Abstract
Vitamin D deficiency has been associated with several common diseases, including cancer and is being investigated as a possible risk factor for these conditions. We reported the striking prevalence of vitamin D deficiency in Scotland. Previous epidemiological studies have reported an association between low dietary vitamin D and colorectal cancer (CRC). Using a case-control study design, we tested the association between plasma 25-hydroxy-vitamin D (25-OHD) and CRC (2,001 cases, 2,237 controls). To determine whether plasma 25-OHD levels are causally linked to CRC risk, we applied the control function instrumental variable (IV) method of the Mendelian randomization (MR) approach using four single nucleotide polymorphisms (rs2282679, rs12785878, rs10741657, rs6013897) previously shown to be associated with plasma 25-OHD. Low plasma 25-OHD levels were associated with CRC risk in the crude model (odds ratio (OR): 0.76, 95% Confidence Interval (CI): 0.71, 0.81, p: 1.4×10−14) and after adjusting for age, sex and other confounding factors. Using an allele score that combined all four SNPs as the IV, the estimated causal effect was OR 1.16 (95% CI 0.60, 2.23), whilst it was 0.94 (95% CI 0.46, 1.91) and 0.93 (0.53, 1.63) when using an upstream (rs12785878, rs10741657) and a downstream allele score (rs2282679, rs6013897), respectively. 25-OHD levels were inversely associated with CRC risk, in agreement with recent meta-analyses. The fact that this finding was not replicated when the MR approach was employed might be due to weak instruments, giving low power to demonstrate an effect (<0.35). The prevalence and degree of vitamin D deficiency amongst individuals living in northerly latitudes is of considerable importance because of its relationship to disease. To elucidate the effect of vitamin D on CRC cancer risk, additional large studies of vitamin D and CRC risk are required and/or the application of alternative methods that are less sensitive to weak instrument restrictions.
Introduction
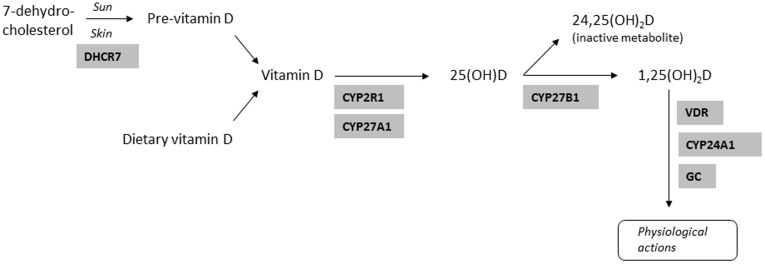
Vitamin D can be ingested or synthesized in the skin from inactive precursors through the action of UV sunlight. Its active form, 1,25(OH)2D (1,25(OH)2D2 and/or 1,25(OH)2D3) is produced after two hydroxylation steps in the liver and kidneys ( Figure 1 ) [1]. The prevalence of vitamin D deficiency in Scotland is high due to high northern latitude, often cloudy weather (lack of sunlight impairs vitamin D synthesis during winter months), indoors oriented lifestyle and poor diet, and so routine vitamin D and calcium supplementation for the housebound (>65 years old) is recommended [2]. In a recent study of over 2000 healthy individuals living in Scotland, we found that 77.5% of the individuals were vitamin D deficient [3]. Although the Reference Nutrient Intake (RNI) of vitamin D by the Scientific Advisory Committee on Nutrition in Scotland for people over 65 years old is 10 ug per day [4], there is a great variation of the recommended daily allowances (RDA) by different research groups and institutions [5]–[8].
Figure 1. Vitamin D metabolic pathway.
Vitamin D has been considered relevant to skeletal disease and calcium metabolism, but there is growing evidence that vitamin D deficiency might be a risk factor for cancer, cardiovascular, metabolic, infectious and autoimmune diseases [3]. In particular, vitamin D may affect colorectal cancer (CRC) risk via its binding to the vitamin D receptor (VDR) [9] influencing cell proliferation, differentiation, apoptosis and angiogenesis [10], [11] or affecting insulin resistance [12]. Results from case-control and cohort studies are inconclusive, but results from cohort studies measuring 25-hydroxy-vitamin D (25-OHD) in the blood or the serum are more consistent indicating an inverse association with CRC [13]–[15].
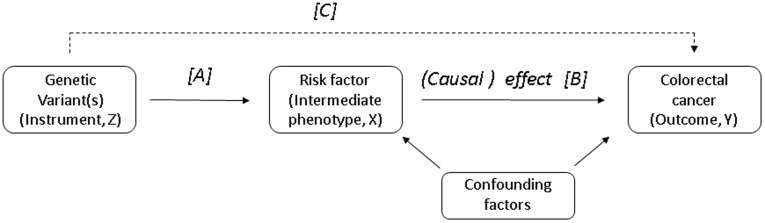
Establishing causal relationships between environmental exposures and common diseases using conventional methods of observational studies is problematic due to unresolved confounding, reverse causation and selection bias [16]. The theory underpinning the Mendelian randomization (MR) approach is based on the random assortment of alleles at the time of gamete formation, which is equivalent to a randomized controlled trial in which people are randomly allocated to therapeutic interventions. The main concept of a MR study is based on three relationships: genotype–intermediate phenotype; intermediate phenotype–disease; genotype–disease [17], [18] and it can be used to identify causal environmental risk factors without the several potential problems of observational epidemiology [19]. The MR approach can also strengthen causal conclusions by limiting reverse causation problems (biological, through exposure assignment, due to reporting bias), selection bias and regression dilution bias [19]. Figure 2 illustrates how this concept is applied to inform causal inference.
Figure 2. Directed acyclic graph (DAG) showing the instrumental variable assumptions underpinning our Mendelian randomisation study (note the instrument is not allowed to have a direct effect on the outcome, hence this line is dashed).
Instrumental variable models use associations C and A to estimate the causal effect of a risk factor on an outcome (B).
The analytic approach employed here for MR is the instrumental variable (IV) model, in which the genetic variant is treated as an instrument which is assumed to be associated with the disease only through its association with the intermediate phenotype [18]. This requires firstly the identification of one or more genetic variants (typically a single nucleotide polymorphism or SNP) as the IV that is known from published data to be associated with the phenotype [18]. The three key assumptions underlying the MR approach are: a) the genotype is associated with the phenotype; b) the genotype is independent of measured and unmeasured confounders; and c) that the effect of genotype on outcome is mediated only through the intermediate phenotype (no pleiotropy) [17], [18].
In this study, we set out to evaluate the relationship between CRC, plasma 25-OHD levels and genotype at 4 genetic loci tagging genes involved in vitamin D metabolism (Table S1) and which have previously been shown to be associated with plasma vitamin D levels in a pooled meta-analysis of Genome Wide Association Studies [20]. In order to estimate whether there is a causal relationship between plasma 25-OHD and CRC risk we applied the control function IV estimator.
