequence identity in NLRs and because it does not contain any known sequence motif. Recently, NAD region has been recognized as a putative regulatory region that possesses ~3-4 independent structure motifs (Proell et al., 2008; Riedl et al., 2005). NAD domain mutations in NOD1 and NOD2 resulted in constitutively active phenotypes, but additional mutations in the first and second LRR motifs down-regulated the activity to a basal level (Tanabe et al., 2004). Similar NAD regulatory regions have also been found in plant NOD-LRR proteins (DeYoung and Innes, 2006). Although LRRNT residues were recognized within the putative LRR domain in an earlier primary sequence analysis of NLRX1 (Moore et al., 2008; Tattoli et al., 2008), our cNLRX1 structure reveals that LRRNT domain is a separate structure unit distinct from the LRRM domain. The contacts between LRRNT and LRRM stabilize the LRR fold and simultaneously involve NLRX1 oligomerization. Given those observations, if we consider that the LRRNT domain is part of the C-terminal region of the NAD domain, we can explain how such functional complementation between NAD-LRR residues in NOD1 and NOD2 could be accomplished by direct NAD-LRR interaction.
The effector domains (ED; pyrin in NLRP proteins and CARD in NOD proteins) of NLRs share common characteristic α-helical structures and mediate homotypic ED-ED dimerization for downstream signaling, but an ED has not been identified in NLRX1 by primary sequence analysis (Moore et al., 2008; Tattoli et al., 2008). Unexpectedly, our cNLRX1 structure contains another structural motif (the LRRCT domain) at its C-terminal end that diagonally buttresses the inner concave face of the LRR modules. The LRRCT exhibits an exclusively α-helical structure and mediates cNLRX1 dimerization via symmetric LRRCT-LRRCT′ interactions, as EDs in other NLRs do. These features allow us to propose that the LRRCT domain would correspond to an effector domain in NLRX1. If so, NLRX1 may adopt an atypical domain organization where the N-terminal MT sequences are required for mitochondrial localization, the central NOD-NAD-LRR domains for ligand recognition and the C-terminal ED for dimerization.
Upon activation, NLRs have been believed to self-oligomerize through NOD domains and heterodimerize via ED-ED interactions. But interactions between the LRR and other domains have been underappreciated. Our cNLRX1 structure reveals that its canonical LRR motifs are unexpectedly flanked by the N- and C-terminal α-helices that appear to play an essential role in stabilizing the LRR fold, as well as the overall oligomeric structure. The contacts between LRRM and LRRCT of cNLRX1 resembles heterodimeric interactions of LRR-containing receptor with protein ligand, which is also mediated through LRR concave side, for example, glycoprotein Ibα with von Willebrand factor A1 (Huizinga et al., 2002), ribonuclease inhibitor with ribonuclease A (Kobe and Deisenhofer, 1995) and mosquito LRR protein LRIM1 with APL1C (Baxter et al., 2010). Furthermore, mutation of the oligomerization interfaces in LRRNT and/or LRRCT of cNLRX1 resulted in no expression of soluble protein. Similar mutations of a full-length NLRX1 construct reduced cellular ROS stimulation in mammalian cells. Thus, the LRRNT and the LRRCT domains are important for protein stability by mediating cNLRX1 trimerization and dimerization, respectively. Such findings may provide a new insight, besides NOD-mediated oligomerization, that NLRs can adopt the various dynamic oligomerization states that should be required for the cellular signaling cascade upon ligand recognition.
