Mast cells exit the bone marrow as immature multilineage cells that circulate in the blood and subsequently undergo full differentiation upon reaching a target organ (reviewed in Gilfillan and Tkaczyk1). Unlike other hematopoietic cells, differentiated mast cells continue to express KIT, a transmembrane receptor with a bipartite intracellular kinase domain.1 The main ligand of KIT is stem cell factor (also known as KIT ligand), which stimulates mast-cell survival, development, maturation and activation.1 Mast cells also express a high-affinity cell-surface IgE receptor, FcεRI, which recognizes the Fc portion of IgE and is required for mast-cell survival via signaling through LYN and SYK kinases.1
Mast-cell leukemia (MCL) is an aggressive form of systemic mastocytosis characterized by the overproliferation of atypical mast cells, promastocytes and blasts that produce, among other substances, tryptase.2 MCL is often linked to somatically acquired activating mutations in the KIT receptor that result in uncontrolled ligand-independent signaling by KIT and hyper-proliferation of mast cells (reviewed in Ustun et al.3). The KIT D816V mutation causes resistance to imatinib therapy.3 Mutations in TET2 and NRAS have also been described in mastocytosis patients and each of them segregates with the KIT D816V mutation,4,5 suggesting that more than one lesion is required to drive leukemogenesis. In order to identify novel MCL determinants, we utilized two approaches to undertake the first comprehensive study of the DNA changes in an MCL patient. This study was approved by the Institutional Review Boards of the North Shore-LIJ Health System and the Cold Spring Harbor Laboratory. The patient gave written informed consent in accordance with the Declaration of Helsinki.
The patient, a 42-year-old female, presented with epigastric pain, fever, weight loss and urticaria. She was found to have splenomegaly, anemia and elevated levels of tryptase and histamine. Marrow findings were consistent with MCL (Supplementary Figures S1 and S2). The karyotype was 46, XX [20] and negative for KIT D816V mutation by PCR. The pertinent treatment details are: induction therapy with cladribine, cytarabine and filgrastim plus daily dasatinib; day 21 re-induction with high-dose cytarabine plus idarubicin; and day 41 imatinib 400 mg daily for 14 days. Bone marrow biopsies on treatment days 21, 41 and 78 revealed persistent MCL. The patient expired 96 days after diagnosis (see Supplementary Information for a detailed summary).
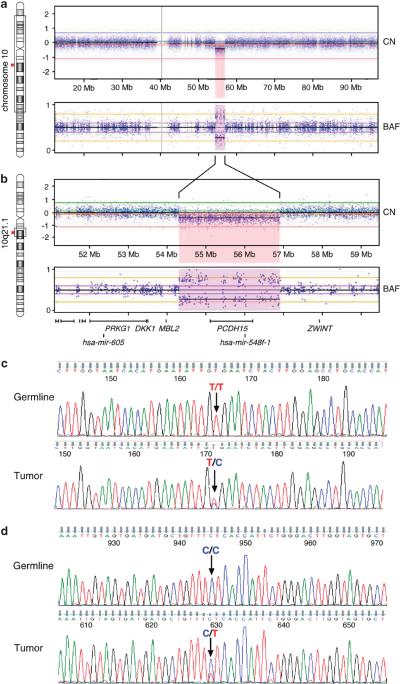
Array comparative genomic hybridization was performed in order to identify chromosomal copy number changes. Tumor and germline DNA were hybridized to Affymetrix 6.0 SNP arrays (Affymetrix; Santa Clara, CA, USA) and data were analyzed and viewed with Nexus Copy Number V6 software (BioDiscovery, El Segundo, CA, USA). Array comparative genomic hybridization revealed two somatic copy number changes in the tumor: copy number neutral loss of heterozygosity on chr1p36.33-p31.1 (81.3 Mb) and a focal hemizygous deletion (2.6 Mb) on chr10q21.1 containing two genes, PCDH15 (a protocadherin) and hsa-mir-548f-1 (Figures 1a and b). Members of the hsa-mir-548 microRNA gene family are differentially expressed in cancer cells6 and have been implicated in tumorigenesis.7
Figure 1.