Methods
Ethics Statement
Ethical approval for the SOCCS study was obtained from the MultiCentre Research Ethics committee for Scotland (reference number 01/0/05) and from the Research and Development Office of NHS Lothian (reference number 2003/W/GEN/05).
Study Population
We studied a subset of 2,001 cases and 2,237 controls from a case-control study of CRC (Study Of Colorectal Cancer in Scotland, SOCCS). We aimed to recruit all incident cases (1999–2006) of adenocarcinoma of colorectum presenting to surgical units in Scotland (18–79 years old). Exclusions were patient death before ascertainment, patient too ill to participate, recurrent cases, or patient unable to give informed consent due to learning difficulties or other medical conditions. We recruited about 40% of all incident cases in Scotland over the study period. During the same period controls were drawn randomly from a population-based register (community health index) and invited to participate. Participation rates among those approached were approximately 58% for cases and an estimated 57% for controls. More than 99% of the study participants were white Caucasian (see [21] for further recruitment details).
The subjects completed one questionnaire with lifestyle and cancer information and they were asked to report their status one year prior diagnosis or recruitment, including information about their general medical history, physical activity, smoking status, intake of any regular intake of aspirin and NSAIDs, height, weight and waist circumference were recorded. Additionally a semi-quantitative food frequency questionnaire (Scottish Collaborative Group FFQ, Version 6.41) was completed by participants (http://www.foodfrequency.org), which consisted of 150 foods and the individuals were asked to describe the amount and frequency of each food on the list they have eaten a year prior to diagnosis or recruitment. Further information about the questionnaires were presented in detail previously [21]. In addition, each recruited cancer subject was assigned an American Joint Committee on Cancer (AJCC) stage derived from a synthesis of clinical, pathological and imaging information [22]. Finally, for a subset of the cancer cases (1,423, of those 1,376 with 25-OHD measured) there was information about the symptoms they had developed before recruitment. We grouped the cases into four categories: (1) no symptoms (190 cases), (2) mild symptoms (290; including: change in bowel habit, constipation, intermittent diarrhoea and constipation, more frequent stools, diarrhoea, loose stools, excess wind, mucus in stool and abdominal discomfort) and (3) severe symptoms (220; including rectal bleeding, vomiting, weight loss, loss of energy, loss of appetite and nausea) and (4) both, mild and severe symptoms (676).
Family history risk was determined according to the Scottish Executive cancer guidelines (http://www.sehd.scot.nhs.uk/), The criteria for high family history risk of colorectal cancer are: 1) at least three family members affected by colorectal cancer or at least two with colorectal cancer and one with endometrial cancer in at least two generations; one affected relative must be <50 years old at diagnosis and one of the relatives must be a first degree relative of the other two; or 2) presence of the HNPCC syndrome; or 3) untested first degree relatives of known gene carriers. The criteria for moderate risk are: 1) one first degree relative affected by colorectal cancer when aged <45 years old; or 2) two affected first degree relatives with one aged <55 years old; or 3) three affected relatives with colorectal or endometrial cancer, who are first degree relatives of each other and one a first degree relative of the consultant. Individuals that do not fulfil all the above criteria are classified as low family history risk (Scottish Executive cancer guidelines). For this analysis family history was coded as low vs. medium/high family history of CRC.
Measurement of Plasma 25-OHD
The liquid chromatography-tandem mass spectrometry (LC-MS/MS) method was used to measure 25-hydroxyvitamin D3 and D2. This paper presents the 25-OHD (the total of 25-OHD2 and 25-OHD3); however, most of our samples contained no D2 (<3 ng/ml). The lower limit of detection with the LC-MS/MS method was 4 ng/ml for D3 [23]. The LC MS/MS method was performed following standard protocols and appropriate quality control procedures (including multiple measurements of the same sample from our cohort and standardization against standard reference material, SRM 972) and it has been rated as the preferred 25-OHD measurement method for population studies by an international panel of experts [23]. More details about this method can be found elsewhere [23], [24]. For the analysis 25-OHD measurements were standardized to remove the prominent effect of the month when blood was taken on the 25-OHD concentration, as described in detail in Zgaga et al [3].
Genotyping Data
DNA samples were accurately quantified by Pico-GreenTM and quality controlled prior to dispatch. Genotyping was undertaken using TaqMan in the Wellcome Trust Clinical Research Facility (WTCRF) in Edinburgh. 2,000 subjects for the rs2282679 were genotyped as part of an array-based candidate gene approach, using the Illumina Infinium I Custom array platform and performed by Illumina (San Diego). Case and control DNA samples were stored, genotyped and analysed in the same way. In addition to avoid potential systematic batch-to-batch variation or bias, samples were anonymised as to affection status and were randomly distributed within plates. Data were subject to Illumina or WTCRF quality control procedures. Assumptions of Hardy-Weinberg Equilibrium (HWE) were tested using a chi-squared test.
Statistical Analysis
The statistical package used was Stata version 11.0 (Stata Corp, College Station, Texas). Participants were divided into quintiles based on the combined distributions of cases and controls. Logistic regression models were used to estimate the strength of association between CRC risk and vitamin D plasma levels. The associations were tested in three logistic regression models (crude model, model I and model II). Model I was corrected for age and sex and Model II was corrected for age, sex, Carstairs Deprivation Index, energy (MJoules/day), smoking (non-smoker, former smoker and current smoker), body mass index (BMI, kg/m2, continuous), regular NSAID intake (yes vs. no), family history (low vs. medium/high) of cancer and physical activity (hours of cycling and other sports activities, 4 groups). We also tested the association after sex, stage of cancer at diagnosis (AJCC), presence of symptoms and time between diagnosis and recruitment stratification. In addition the association between CRC and rs2282679, rs12785878, rs10741657 and rs6013897 was tested. Dataset for this analysis was larger, and it comprised all SOCCS study participants for whom genotyping of selected SNPs was successful (up to 5,449). We also tested the interaction between genotype and vitamin D plasma levels on CRC by comparing a model with and without an interaction term between the two variables, using a likelihood ratio test. Assumptions of Hardy-Weinberg Equilibrium were tested using a chi-squared test.
To estimate the causal odds ratio we applied the control function IV estimator for a 3-level categorical instrument Z coded 0, 1, 2 (SNP) a continuous intermediate phenotype X (plasma 25-OHD3) and a binary outcome Y (CRC). The first stage of the control function is a linear regression of the intermediate phenotype (X) on the instrument(s) (Z), which generates predicted values for the intermediate phenotype. The second stage is a logistic regression of the outcome (Y) on the predicted values of the intermediate phenotype including the estimated residuals from the first-stage linear regression in the second-stage logistic regression [25]. The rationale is that the first-stage residuals may be correlated with unmeasured confounding factors. In addition to a crude model, we also adjusted for age and sex. The strength of the applied instruments were evaluated using the F statistics from the first stage linear regression, with values lower than 10 taken as evidence of a weak instrument [26]. Finally, we applied four additional IV estimators which are presented and described in the supplementary material (Methods S1).
