Abstract
Human manganese superoxide dismutase (Sod2p) has been expressed in yeast and the protein purified from isolated yeast mitochondria, yielding both the metallated protein and the less stable apoprotein in a single chromatographic step. At 30 °C growth temperature, more than half of the purified enzyme is apoprotein that can be fully activated following reconstitution, while the remainder contains a mixture of manganese and iron. In contrast, only fully metallated enzyme was isolated from a similarly constructed yeast strain expressing the homologous yeast manganese superoxide dismutase. Both the manganese content and superoxide dismutase activity of the recombinant human enzyme increased with increasing growth temperatures. The dependence of in vivo metallation state on growth temperature resembles the in vitro thermal activation behavior of human manganese superoxide dismutase observed in previous studies. Partially metallated human superoxide dismutase is fully active in protecting yeast against superoxide stress produced by addition of paraquat to the growth medium. However, a splice variant of human manganese superoxide dismutase (isoform B) is expressed as insoluble protein in both Escherichia coli and yeast mitochondria and did not protect yeast against superoxide stress.
Keywords: Manganese, Superoxide dismutase, Thermal activation, Metallation, Splice variant, Mitochondria, Isoform
1. Introduction
Manganese superoxide dismutases (MnSOD) serves as the front-line defense against oxidative stress in living cells, protecting against damage by toxic superoxide free radical formed as an unavoidable by-product of respiration (1–3). Considering that only the manganese complex is catalytically active, MnSODs are surprisingly unselective in metal binding. Both yeast mitochondrial MnSOD (4) and its prokaryotic (E. coli) counterpart (5, 6) can bind either Fe or Mn in vivo, with the metal that is bound being determined by availability. Unlike eukaryotic Cu, Zn-SOD, whose metallocofactor is delivered via well-characterized copper trafficking pathways (7,8), the cellular processes involved in activation of apo-MnSOD are presently unknown.
Human MnSOD (hMnSOD) is an essential antioxidant defense enzyme, which, like the yeast counterpart, is encoded in the nucleus and the preprotein is imported into the mitochondria, where the targeting signal is cleaved and metal activation occurs (9). A homozygous MnSOD knock-out is lethal in mice (10), and decreased expression of MnSOD is associated with malignancies in man (11). The low metal binding selectivity of MnSOD is reflected in misincorporation of iron for manganese in MnSOD when iron homeostasis is disrupted, producing inactive enzyme and exacerbating the effects of iron overload in the mitochondria (4,12). Human apo-MnSOD (apo-hMnSOD), prepared by denaturation, chelation, and refolding, also exhibits unspecific metal binding behavior in vitro (13). Like the dimeric E. coli apo-MnSOD, the tetrameric apo-hMnSOD binds metal in a thermally activated, pH-dependent process, but with a distinct kinetic profile (14, 15). While thermal activation of metal binding by E. coli apo-MnSOD has been demonstrated both in vitro and in vivo (16), only in vitro metal uptake has previously been studied for human apo-hMnSOD.
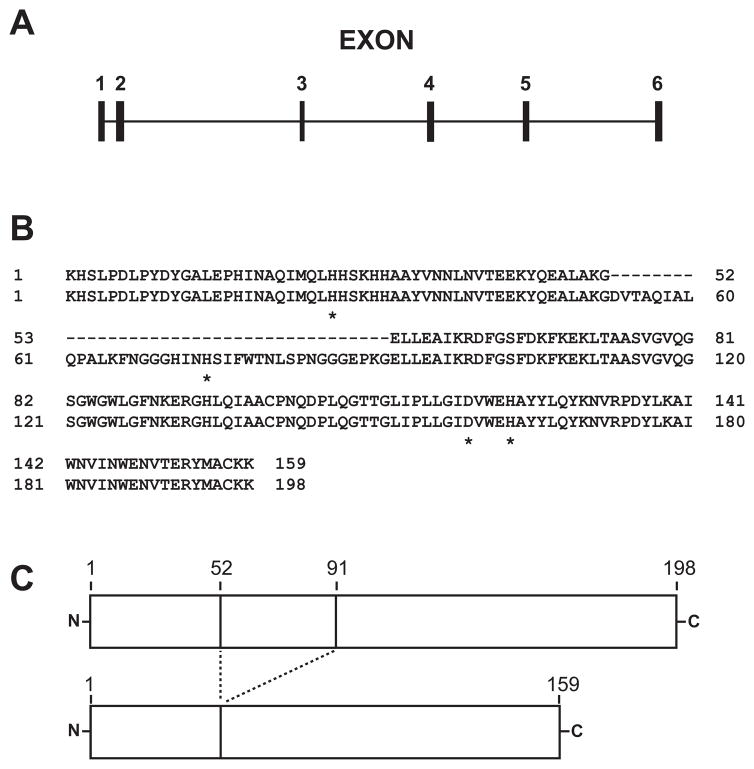
The majority of research on hMnSOD has focused on the full-length wild type protein and mutational polymorphisms that appear to be linked to a variety of human diseases including cardiomyopathy, neurodegeneration, and cancer (9,11,17–19). hMnSOD is encoded by the SOD2 locus on chromosome 6 (18), which produces an intron-containing pre-mRNA that must be spliced to generate a functional transcript (Fig. 1A). Variations in nucleotide sequences in the flanking regions upstream and downstream of the MnSOD gene locus, in the coding regions of the exons, or even within the introns all have been linked altered expression levels of the protein that can be biomedically significant (10,11,18,19). Further, alternative splicing of the intron-containing hMnSOD pre-mRNA can give rise to distinct mRNAs encoding different protein isoforms (20–22). One of these splice variants (variant 