Since the first discovery of the NLR family (Inohara et al., 1999), it has been hotly debated how NLR recognizes its ligands due to a lack of direct evidence for NLR interaction with its cognate ligands. It is noteworthy that there have been discrepancies and differences in the assigned function of NLRX1, either as antiviral response activator (Tattoli et al., 2008) or as a negative immune regulator of RIG-1 and TLRs (Allen et al., 2011; Moore et al., 2008; Xia et al., 2011), and in its sub-cellular localizations, either on the outer mitochondria membrane (Moore et al., 2008) or inside the mitochondrial matrix (Arnoult et al., 2009), thus requiring further clarification to decipher how NLRX1 could engage in such various functions. Another recent finding is that NLRX1 is also present to some extent in the cytoplasm and its association with the IKK complex varies on cellular activation (Xia et al., 2011). Collectively, the different cellular functions of NLRX1 could be attributable to differences in its sub-cellular localizations or associated signaling molecules. Despite these proposals of diverse functions, it is evident that the NLRX1 activities are exerted in response to intracellular viral RNA and poly I:C (Arnoult et al., 2009; Tattoli et al., 2008). Here, for the first time, we demonstrate a direct physical interaction of purified cNLRX1 (residues 629-975) with poly I:C, ssRNAs and dsRNAs. Based on electrostatic surface analysis of cNLRX1, we propose two basic patches as potential biding RNA binding sites. Indeed, a patch-A residue, Arg699 plays a critical role in RNA binding and cellular ROS response. Nevertheless, it is highly likely that the NLRX1-RNA recognition would occur using more than a single residue, as for TLR3 that uses its opposite ends, N- and C-terminal sites, for the RNA binding (Liu et al., 2008). Therefore, we conclude that C-terminal region of NLRX1 is indispensible not only for RNA binding (Arg699), but also for oligomerization of the full-length NLRX1 protein.
Finally, from the structure of a NLR distant homologue, apoptotic protease activating factor 1 (Apaf-1), we propose a model for the full-length NLRX1 hexamer. Our cNLRX1 structure lacks the NOD and the N-terminal region of the NAD domains that are present in the Apaf-1 structure. In the full-length NLRX1 model, the NOD-NAD domains extend from the LRR domain and decorate the exterior surface of the NLRX1 hexamer. The putative RNA binding regions, patches-A and -B, are still exposed to solvent and accessible to binding by RNA. Since the NOD domain is also exposed in the full-length NLRX1 hexamer model, the NOD domain would be available to self-oligomerize to a ring structure as well as to directly interact with other interacting partners, such as MAVS. To complete our understanding of the NLRX1 activation mechanism, crystal structures of NLRX1 complexed with its RNA ligand or with its interacting partners now need to be elucidated.
EXPERIMENTAL PROCEDURES
Expression and purification of cNLRX1
Numerous NLR constructs were generated and tested in Escherichia coli, Drosophila and baculovirus expression systems to obtain soluble and homogeneous recombinant proteins for structural studies of NLR members. One construct (669-975) that encodes a putative LRR domain of human NLRX1 provided sufficient soluble, homogeneous material for structural studies. However, proteins from the initial construct were relatively unstable and aggregated during purification. Crystals were obtained, but only diffracted to 6 Å resolution. The original construct was further modified to improve protein stability and crystal packing for improved x-ray diffraction. The optimized construct contains human NLRX1 residues 629 to 975 (cNLRX1). The cNLRX1 coding region was amplified by PCR with C-terminal Strep-Tactin II and His6 tags and ligated into a pAcGP67 baculovirus transfer vector (BD Biosciences). Recombinant baculovirus of cNLRX1 was obtained by transfecting SF9 insect cells with the cloned plasmid DNA and a linearized baculovirus DNA (AB vector). cNLRX1 was expressed in Hi5 insect cells for 2 days with amplified recombinant virus. The cells were harvested, resuspended in PBS, and lysed with a high pressure homogenizer (Avestin). Supernatant containing recombinant cNLRX1 protein was cleared by centrifugation at 10,000 rpm followed by filtration. cNLRX1 protein was purified by three-step column procedure using Ni-NTA (Qiagen), Strep-Tactin affinity (IBA), and gel filtration (GE Healthcare) chromatography. The purified recombinant cNLRX1 protein was 99% pure with a final yield of ~ 0.9 mg per 10 liter culture and its identity was confirmed by tandem MS/MS (TSRI Center for Mass Spectrometry).