Results
Levels of Plasma 25-OHD and CRC
Colorectal cancer risk was associated with lower levels of plasma 25-OHD in the crude model (Odds ratio (OR): 0.76, 95% Confidence Interval (CI): 0.71, 0.81, p: 1.4×10−14), after adjusting for age and sex (OR: 0.75, 95% CI: 0.70, 0.81, p: 9.1×10−15) and after adjusting for age, sex, Carstairs Deprivation Index, energy, smoking, BMI, regular NSAID intake, family history of cancer and physical activity (OR: 0.75, 95% CI: 0.69, 0.81, p: 4.6×10−12) ( Table 1 ). The 25-OHD CRC association was stronger for men (crude model: OR: 0.68, 95% CI: 0.62, 0.75, p: 1.1×10−13) than women (crude model: OR: 0.84, 95% CI: 0.76, 0.93, p: 0.0009) (Table S2). The 25-OHD CRC association was similar for early (crude model: OR: 0.79, 95% CI: 0.72, 0.86, p: 2.0×10−8) versus late AJCC stage (crude model: OR: 0.74, 95% CI: 0.68, 0.80, p: 5.8×10−12; Table S3). Furthermore, the 25-OHD CRC association was weaker for those CRC patients that had no symptoms at the time of the diagnosis (crude model: OR: 0.87, 95% CI: 0.73, 1.02, p: 0.09), when compared to those with mild (crude model: OR: 0.80, 95% CI: 0.70, 0.91, p: 0.001) or severe symptoms (crude model: OR: 0.78, 95% CI: 0.67, 0.90, p: 0.001; Table S4). Finally, the 25-OHD CRC association was similar for those CRC patients that were recruited soon after diagnosis (crude model: OR: 0.77, 95% CI: 0.71, 0.83, p: 7.8×10−5), when compared to those who were recruited later (crude model: OR: 0.76, 95% CI: 0.70, 0.83, p: 1.3×10−10; Table S5).
Table 1. Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk.
| N | Crude model | Model I* | Model II† | ||||||||
| Standard logistic regression analysis | Cases | Controls | OR | 95% CI | p-value | OR | 95% CI | p-value | OR | 95% CI | p-value |
| 25-OHD (continuous; ng/ml) | 2001 | 2237 | 0.76 | 0.71, 0.81 | 1.4×10−14 | 0.75 | 0.70, 0.81 | 9.13×10−15 | 0.75 | 0.69, 0.81 | 4.6×10−12 |
| 25-0HD (binary) | |||||||||||
| <10 ng/ml | 973 | 826 | 1.00 | 1.00 | 1.00 | ||||||
| ≥10 ng/ml | 1028 | 1411 | 0.62 | 0.55, 0.70 | 1.2×10−14 | 0.61 | 0.54, 0.69 | 7.1×10−15 | 0.62 | 0.54, 0.72 | 6.1×10−11 |
| 25-0HD (quintiles) | |||||||||||
| <5.31 | 486 | 366 | 1.00 | 1.00 | 1.00 | ||||||
| 5.31–9.39 | 461 | 425 | 0.82 | 0.68, 0.99 | 0.04 | 0.82 | 0.68, 0.99 | 0.04 | 0.75 | 0.61, 0.94 | 0.01 |
| 9.39–13.20 | 376 | 430 | 0.66 | 0.54, 0.80 | <0.0005 | 0.66 | 0.54, 0.80 | <0.0005 | 0.67 | 0.53, 0.83 | <0.0005 |
| 13.20–18.36 | 358 | 505 | 0.53 | 0.44, 0.65 | <0.0005 | 0.53 | 0.44, 0.64 | <0.0005 | 0.49 | 0.39, 0.61 | <0.0005 |
| ≥18.36 | 320 | 511 | 0.47 | 0.39, 0.57 | <0.0005 | 0.46 | 0.38, 0.56 | <0.0005 | 0.45 | 0.35, 0.56 | <0.0005 |
| p-value trend | 2.2×10−18 | 7.8×10−19 | 3.6×10−15 | ||||||||
Adjusted for age and sex.
Adjusted for age, sex, Carstairs Deprivation Index, energy (MJoules/ day), smoking (non-smoker, former smoker and current smoker), body mass index (BMI, kg/m2, continuous), regular Non Steroidal Anti-Inflammatory Drug (NSAID) intake (yes vs. no), family history of cancer and physical activity (hours of cycling and other sports activities, 4 groups).
Genotype and Plasma 25-OHD Levels
There was no evidence for departure from HWE for all four SNPs: rs2282679 p-value = 0.25, rs12785878 p-value = 0.78, rs10741657 p-value = 0.07 and rs6013897 p-value = 0.52. The A allele of rs2282679 and the T allele of rs12785878 were associated with higher levels of plasma 25-OHD ( Table 2 ). In particular, we found that rs2282679 and rs12785878 genotypes were associated with a decreased risk of 25-OHD deficiency defined as <10 ng/ml (rs2282679: for each A allele OR = 0.88, 95% CI 0.80, 0.98, p = 0.02; rs12785878: for each T allele OR = 0.89, 95% CI 0.79, 1.00, p = 0.05; rs10741657; Table 2 ). These associations were not different when we restricted the analysis only in the controls (data not shown).
Table 2. Association between plasma 25-0HD levels and rs2282679, rs12785878, rs10741657 and rs6013897.
| 25-0HD levels | Crude model | Model I* | ||||||
| Standard logistic regression analysis | <10 ng/ml | ≥10 ng/ml | OR | 95% CI | p-value | OR | 95% CI | p-value |
| rs2282679 | ||||||||
| CC | 133 | 165 | 1.00 | 1.00 | ||||
| AC | 698 | 905 | 0.96 | 0.75, 1.23 | 0.73 | 0.95 | 0.74, 1.22 | 0.69 |
| AA | 767 | 1163 | 0.82 | 0.64, 1.05 | 0.11 | 0.80 | 0.63, 1.03 | 0.09 |
| per A allele | 0.88 | 0.80, 0.98 | 0.02 | 0.87 | 0.79, 0.97 | 0.01 | ||
| rs12785878 | ||||||||
| GG | 72 | 66 | 1.00 | 1.00 | ||||
| TG | 470 | 620 | 0.69 | 0.49, 0.99 | 0.04 | 0.69 | 0.48, 0.99 | 0.04 |
| TT | 980 | 1366 | 0.65 | 0.47, 0.93 | 0.02 | 0.65 | 0.46, 0.92 | 0.02 |
| per T allele | 0.89 | 0.79, 1.00 | 0.05 | 0.89 | 0.79, 1.00 | 0.05 | ||
| rs10741657 | ||||||||
| GG | 512 | 672 | 1.00 | 1.00 | ||||
| GA | 385 | 925 | 0.97 | 0.84, 1.13 | 0.71 | 0.97 | 0.83, 1.13 | 0.71 |
| AA | 211 | 318 | 0.87 | 0.71, 1.07 | 0.19 | 0.88 | 0.71, 1.08 | 0.22 |
| per A allele | 0.94 | 0.84, 1.04 | 0.23 | 0.94 | 0.85, 1.04 | 0.25 | ||
| rs6013897 | ||||||||
| AA | 47 | 72 | 1.00 | 1.00 | ||||
| TA | 470 | 599 | 1.20 | 0.82, 1.77 | 0.35 | 1.19 | 0.82, 1.75 | 0.39 |
| TT | 968 | 1319 | 1.12 | 0.77, 1.64 | 0.54 | 1.11 | 0.76, 1.62 | 0.60 |
| per T allele | 0.98 | 0.87, 1.11 | 0.73 | 0.98 | 0.86, 1.10 | 0.70 | ||
Adjusted for age and sex.