Hemizygous deletion of chromosome 10q21.1 detected by high-resolution SNP array in leukemic cells and independent sequence confirmation for two non-synonymous somatic tumor variants. (a) The majority of chromosome 10 has 2N copy number (CN) and heterozygous B allele frequency (BAF), except for a hemizygous deletion (b) of chromosome 10q21.1; 2.6 Mb (chr10:54,305,820 – 56,908,099). This region encompasses one protein coding gene, PCDH15, and one microRNA gene, hsa-mir-548f-1. (c and d) Germline (saliva DNA) and tumor (leukemic) DNA were PCR amplified for (c) KIT (top two panels) and (d) MS4A2 (bottom two panels), and capillary sequenced. The germline genotype for KIT is homozygous T/T; GTG encodes for Valine 654. The tumor is heterozygous T/C; GCG encodes for Alanine. The germline genotype for MS4A2 is homozygous C/C; CTC encodes for Leucine 188. The tumor is heterozygous C/T; TTC encodes for Phenylalanine.
In order to identify tumor-specific single-nucleotide variants, tumor and germline DNA were enriched for exonic regions (SeqCap EZ Human Exome Library v2.0; Roche NimbleGen, Madison, WI, USA) and 76 bp paired-end-sequenced on a Genome Analyzer IIX platform (Illumina, San Diego, CA, USA). Paired-end reads were aligned and single-nucleotide variants in each genome were identified (Supplementary Information).
Thirty-eight tumor-specific single-nucleotide variants were identified in the tumor genome: 5 variants outside a coding region (intron, untranslated region or intergenic), 12 synonymous coding changes and 21 non-synonymous coding changes, the latter of which were further evaluated. The amino acid changes were filtered through PolyPhen-2 and annotated for whether the amino acid change is predicted to be benign or damaging to the function of the protein (Table 1). Nine of the non-synonymous variants occurred in genes within the chr1p uniparental disomy (UPD) region and underwent loss of heterozygosity. However, eight of these were not evaluated further because they either retained the reference allele in the tumor or were present in dbSNP130 at high allele frequencies and are predicted by PolyPhen-2 to be benign. The remaining 13 candidate disease-relevant missense mutations that caused protein-coding changes were evaluated further (Table 1). Of these, 11 single-nucleotide variants were validated independently using Sanger sequencing of PCR products generated from the original patient DNA (two of these are shown in Figures 1c and d). The majority of these genes are annotated as targets of cancer-associated somatic mutations (COSMIC database http://www.sanger.ac.uk), although not all of them in hematologic cancers; some have been linked to cancer via changes in copy number or expression levels (Table 1 and Supplementary Information). Of greatest interest are the variants we found in two genes, KIT and MS4A2.
Table 1.
Loss or gain of heterozygosity and missense coding change in tumor
| Chr | Position | Reference ollele |
Germline genotype |
Tumor genotype |
Gene | Amino- acid change |
Protein position |
PolyPhen-2 prediction |
Sanger confirm? |
Gene listed as mutated in COSMIC |
Gene function; cancer-relation | Reference in supplement |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7887756 | C | C/A | A/A-LOHa | PER3 | PRO,THR | 915/1202 | Damaging | Yes | O,P | CHK2 activation/apoptosis upon DNA damage | 5,6 |
| 1 | 182835714 | A | A/A | A/G | DHX9 | THR,ALA | 490/1271 | Damaging | Yes | 1 (silent) | ATP-dependent RNA helicase | 20 |
| 2 | 8891719 | C | C/C | C/T | KIDINS220 | GLY,ARG | 1023/1772 | Damaging | NP | 4 (silent) | MEK/ERK signaling; overexpressed in melanoma | 7 |
| 2 | 141004724 | T | T/T | T/C | LRP1B | THR,ALA | 4419/4600 | Benign | Yes | N,K,L, Lu,O,P | Regulates extracellular proteolytic activity and intracellular signaling; deleted or underexpressed in multiple tumor types | 9 |
| 4 | 55594258 | T | T/T | T/C | KIT | VAL,ALA | 654/977 | Damaging | Yes | H+L,ST,etc | Receptor tyrosine kinase; activating mutants in multiple tumor types | |