Crystallization and data collection
cNLRX1 was crystallized at 23°C by sitting drop vapor diffusion. cNLRX1 crystals were obtained by mixing 0.5 μl of protein solution (9 mg/ml) with 0.7 μl of reservoir solution containing 18% PEG 1000 and 200 mM Tris, pH 8.0. Crystals were soaked in 5 mM potassium tetranitroplatinate for experimental phasing and cryo-protected in 35% PEG 1000 and 250 mM Tris pH 8.0. Native and platinum derivative x-ray diffraction data of cNLRX1 were collected at the Argonne Advanced Photon Source (APS) beamline 23ID-B, and the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 11-1, respectively. X-ray data were processed with MOSFLM (Leslie, 1992) and Scala (Evans, 1997). Data collection statistics are summarized in Table 1.
Structure determination and refinement
The cNLRX1 structure was determined by the MAD method (Table 1). Using intensity data from three wavelengths, four of heavy atom sites were located by SOLVE (Terwilliger and Berendzen, 1999). After solvent flipping (27% solvent content), as implemented in the crystallography & NMR system software (CNS) (Brünger et al., 1998), the map revealed clear density for α-helices and bulky side-chains, which were used to define the register of the residues with the amino-acid sequence. The cNLRX1 structure was iteratively built using COOT (Emsley and Cowtan, 2004) and refined with REFMAC5 using NCS restraints and TLS corrections (Vagin et al., 2004). Since the data sets collected from a platinum derivative crystal showed the best diffraction to 2.65 Å resolution, the final cNLRX1 structure was refined with the platinum peak wavelength data. The native structure at 2.9 Å resolution was also solved by molecular replacement with MolRep (CCP4, 1994), using the refined cNLRX1 structure as a search model. An overlay of these structures resulted in an r.m.s.d. of 0.1 Å, underscoring their structural identity regardless of platinum derivatization. Refinement statistics are summarized in Table 1. The model of cNLRX1 has excellent stereochemistry with just one outlier (Thr792 in the LRR4 loop, but in good agreement with its corresponding electron density) in the Ramachandran plot calculated by MolProbity (http://molprobity.biochem.duke.edu). Several regions were not built in the final model due to poor electron density that include the N-and C-terminal regions (residues 629-666 and 971-975), a LRR7-8 loop (887-893), a connecting loop between the LRRM and LRRCT domains (residues 902-905), and loops in LRRCTC (Figure S2). The overall Wilson B factor was estimated to 74.5 Å2, confirming that portions of the cNLRX1 structure are relatively flexible. Five cysteines are present in cNLRX1, but no disulfide bonds are observed, as expected for an intracellular protein. The cNLRX1 structure includes 4 platinum sites and 8 ordered water molecules bound to cNLRX1 protein residues.
The atomic coordinates and structure factors for cNLRX1 (PDB ID code 3UN9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University (www.rcsb.org). All figures were made using Pymol (DeLano, 2002) and electrostatic potentials were calculated with APBS
Measurement of cellular ROS production
Mammalian cell expression constructs of full-length NLRX1 (residues 1-975) with a C-terminal Flag tag were generated by PCR and ligated into pcDNA3.1A (Invitrogen). Matagenesis was done by QuickChange™ site-Directed mutagenesis kit (Stratagene). Cellular ROS measurement was carried out as described previously (Tattoli et al., 2008). Briefly, HeLa cells were transiently transfected with NLRX1 expression constructs or a negative control, pcDNA3.1A using FuGENE (Roche) for 18 hours. Subsequently, the transfected cells were stimulated by poly I:C (Invivogen). Intracellular ROS accumulation was detected by using the fluorescent probe (CM-H2DCFDA; Invitrogen) at 3 hours after stimulation.
Fluorescence polarization analysis
Fluorescence polarization analysis (Lundblad et al., 1996) was applied to measure the binding affinity of cNLRX1 for RNA ligands utilizing Tecan plate reader (Tecan Group Ltd.). Binding was monitored as a function of the increased polarization/anisotropy of fluoresceinated 16-, 21- and 46-mer RNAs (Oligosetc), when cNLRX1 was titrated into a 100 μL solution. Binding assays were carried out in triplicate in 25 mM Hepes, pH 7.4, 150 mM NaCl and 1 mM β-mercaptoenthanol at 25°C.
Supplementary Material
HIGHLIGHTS.