Genotype and CRC
Overall there was no evidence of an association between any of the four SNPs and CRC risk ( Table 3 ). When we stratified according to plasma 25-OHD levels, the rs10741657 SNP was associated with a decreased CRC risk for those of low plasma 25-OHD levels (per A allele OR = 0.88, 95% CI 0.75, 1.02, p = 0.09) and with an increased CRC risk for those of high plasma 25-OHD levels (OR = 1.12, 95% CI 0.98, 1.27, p = 0.09), with a p-value of interaction (p = 0.05).
Table 3. Association between colorectal cancer risk and rs2282679, rs12785878, rs10741657 and rs6013897.
| Standard logistic regression analysis* | Case/control status | Mean (SD) of25-0HD (ng/ml) | Crude model | Model I† | ||||||
| All-samples | Cases | Controls | Cases | Controls | OR | 95% CI | p-value | OR | 95% CI | p-value |
| rs2282679 | ||||||||||
| CC | 216 | 218 | 11.63 (7.45) | 12.59 (8.45) | 1.00 | 1.00 | ||||
| AC | 1130 | 1160 | 11.01 (7.82) | 13.29 (8.24) | 0.98 | 0.80, 1.21 | 0.87 | 0.99 | 0.81, 1.21 | 0.92 |
| AA | 1325 | 1400 | 12.00 (7.86) | 14.01 (8.65) | 0.96 | 0.78, 1.17 | 0.66 | 0.96 | 0.78, 1.17 | 0.68 |
| per A allele | 0.97 | 0.90, 1.06 | 0.55 | 0.97 | 0.90, 1.06 | 0.55 | ||||
| rs12785878 | ||||||||||
| GG | 81 | 112 | 9.16 (7.29) | 10.67 (7.11) | 1.00 | 1.00 | ||||
| TG | 749 | 798 | 10.89 (7.50) | 13.50 (8.20) | 1.30 | 0.96, 1.76 | 0.09 | 1.30 | 0.96, 1.76 | 0.09 |
| TT | 1667 | 1713 | 11.69 (7.72) | 13.59 (8.59) | 1.35 | 1.00, 1.80 | 0.05 | 1.35 | 1.00, 1.81 | 0.05 |
| per T allele | 1.08 | 0.98, 1.20 | 0.11 | 1.08 | 0.98, 1.20 | 0.11 | ||||
| rs10741657 | ||||||||||
| GG | 851 | 855 | 11.15 (7.58) | 13.82 (8.88) | 1.00 | 1.00 | ||||
| GA | 1082 | 1201 | 11.54 (7.88) | 13.19 (8.25) | 0.91 | 0.80, 1.03 | 0.12 | 0.90 | 0.79, 1.02 | 0.11 |
| AA | 402 | 371 | 11.82 (7.62) | 13.78 (8.51) | 1.09 | 0.92, 1.29 | 0.33 | 1.08 | 0.91, 1.29 | 0.35 |
| per A allele | 1.01 | 0.94, 1.10 | 0.72 | 1.01 | 0.93, 1.10 | 0.77 | ||||
| rs6013897 | ||||||||||
| AA | 92 | 83 | 12.44 (8.83) | 13.89 (11.98) | 1.00 | 1.00 | ||||
| TA | 739 | 806 | 11.45 (7.29) | 12.84 (8.44) | 0.83 | 0.60, 1.13 | 0.24 | 0.84 | 0.61, 1.15 | 0.27 |
| TT | 1615 | 1648 | 11.32 (7.83) | 13.72 (8.34) | 0.88 | 0.65, 1.20 | 0.43 | 0.90 | 0.66, 1.21 | 0.48 |
| per T allele | 1.02 | 0.92, 1.13 | 0.71 | 1.02 | 0.92, 1.13 | 0.68 | ||||
| Allele score | 2815 | 2924 | 1.00 | 0.97, 1.04 | 0.87 | 1.01 | 0.97, 1.04 | 0.69 | ||
| “Synthesis” allele score | 2630 | 2761 | 1.04 | 0.98, 1.10 | 0.17 | 1.04 | 0.99, 1.10 | 0.14 | ||
| “Metabolism” allele score | 2779 | 2884 | 0.99 | 0.94, 1.05 | 0.79 | 0.99 | 0.94, 1.05 | 0.82 | ||
| <10ng/ml 25-0HD levels | Cases | Controls | OR | 95% CI | p-value | OR | 95% CI | p-value | ||
| rs2282679 | ||||||||||
| CC | 66 | 67 | 6.01 (2.84) | 5.71 (3.05) | 1.00 | 1.00 | ||||
| AC | 392 | 306 | 5.21 (2.90) | 5.54 (2.97) | 1.30 | 0.90, 1.89 | 0.17 | 1.29 | 0.89, 1.88 | 0.17 |
| AA | 399 | 368 | 5.47 (2.78) | 5.81 (2.79) | 1.10 | 0.76, 1.59 | 0.61 | 1.10 | 0.76, 1.59 | 0.61 |
| per A allele | 0.96 | 0.82, 1.12 | 0.58 | 0.96 | 0.82, 1.12 | 0.60 | ||||
| rs12785878 | ||||||||||
| GG | 32 | 40 | 3.98 (2.79) | 5.18 (3.12) | 1.00 | 1.00 | ||||
| TG | 249 | 221 | 5.37 (2.87) | 5.88 (2.81) | 1.41 | 0.86, 2.32 | 0.18 | 1.41 | 0.86, 2.32 | 1.18 |
| TT | 526 | 454 | 5.52 (2.83) | 5.66 (2.90) | 1.45 | 0.89, 2.34 | 0.13 | 1.46 | 0.90, 2.36 | 0.13 |
| per T allele | 1.10 | 0.93, 1.31 | 0.27 | 1.11 | 0.93, 1.32 | 0.25 | ||||
| rs10741657 | ||||||||||
| GG | 296 | 216 | 5.55 (2.84) | 5.60 (2.99) | 1.00 | 1.00 | ||||
| GA | 346 | 339 | 5.45 (2.81) | 5.74 (2.85) | 0.74 | 0.59, 0.94 | 0.01 | 0.74 | 0.59, 0.94 | 0.01 |
| AA | 113 | 98 | 5.11 (2.73) | 5.61 (2.79) | 0.84 | 0.61, 1.16 | 0.29 | 0.84 | 0.61, 1.17 | 0.31 |
| per A allele | 0.88 | 0.75, 1.02 | 0.09 | 0.88 | 0.75, 1.02 | 0.10 | ||||
| rs6013897 | ||||||||||
| AA | 23 | 24 | 5.00 (2.62) | 4.55 (2.71) | 1.00 | 1.00 | ||||
| TA | 237 | 233 | 5.66 (2.74) | 5.36 (3.06) | 1.06 | 0.58, 1.93 | 0.85 | 1.07 | 0.59, 1.96 | 0.82 |
| TT | 531 | 437 | 5.35 (2.84) | 5.88 (2.78) | 1.27 | 0.71, 2.28 | 0.43 | 1.29 | 0.72, 2.31 | 0.40 |
| per T allele | 1.17 | 0.97, 1.41 | 0.10 | 1.18 | 0.98, 1.42 | 0.09 | ||||
| ≥10 ng/ml 25-0HD levels | Cases | Controls | OR | 95% CI | p-value | OR | 95% CI | p-value | ||
| rs2282679 | ||||||||||
| CC | 69 | 96 | 17.01 (6.46) | 17.34 (7.68) | 1.00 | 1.00 | ||||