| 4 | 79792056 | C | C/C | C/G | BMP 2K | LEU,VAL | 451/1162 | Unknown | Yes | 1 in Lu | Putative serine/threonine protein kinase | 21, 22, 23 |
| 7 | 157369462 | C | C/C | C/A | PTPRN2 | VAL,PHE | 876/1016 | Damaging | Yes | O | Phosphatase; underexpressed in lung cancer | 19 |
| 9 | 32633225 | G | G/G | G/A | TAF1 L | ARG,TRP | 785/1827 | Damaging | Yes | Lu,O,B,N, | Transcription | 24 |
| 10 | 55755491b | C | C/T | T-LOHc | PCDH 15 | ARG,GLN | 929/1956 | Damaging | Yes | O,P | Protocadherin | 17 |
| 11 | 59861461 | C | C/C | C/T | MS4A2 | LEU,PHE | 188/245 | Damaging | Yes | 1 in O | β chain of IgE mast cell receptor that is required for mast cell survival | |
| 12 | 81242049 | C | C/C | C/T | LIN7A | ARG,HIS | 85/234 | Damaging | Yes | 1 in O | Establish/maintain receptor distribution | 8 |
| 17 | 62034632 | G | G/G | G/A | SCN4A | ARG,CYS | 756/1837 | Damaging | Yes | O,Sk | Voltage-gated sodium channel; overexpressed in ovarian cancer | 16 |
| X | 19449574 | C | C/C | C/T | MAP3K15 | ARG,HIS | 383/1314 | Damaging | ND | Lu,O,Sk | Mitogen-activated protein kinase | 18 |
Abbreviations: B, breast; Chr, chromosome; H+L, hemato+lymph; K, kidney; L, liver; Lu, lung; N, central nervous system; ND, not done; NP, not possible because PCR product is not obtainable; O, ovary; P, pancreas; Sk, skin; ST, soft tissue. UPD region of chr 1.
In dbSNP; rs2135720.
Hemizygous region of chr 10. PolyPhen-2 prediction: damaging includes possibly and probably.
As part of the clinical evaluation, this patient was screened for the KIT D816V mutation and was found to be negative. However, exome sequencing revealed a mutation in KIT, V654A, previously reported in gastrointestinal stromal tumor (GIST),8 but not in leukemia. This mutation may confer resistance to imatinib treatment as the V654A substitution directly affects the binding of imatinib to the receptor.9 Prior knowledge of this mutation would have altered the decision to administer imatinib, to which the patient showed no response. Our results suggest that sequencing of larger regions of the KIT gene may be informative.
We also identified a missense mutation in MS4A2, the gene that encodes the b chain of the high-affinity tetrameric cell-surface IgE (FcεRI) mast-cell receptor. To date, a mutation in this gene has only been reported in ovarian carcinoma (COSMIC). The mutation found here, L188F, is within the last of the four transmembrane domains and is predicted to be damaging to protein function. In addition to its role in immunologic responses, FcεRI has antigen-independent effects, in particular enhancing mast-cell survival in the absence of allergen (reviewed in Gilfillan and Tkaczyk1, and Kraft and Kinet10). Cell-surface expression of the tetrameric receptor (α, β and two γ chains) is regulated at several levels and, once engaged, sets off a complex intracellular signaling cascade via the interaction of the β chain with LYN and SYK kinases.10 The full-length β chain acts as an amplifier of surface-tetramer expression as it promotes the maturation and transport of the tetramer subunits to the cell surface.10 In addition, tetrameric cell-surface expression is regulated by cell-type specific alternative splicing of the β chain that produces two truncated isoforms of the protein.11,12 Each truncated isoform antagonizes the function of the full-length protein resulting in less surface expression and less downstream signaling, which limits cell proliferation/survival. This mutation identified in the FcεRI β chain is an intriguing finding as the mast cell tetrameric receptor complex is involved in enhancing mast-cell survival.10 In principle, increasing the levels of mast-cell receptor signaling could lead to prolonged or aberrant mast-cell survival, increasing the likelihood that the dividing cells acquire additional mutations, including those that block differentiation and promote leukemogenesis. This is particularly relevant for mast cells as they spend a majority of their lives not fully differentiated.