-The C-terminal fragment of NLRX1 (cNLRX1) assembles into a hexamer.
-cNLRX1 hexamerization occurs through trimerization of three dimers.
-The N- and C-terminal caps of cNLRX1 play key roles in its hexamerization.
- cNLRX1 can directly interact with RNA.
ACKNOWLEDGEMENTS
This work was supported by NIH grant AI042266 (IAW) and the Skaggs Institute for Chemical Biology. X-ray diffraction datasets were collected at SSRL beamline 11-1 and APS beamline 23ID-B. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Use of the APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The APS beamline 23ID-B has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Sciences (Y1-GM-1104). We thank Henry Tien and David Marciano (Robotics Core of the Joint Center for Structural Genomics) for automated crystal screening, Xiaoping Dai and Marc A. Elsliger (The Scripps Research Institute) for expert crystallographic and computational support, and Robyn Stanfield (The Scripps Research Institute) and Kate J. Newberry (The University of Texas M. D. Anderson Cancer Center) for helpful discussions. We also thank Dr. Andrey Bobkov (Protein production analysis facility, Sanford-Burnham Medical Research Institute) for the analytical ultracentrifugation analysis. This is manuscript no. 20843 from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicting financial interests.
REFERENCES
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter RH, Steinert S, Chelliah Y, Volohonsky G, Levashina EA, Deisenhofer J. A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:16817–16822. doi: 10.1073/pnas.1010575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira S. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Brickey WJ, Barker BR, Kwan M, Taxman DJ, Accavitti-Loper MA, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186:1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nature Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieude P, Guedj M, Wipff J, Ruiz B, Riemekasten G, Airo P, Melchers I, Hachulla E, Cerinic MM, Diot E, et al. NLRP1 influences the systemic sclerosis phenotype: a new clue for the contribution of innate immunity in systemic sclerosis-related fibrosing alveolitis pathogenesis. Ann Rheum Dis. 2011;70:668–674. doi: 10.1136/ard.2010.131243. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scala. Joint CCP4 and ESF-EACBM Newsletter. 1997;33:22–24. [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Jehanno M, Mengin-Lecreulx D, Sansonetti PJ, Alzari PM, Philpott DJ. Identification of the critical residues involved in peptidoglycan detection by Nod1. J Bio Chem. 2005;280:38648–38656. doi: 10.1074/jbc.M509537200. [DOI] [PubMed] [Google Scholar]
- Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Hillig RC, Renault L, Vetter IR, Drell TT, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Ann Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Bio Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Kobe B. Crystallization and crystal structure determination of ribonuclease A-ribonuclease inhibitor protein complex. Methods Mol Biol. 2001;160:201–211. doi: 10.1385/1-59259-233-3:201. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Leslie A. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WG, Zou YF, Feng XL, Yuan FL, Gu YL, Li X, Li CW, Jin C, Li JP. Association of NOD1 (CARD4) insertion/deletion polymorphism with susceptibility to IBD: a meta-analysis. World J Gastroenterol. 2010;16:4348–4356. doi: 10.3748/wjg.v16.i34.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Molnar T, Hofner P, Nagy F, Lakatos PL, Fischer S, Lakatos L, Kovacs A, Altorjay I, Papp M, Palatka K, et al. NOD1 gene E266K polymorphism is associated with disease susceptibility but not with disease phenotype or NOD2/CARD15 in Hungarian patients with Crohn's disease. Dig Liver Dis. 2007;39:1064–1070. doi: 10.1016/j.dld.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Pontillo A, Vendramin A, Catamo E, Fabris A, Crovella S. The missense variation Q705K in CIAS1/NALP3/NLRP3 gene and an NLRP1 haplotype are associated with celiac disease. Am J Gastroenterol. 2011;106:539–544. doi: 10.1038/ajg.2010.474. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PloS one. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Rosenstiel P, Till A, Schreiber S. NOD-like receptors and human diseases. Microbes Infect. 2007;9:648–657. doi: 10.1016/j.micinf.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Tanabe, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO reports. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Davis BK. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annual review of immunology. 2005;23:387–414. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, et al. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nature Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.