| AC | 380 | 525 | 17.00 (6.71) | 17.80 (6.86) | 1.01 | 0.72, 1.41 | 0.97 | 1.01 | 0.72, 1.42 | 0.94 |
| AA | 487 | 676 | 17.35 (6.53) | 18.48 (7.40) | 1.00 | 0.72, 1.39 | 0.99 | 1.00 | 0.72, 1.39 | 0.99 |
| per A allele | 1.00 | 0.87, 1.14 | 0.98 | 0.99 | 0.87, 1.14 | 0.93 | ||||
| rs12785878 | ||||||||||
| GG | 23 | 43 | 16.36 (5.11) | 15.78 (5.86) | 1.00 | 1.00 | ||||
| TG | 246 | 374 | 16.47 (6.55) | 18.00 (6.91) | 1.23 | 0.72, 2.09 | 0.45 | 1.24 | 0.73, 2.11 | 0.43 |
| TT | 584 | 782 | 17.24 (6.41) | 18.21 (7.34) | 1.40 | 0.83, 2.34 | 0.21 | 1.40 | 0.83, 2.35 | 0.20 |
| per T allele | 1.15 | 0.98, 1.35 | 0.09 | 1.15 | 0.98, 1.35 | 0.09 | ||||
| rs10741657 | ||||||||||
| GG | 271 | 401 | 17.27 (6.30) | 18.26 (7.78) | 1.00 | 1.00 | ||||
| GA | 383 | 542 | 17.04 (6.87) | 17.85 (7.00) | 1.05 | 0.85, 1.28 | 0.67 | 1.04 | 0.85, 1.27 | 0.70 |
| AA | 148 | 170 | 16.94 (5.99) | 18.50 (6.98) | 1.29 | 0.98, 1.69 | 0.07 | 1.28 | 0.98, 1.68 | 0.07 |
| per A allele | 1.12 | 0.98, 1.27 | 0.09 | 1.11 | 0.98, 1.27 | 0.10 | ||||
| rs6013897 | ||||||||||
| AA | 35 | 37 | 17.33 (8.01) | 19.95 (11.77) | 1.00 | 1.00 | ||||
| TA | 249 | 350 | 16.96 (5.86) | 17.82 (7.09) | 0.75 | 0.46, 1.23 | 0.25 | 0.76 | 0.47, 1.24 | 0.28 |
| TT | 545 | 774 | 17.13 (6.69) | 18.15 (7.07) | 0.74 | 0.46, 1.20 | 0.22 | 0.75 | 0.47, 1.21 | 0.24 |
| per T allele | 0.94 | 0.80, 1.10 | 0.44 | 0.94 | 0.80, 1.10 | 0.44 | ||||
Before applying the MR approach we assessed the IV assumptions. The first (that genotype is associated with the phenotype) was fulfilled since we selected four SNPs that were found to be linked to plasma 25-OHD levels in a pooled meta-analysis of Genome Wide Association Studies [20]. The second (genotype is independent of measured and unmeasured confounders) was tested by investigating whether the instruments were associated with any of the measured confounding factors that might influence the relationship between plasma 25-OHD levels and CRC (Table S6) and, as expected [27], there was no evidence for an association between these confounding factors and the genotypes. Finally, the third assumption (effect of genotype on outcome is mediated only through the intermediate phenotype) was tested by interrogating of pleiotropic links of genes and SNPs that we recently created [28]. For all SNPs there was no evidence of pleiotropy and they were only found to be linked to plasma vitamin D levels.
Using the rs2282679 as the IV, the estimated causal effect of plasma 25-0HD on CRC risk was 0.94 (95% CI 0.49, 1.83), and the F-statistic for the rs2282679 from the first stage of the IV analysis was 15.80 in the age and sex adjusted analysis ( Table 4 ). Using the rs12785878 as the IV the causal effect was 1.23 (95% CI 0.60, 2.53), and the F-statistic for the rs12785878 from the first stage of the IV analysis was 13.50 ( Table 4 ). Using the rs10741657 as the IV the causal effect was 0.89 (95% CI 0.40, 1.98), and the F-statistic for the rs10741657 from the first stage of the IV analysis was 10.89 ( Table 4 ). Finally, using the rs6013897 as the IV the causal effect was 0.99 (95% CI 0.40, 2.45), and the F-statistic for the rs6013897 from the first stage of the IV analysis was 0.98 ( Table 4 ). The results of the other IV estimators are presented in Tables S7, S8, S9, and S10.
Table 4. Control function instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
| Model | plasma 25-0HD (continuous, ng/ml) | ||
| rs2282679 | OR | 95% CI | F statistic |
| Unadjusted | 0.58 | 0.11, 3.10 | 7.29 |
| Adjusted for age and sex | 0.94 | 0.49, 1.83 | 15.80 |
| rs12785878 | |||
| Unadjusted | 4.03 | 0.93, 17.41 | 9.70 |
| Adjusted for age and sex | 1.23 | 0.60, 2.53 | 13.50 |
| rs10741657 | |||
| Unadjusted | 1.15 | 0.00, 762.13 | 0.49 |
| Adjusted for age and sex | 0.89 | 0.40, 1.98 | 10.89 |
| rs6013897 | |||
| Unadjusted | 2.26 | 0.06, 86.31 | 1.58 |
| Adjusted for age and sex | 0.99 | 0.40, 2.45 | 0.98 |
| Allele score * | |||
| Unadjusted | 1.50 | 0.42, 5.35 | 12.95 |
| Adjusted for age and sex | 1.16 | 0.60, 2.23 | 16.52 |
| Allele score (upstream) † | |||
| Unadjusted | 1.83 | 0.22, 15.19 | 4.63 |
| Adjusted for age and sex | 0.94 | 0.46, 1.91 | 7.87 |
| Allele score (downstream) ‡ | |||
| Unadjusted | 0.83 | 0.34, 2.05 | 14.89 |
| Adjusted for age and sex | 0.93 | 0.53, 1.63 | 12.67 |
Allele score that combines all 4 SNPs: rs12785878, rs10741657, rs2282679 and rs6013897.