Although we cannot rule out the possibility that the mutant β chain of the FcεRI receptor identified in this patient is inactivating or inert, we hypothesize that this mutation results in increased signaling/survival in one of the following ways: (1) the mutant protein may have a higher affinity for the immature α chain and as such can more efficiently transport it to the Golgi for processing, which would result in increased surface expression, (2) the mutant protein is mislocalized in the cell, yet can function as an aberrant signal transducer in a different cellular compartment, similar to what has been shown for mutant FLT3 and other receptor tyrosine kinases13 or (3) the mutation causes the surface-bound tetramer to be locked in a constitutively active form that can very efficiently signal to downstream effectors–either without requiring ligand or by more efficiently interacting with LYN/SYK. Future functional studies are required to determine the role of this mutant receptor in leukemogenesis. If SYK is activated, we propose that the recently described SYK inhibitors, which showed efficacy in lymphoma14 and promoted differentiation of AML cells,15 should be evaluated for their potential therapeutic value in MCLs harboring mutations of the components of the IgE mast-cell receptor.
Supplementary Material
ACKNOWLEDGEMENTS
This research was funded by The Don Monti Memorial Research Foundation (SLA and SWL) and by The Ryan Gibson Foundation (MSS). SWL is a Howard Hughes Medical Institute Investigator. We gratefully acknowledge Dr W Richard McCombie for guidance with sequencing, Dr Shiroo Parshad for assistance with patient medical care, Dr Judith Brody for bone marrow interpretation, Dr Saul Teichberg for electron microscopy, Dr Piers Patten for scientific discussion, Candace Schiffer and Erin Boyle for patient sample collection and the members of the Laboratory of Experimental Immunology of the Feinstein Institute for Medical Research for their assistance in cryopreserving cells. Drs Chris Vakoc, Iris Applemann and Zhen Zhao are thanked for their critical reading of the manuscript.
Footnotes
CONFLICT OF INTEREST The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nat Rev Immunol. 2006;6:218–230. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- 2.Valent P, Horny HP, Escribano L, Longley BJ, Li CY, Schwartz LB, et al. Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res. 2001;25:603–625. doi: 10.1016/s0145-2126(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 3.Ustun C, Deremer DL, Akin C. Tyrosine kinase inhibitors in the treatment of systemic mastocytosis. Leuk Res. 2011;35:1143–1152. doi: 10.1016/j.leukres.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson TM, Maric I, Simakova O, Bai Y, Chan EC, Olivares N, et al. Clonal analysis of NRAS activating mutations in KIT-D816V systemic mastocytosis. Haematologica. 2011;96:459–463. doi: 10.3324/haematol.2010.031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann K, Wardelmann E, Ma Y, Merkelbach-Bruse S, Preussner LM, Woolery C, et al. Novel germline mutation of KIT associated with familial gastrointestinal stromal tumors and mastocytosis. Gastroenterology. 2005;129:1042–1046. doi: 10.1053/j.gastro.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 9.Roberts KG, Odell AF, Byrnes EM, Baleato RM, Griffith R, Lyons AB, et al. Resistance to c-KIT kinase inhibitors conferred by V654A mutation. Mol Cancer Ther. 2007;6:1159–1166. doi: 10.1158/1535-7163.MCT-06-0641. [DOI] [PubMed] [Google Scholar]
- 10.Kraft S, Kinet JP. New developments in FcεRI regulation, function and inhibition. Nat Rev Immunol. 2007;7:365–378. doi: 10.1038/nri2072. [DOI] [PubMed] [Google Scholar]
- 11.Donnadieu E, Jouvin MH, Rana S, Moffatt MF, Mockford EH, Cookson WO, et al. Competing functions encoded in the allergy-associated FcεRIbeta gene. Immunity. 2003;18:665–674. doi: 10.1016/s1074-7613(03)00115-8. [DOI] [PubMed] [Google Scholar]
- 12.Cruse G, Kaur D, Leyland M, Bradding P. A novel FcεRIbeta-chain truncation regulates human mast cell proliferation and survival. FASEB J. 2010;24:4047–4057. doi: 10.1096/fj.10-158378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell. 2009;36:326–339. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg JW, Sharman J, Sweetenham J, Johnston PB, Vose JM, LaCasce A, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115:2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn CK, Berchuck JE, Ross KN, Kakoza RM, Clauser K, Schinzel AC, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.