Allele score that combines 2 upstream (synthesis) SNPs: rs12785878, rs10741657.
Allele score that combines 2 “metabolism” SNPs: rs2282679 and rs6013897.
Furthermore, we combined these four SNPs to form three allele scores: 1) one allele score that combined all four SNPs, 2) an upstream allele score that combined the SNPs rs12785878 and rs10741657 and 3) a downstream allele score that combined the SNPs rs2282679 and rs6013897. We then used this allele scores as the IV. The causal effect for the overall allele score was 1.16 (95% CI 0.60, 2.23; F-statistic 16.52), for the upstream allele score was 0.94 (95% CI 0.46, 1.91; F-statistic 7.87) and for the downstream allele score was 0.93 (95% CI 0.53, 1.63; F-statistic 12.67) ( Table 4 ).
Discussion
Levels of Plasma 25-OHD and CRC
In this study, low levels of plasma 25-OHD were associated with a higher risk of CRC in the whole sample and after stratification for sex, tumour stage and severity of symptoms at presentation. These results are in accordance with two recent meta-analyses of serum or plasma prospective studies [14], [29]. In addition a systematic review and meta-analysis on colorectal adenoma (CRA) showed a decreased risk with both incidence and recurrence of CRA for an increase of 25-OHD by 20 ng/ml [30]. However, a randomised clinical trial (RCT) (2686 average risk subjects) found no effect of vitamin D supplementation and incidence of CRC [31]. Similarly, a second RCT (Women’s Health Initiative –WHI) investigating the effects of daily calcium and vitamin D supplementation for seven years showed no effect on CRC incidence among postmenopausal women [32]. However, it should be noted that neither of these RCTs were designed and powered for cancer as the primary outcome. In addition, re-analysis of the WHI RCT found that concurrent oestrogen therapy was an effect modifier of calcium and vitamin D supplementation and for women that were not assigned to oestrogen therapy calcium and vitamin D supplementation decreased CRC risk [33]. Data from case-control and cohort studies examining the associations between dietary vitamin D intake and CRC are inconclusive [13].
Genotype, Plasma 25-OHD Levels and CRC
The A allele of rs2282679 and the T allele of rs12785878 were associated with higher levels of plasma 25-OHD. These results are in accordance with a GWAS investigating genetic determinants of vitamin D deficiency [20]. The rs2282679 SNP is located in the GC gene, which encodes a vitamin D binding protein that binds and transports vitamin D ( Figure 1 ) [20]. The rs12785878 SNP is located in the DHCR7 gene that encodes the enzyme 7-dehydrocholesterol (7-DHC) reductase, which converts 7-DHC to cholesterol. 7-DHC is a precursor of vitamin D3. Mutations in the DHCR7 may lead to a decreased activity of the 7-DHC reductase and therefore to high levels of 7-DHC and vitamin D3 ( Figure 1 ) [20]. However the other two SNPs (rs10741657, rs6013897) that were also found to be strongly associated with vitamin D levels in the vitamin D genome wide association study were not associated with vitamin D status in our cohort. rs10741657 is located in the CYP2R1 gene, which encodes an enzyme thought to be involved in the 25-hydroxylation of vitamin D3 to 25(OHD) [20]. rs6013897 is located in the CYP24A1 gene, which encodes an enzyme that initiates the degradation of 1,25(OH)2D [20]. The evidence for the role of the enzymes coded by CYP2R1and CYP24A1 is limited and not replicated in other candidate studies [20].
None of the four SNPs were associated with CRC risk, although we found an interaction between 25-OHD levels and rs10741657. The results of the MR analysis did not support a causal relationship between plasma 25-OHD and CRC risk. Although not significant, the inverse relationship was noted when rs2282679 or rs10741657 were used as instrument, but not when rs12785878 or rs6013897. The results remained inconsistent when the three allele scores of the four SNPs was used. The fact that the inverse association that was observed when we applied the conventional epidemiological methods was not replicated when the MR approach was used might be due to several reasons. It is possible that unmeasured or latent variables confounded the associations and there is no true effect of vitamin D on CRC. An alternative explanation is that there might be reverse causality between vitamin D and CRC, given that the plasma of the cases was collected after diagnosis: it cannot be excluded that low plasma 25-OHD is the consequence of disease or a result of patients being bedbound and lacking the exposure to sun. However, when we looked at cases with very mild or no symptoms or cases at the very early stages of the disease we still observed an inverse association between 25-OHD levels and CRC. In addition, given the biological potential of vitamin D having a causal link with cancer, factors that affect the performance of the IV estimators might also explain these findings.
Major limitations of conventional instrumental variable approaches result from the strict assumptions that need to be satisfied for method to be reliable. It is true that genotype is associated with phenotype, however, SNPs that have been used as instruments are only weakly associated to phenotype and explain only a small portion of trait variance. While we tested for common confounders, it is possible that hidden confounding from unmeasured variables affects the analysis. While we see no association between genotype and the outcome, weaker pleiotropic links cannot be excluded with certainty.
We are currently working on new instrumental variable methods for assessing causality between 25-OHD concentrations and CRC, based on the platform for the Bayesian analysis of complex statistical models using Markov chain Monte Carlo methods. This method improves on the classical Mendelian Randomisation approach as it allows for pleiotropic links between components of the model, enables easy inclusion of other covariates and does not depend as much on the strength of the instruments.
The wide application of the genome wide association studies (GWAS), has allowed for the MR studies to become more feasible now, as common SNPs linked to various intermediate phenotypes have been identified [34]. To date in 2011, several applied MR studies were published investigating causal relationships in a wide range of diseases including cancer [35], coronary heart disease [36], [37], diabetes [38], [39], mental disorders[40]–[42], lung function [43] and other diseases [44], [45]. Some of these studies replicated the results of the conventional epidemiological methods and confirmed causality (or no causality) of the intermediate phenotype on the outcome [40], [44], [45]. One of the main reasons behind this lack of replication is the fact that the genetic instruments that are employed are generally weak and therefore the power of the MR studies will be inadequate [34].
According to a recent study on the power and sample size requirements of MR studies based on the strength of the instruments, found that MR studies will require large (n>1000) and often very large (n>10000) sample sizes to draw causal conclusions that are statistically significant [34]. Based on their findings from simulated analyses most of the published MR studies are under-powered (see Table S11 for a review of the 11 MR studies published in 2011). Using simulations presented in the paper of Pierce et al [34] and accounting for sample size, strength of the employed instruments and observed effect size, we estimate that the sample size in this study gave power of ∼0.35.
Conclusion
This study shows that higher plasma 25-OHD levels are associated with a lower CRC risk, a finding that is consistent with recent meta-analyses of prospective studies. However, this finding was not replicated when the MR approach was employed. This finding might be due to a lack of a true effect of vitamin D on CRC or due to reverse causation. It may also be due to weak instruments and limited statistical power. The lack of power is a common characteristic of many MR studies and therefore a careful selection of instruments plus an adequate sample size are deemed necessary for this method to be able to make causal conclusions. Given the extent of vitamin D deficiency among individuals living in high latitudes, a large consortium of similar vitamin D and CRC studies and/or the application of alternative methods that are less sensitive to weak instrument bias [46] are necessary to refine the effect of vitamin D on CRC cancer and other chronic diseases.
Supporting Information
Information about the SNPs that were used as Instrumental variables in the MR analysis.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after sex stratification.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stage stratification.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stratification for presence of symptoms.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stratification based on the time between diagnosis and recruittment (TDR).
(DOC)
Distribution of possible confounding factors and the instruments.
(DOC)
Wald/ratio instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Two stage instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Multiplicative structural mean models instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Logistic structural mean models instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Review of the characteristics of the MR studies published in 2011.
(DOC)
Information about the following Instrumental Variable estimators: Wald (ratio) estimator, Two stage least squares estimator, Multiplicative structural mean models, Logistic structural mean models.
(DOC)
Acknowledgments
We thank Mrs Gisela Johnstone, Stephanie Scott and Rosa Bisset for their administrative support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the following grants: Cancer Research UK Programme grant (C348/A12076), Medical Research Council (G0000657-53203) and Scottish Executive Chief Scientist’s Office (CZH/4/529, CZB/4/449), and a Centre Grant from CORE as part of the Digestive Cancer Campaign. Dr. Theodoratou is funded by Cancer Research UK Fellowship C31250/A10107. Dr. Palmer is funded by MRC centre grant G0600705. Dr. Zgaga is supported by the United States National Institute for Health- National Cancer Institute U19 programme grant (1U19CA148107-01) as part of the CORECT consortium. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jimenez-Lara AM. Colorectal cancer: potential therapeutic benefits of Vitamin D. Int J Biochem Cell Biol. 2007;39:672–677. doi: 10.1016/j.biocel.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Burleigh E, Potter J. Vitamin D deficiency in outpatients:–a Scottish perspective. Scott Med J. 2006;51:27–31. doi: 10.1258/RSMSMJ.51.2.27. [DOI] [PubMed] [Google Scholar]
- 3.Zgaga L, Theodoratou E, Farrington SM, Agakov F, Tenesa A, et al. J Nutr; 2011. Diet, Environmental Factors, and Lifestyle Underlie the High Prevalence of Vitamin D Deficiency in Healthy Adults in Scotland and Supplementation Reduces the Proportion That Are Severely Deficient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Report of the Panel on DRVs of the Committee on Medical Aspectsof Food Policy (COMA) Dietary Reference Values for Food Energy and Nutrients for the United Kingdom. 1991. [PubMed]
- 5.Pearce SH, Cheetham TD. Diagnosis and management of vitamin D deficiency. BMJ. 2010;340:b5664. doi: 10.1136/bmj.b5664. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff-Ferrari HA, Giovannucci E, Willett WC, Dietrich T, Dawson-Hughes B. Estimation of optimal serum concentrations of 25-hydroxyvitamin D for multiple health outcomes. Am J Clin Nutr. 2006;84:18–28. doi: 10.1093/ajcn/84.1.18. [DOI] [PubMed] [Google Scholar]
- 7.Vieth R, Bischoff-Ferrari H, Boucher BJ, Dawson-Hughes B, Garland CF, et al. The urgent need to recommend an intake of vitamin D that is effective. Am J Clin Nutr. 2007;85:649–650. doi: 10.1093/ajcn/85.3.649. [DOI] [PubMed] [Google Scholar]
- 8.Wagner CL, Greer FR. Prevention of rickets and vitamin D deficiency in infants, children, and adolescents. Pediatrics. 2008;122:1142–1152. doi: 10.1542/peds.2008-1862. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Lara AM. Colorectal cancer: potential therapeutic benefits of Vitamin D. Int J Biochem Cell Biol. 2007;39:672–677. doi: 10.1016/j.biocel.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Park SY, Murphy SP, Wilkens LR, Nomura AM, Henderson BE, et al. Calcium and vitamin D intake and risk of colorectal cancer: the Multiethnic Cohort Study. Am J Epidemiol. 2007;165:784–793. doi: 10.1093/aje/kwk069. [DOI] [PubMed] [Google Scholar]
- 11.Gross MD. Vitamin D and calcium in the prevention of prostate and colon cancer: new approaches for the identification of needs. J Nutr. 2005;135:326–331. doi: 10.1093/jn/135.2.326. [DOI] [PubMed] [Google Scholar]
- 12.Slattery ML, Samowitz W, Hoffman M, Ma KN, Levin TR, et al. Aspirin, NSAIDs, and colorectal cancer: possible involvement in an insulin-related pathway. Cancer Epidemiol Biomarkers Prev. 2004;13:538–545. [PubMed] [Google Scholar]
- 13.Theodoratou E, Farrington SM, Tenesa A, McNeill G, Cetnarskyj R, et al. Modification of the inverse association between dietary vitamin D intake and colorectal cancer risk by a FokI variant supports a chemoprotective action of Vitamin D intake mediated through VDR binding. Int J Cancer. 2008;123:2170–2179. doi: 10.1002/ijc.23769. [DOI] [PubMed] [Google Scholar]
- 14.Lee JE, Li H, Chan AT, Hollis BW, Lee IM, et al. Circulating levels of vitamin D and colon and rectal cancer: the Physicians’ Health Study and a meta-analysis of prospective studies. Cancer Prev Res (Phila) 2011;4:735–743. doi: 10.1158/1940-6207.CAPR-10-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng K, Sargent DJ, Goldberg RM, Meyerhardt JA, Green EM, et al. Vitamin D status in patients with stage IV colorectal cancer: findings from Intergroup trial N9741. J Clin Oncol. 2011;29:1599–1606. doi: 10.1200/JCO.2010.31.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey-Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 17.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16:309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 18.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Stat Med; 2007. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. [DOI] [PubMed] [Google Scholar]
- 19.Davey-Smith G, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 20.Wang TJ, Zhang F, Richards JB, Kestenbaum B, Van Meurs JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theodoratou E, Kyle J, Cetnarskyj R, Farrington SM, Tenesa A, et al. Dietary flavonoids and the risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:684–693. doi: 10.1158/1055-9965.EPI-06-0785. [DOI] [PubMed] [Google Scholar]
- 22.Din FV, Theodoratou E, Farrington SM, Tenesa A, Barnetson RA, et al. Effect of aspirin and NSAIDs on risk and survival from colorectal cancer. Gut. 2010;59:1670–1679. doi: 10.1136/gut.2009.203000. [DOI] [PubMed] [Google Scholar]
- 23.Wallace AM, Gibson S, de la HA, Lamberg-Allardt C, Ashwell M. Measurement of 25-hydroxyvitamin D in the clinical laboratory: current procedures, performance characteristics and limitations. Steroids. 2010;75:477–488. doi: 10.1016/j.steroids.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Knox S, Harris J, Calton L, Wallace AM. A simple automated solid-phase extraction procedure for measurement of 25-hydroxyvitamin D3 and D2 by liquid chromatography-tandem mass spectrometry. Ann Clin Biochem. 2009;46:226–230. doi: 10.1258/acb.2009.008206. [DOI] [PubMed] [Google Scholar]
- 25.Palmer TM, Sterne JA, Harbord RM, Lawlor DA, Sheehan NA, et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in mendelian randomization analyses. Am J Epidemiol. 2011;173:1392–1403. doi: 10.1093/aje/kwr026. [DOI] [PubMed] [Google Scholar]
- 26.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 27.Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, et al. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, et al. Am J Hum Genet in press; 2011. Abundant pleiotropy in human complex diseases and traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touvier M, Chan DS, Lau R, Aune D, Vieira R, et al. Meta-analyses of vitamin D intake, 25-hydroxyvitamin D status, vitamin D receptor polymorphisms, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20:1003–1016. doi: 10.1158/1055-9965.EPI-10-1141. [DOI] [PubMed] [Google Scholar]
- 30.Yin L, Grandi N, Raum E, Haug U, Arndt V, et al. Meta-analysis: Serum vitamin D and colorectal adenoma risk. Prev Med. 2011;53:10–16. doi: 10.1016/j.ypmed.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi DP, Doll R, Khaw KT. Effect of four monthly oral vitamin D3 (cholecalciferol) supplementation on fractures and mortality in men and women living in the community: randomised double blind controlled trial. BMJ. 2003;326:469. doi: 10.1136/bmj.326.7387.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wactawski-Wende J, Kotchen JM, Anderson GL, Assaf AR, Brunner RL, et al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. N Engl J Med. 2006;354:684–696. doi: 10.1056/NEJMoa055222. [DOI] [PubMed] [Google Scholar]
- 33.Ding EL, Mehta S, Fawzi WW, Giovannucci EL. Interaction of estrogen therapy with calcium and vitamin D supplementation on colorectal cancer risk: reanalysis of Women’s Health Initiative randomized trial. Int J Cancer. 2008;122:1690–1694. doi: 10.1002/ijc.23311. [DOI] [PubMed] [Google Scholar]
- 34.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benn M, Tybjaerg-Hansen A, Stender S, Frikke-Schmidt R, Nordestgaard BG. Low-density lipoprotein cholesterol and the risk of cancer: a mendelian randomization study. J Natl Cancer Inst. 2011;103:508–519. doi: 10.1093/jnci/djr008. [DOI] [PubMed] [Google Scholar]
- 36.Breitling LP, Koenig W, Fischer M, Mallat Z, Hengstenberg C, et al. Type II secretory phospholipase A2 and prognosis in patients with stable coronary heart disease: mendelian randomization study. PLoS One. 2011;6:e22318. doi: 10.1371/journal.pone.0022318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, et al. Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Silva NM, Freathy RM, Palmer TM, Donnelly LA, Luan J, et al. Mendelian randomization studies do not support a role for raised circulating triglyceride levels influencing type 2 diabetes, glucose levels, or insulin resistance. Diabetes. 2011;60:1008–1018. doi: 10.2337/db10-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfister R, Barnes D, Luben R, Forouhi NG, Bochud M, et al. No evidence for a causal link between uric acid and type 2 diabetes: a Mendelian randomisation approach. Diabetologia. 2011;54:2561–2569. doi: 10.1007/s00125-011-2235-0. [DOI] [PubMed] [Google Scholar]
- 40.Kivimaki M, Jokela M, Hamer M, Geddes J, Ebmeier K, et al. Examining overweight and obesity as risk factors for common mental disorders using fat mass and obesity-associated (FTO) genotype-instrumented analysis: The Whitehall II Study, 1985–2004. Am J Epidemiol. 2011;173:421–429. doi: 10.1093/aje/kwq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawlor DA, Harbord RM, Tybjaerg-Hansen A, Palmer TM, Zacho J, et al. Using genetic loci to understand the relationship between adiposity and psychological distress: a Mendelian Randomization study in the Copenhagen General Population Study of 53,221 adults. J Intern Med. 2011;269:525–537. doi: 10.1111/j.1365-2796.2011.02343.x. [DOI] [PubMed] [Google Scholar]
- 42.Lewis SJ, Araya R, Smith GD, Freathy R, Gunnell D, et al. Smoking is associated with, but does not cause, depressed mood in pregnancy–a mendelian randomization study. PLoS One. 2011;6:e21689. doi: 10.1371/journal.pone.0021689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahl M, Vestbo J, Zacho J, Lange P, Tybjaerg-Hansen A, et al. C reactive protein and chronic obstructive pulmonary disease: a Mendelian randomisation approach. Thorax. 2011;66:197–204. doi: 10.1136/thx.2009.131193. [DOI] [PubMed] [Google Scholar]
- 44.Kivimaki M, Magnussen CG, Juonala M, Kahonen M, Kettunen J, et al. Conventional and Mendelian randomization analyses suggest no association between lipoprotein(a) and early atherosclerosis: the Young Finns Study. Int J Epidemiol. 2011;40:470–478. doi: 10.1093/ije/dyq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumby HS, Elks CE, Li S, Sharp SJ, Khaw KT, et al. J Obes 2011: 180729; 2011. Mendelian Randomisation Study of Childhood BMI and Early Menarche. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKeigue PM, Campbell H, Wild S, Vitart V, Hayward C, et al. Bayesian methods for instrumental variable analysis with genetic instruments (‘Mendelian randomization’): example with urate transporter SLC2A9 as an instrumental variable for effect of urate levels on metabolic syndrome. Int J Epidemiol. 2010;39:907–918. doi: 10.1093/ije/dyp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information about the SNPs that were used as Instrumental variables in the MR analysis.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after sex stratification.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stage stratification.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stratification for presence of symptoms.
(DOC)
Logistic regression analysis for the association between plasma 25-0HD on colorectal cancer risk after stratification based on the time between diagnosis and recruittment (TDR).
(DOC)
Distribution of possible confounding factors and the instruments.
(DOC)
Wald/ratio instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Two stage instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Multiplicative structural mean models instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Logistic structural mean models instrumental variable estimator of the causal odds ratio for the effect of plasma 25(0H)D on colorectal cancer risk.
(DOC)
Review of the characteristics of the MR studies published in 2011.
(DOC)
Information about the following Instrumental Variable estimators: Wald (ratio) estimator, Two stage least squares estimator, Multiplicative structural mean models, Logistic structural mean models.
(DOC)