3) (GenBank Reference Sequence NM_001024466.1) lacks exon 3, and encodes a polypeptide defined as hMnSOD isoform B (GenBank Reference Sequence NP_001019637.1) (Fig. 1). This mRNA splice variant has been identified in cDNA expressed sequence tag (EST) libraries from a variety of tissues (Table 1). The isoform B polypeptide is 39 amino acids shorter than the normal hMnSOD (isoform A) (Fig. 1B&C), eliminating a region within the N-terminal domain (residues 53–91) that includes one of the four metal ligating residues. A possible link between expression of hMnSOD isoform B and a variety of disease states has made the protein potentially interesting in a clinical context (20). However, isoform B appears to be expressed in both healthy and diseased tissue, at all stages of development (Table 1). Unlike hMnSOD isoform A, very little experimental work has previously been reported for hMnSOD isoform B (20,21). The “rogue” M2 isoform of the terminal glycolytic enzyme pyruvate kinase, present in fetal tissue and some cancers (24), provides an example of how the expression of protein isoforms can be an important factor in health and disease. In the present work, thermally activated metal uptake by apo-hMnSOD was investigated in vivo in a yeast model. Human manganese superoxide dismutase isoform B was also studied, and the recombinant proteins were evaluated for biological function in yeast using a superoxide stress test.
Fig. 1.
Organization of the human SOD2 pre-mRNA and comparison of hMnSOD isoforms A and B. (A) Predicted exon/intron organization of the human SOD2 pre-mRNA. Exon 3 is retained in the variant 1 splicing product, encoding hMnSOD isoform A, but is eliminated from the variant 3 splicing product, encoding hMnSOD isoform B. (B) Alignment of mature protein sequences for hMnSOD isoforms A (bottom) and B (top). Residues that serve as metal ligands in hMnSOD isoform A are indicated by an asterisk (*). (C) Comparison of hMnSOD isoform A (top) and B (bottom) mature proteins, illustrating the different polypeptide lengths.
Table 1.
Curated human EST sequence data for hMnSOD splice variant 3 (isoform B) cDNA
| GenBank Accession Number1 | Tissue |
|---|---|
| DC346416.1 | tongue tumor tissue |
| DC410435.1 | thymus |
| DC415774.1 | trachea |
| DC418138.1 | trachea |
| DC300755.1 | bladder |
| DC308373.1 | cerebellum |
| DC369972.1 | teratocarcinoma cell line NT2 |
| DC297425.1 | CD34+ embryonic stem cells |
| DA415123.1 | thalamus |
| CA312442.1 | human lung epithelial cells |
| CA307772.1 | alveolar Macrophage |
| BU741675.1 | fetal eye |
| BM976348.1 | primary lung cystic fibrosis epithelial cells |
| BI869849.1 | adenocarcinoma cell line |
| DB467363.1 | hippocampus |
| BI899373.1 | Islets of Langerhans |
Subject sequences returned by a BLAST (Ref. 20) search of the GenBank human EST library database, using a 60 nt sequence spanning the exon 2/exon 4 splice site as the query.
2. Material and Methods
2.1. Biological materials
Saccharomyces cerevisiae BY4700 (MATa Δura3) was from ATCC (Manassas, VA). S. cerevisiae BY4230 (MATa Δleu2 Δmet5 Δura3 Δsod2) was from Invitrogen (Carlsbad, CA). E. coli BL21-AI was from Invitrogen. E. coli C41(DE3) was from Lucigen (Middleton, WI). E. coli BW25113 ΔsodA was prepared by phage lambda red recombineering methods (15).
2.2. Plasmid Construction
2.2.1 Homologous expression of yeast mitochondrial MnSOD in S. cerevisiae
DNA encoding full-length S. cerevisiae MnSOD (yMnSOD) polypeptide (including the yeast MnSOD mitochondrial targeting sequence (yMTS)) together with the PSOD2 promoter was amplified from yeast genomic DNA using primers PSOD-1 and YSOD-2 (Table 2) and the product was digested with AgeI and XbaI. pYES2 (Invitrogen, Carlsbad, CA) was digested with AgeI and XbaI to remove the PGAL1 promoter and ligated with the insert to generate the plasmid pYEPSOD-YSOD. After verifying the sequence of the insert by nucleotide sequence analysis (Molecular Biology Core, OHSU), the plasmid was transformed into yeast by electrotransformation.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| PSOD-1 | 5′-CGACCGGTAGTTGGGCTAAAGTGTGACGAGATGGATTGTGTTCATTC-3′ |
| YSOD-2 | 5′-CGATCTAGATCAGATCTTTCCTGCATCGAATCTTCTGGATGC-3′ |
| YSOD-3 | 5′-GCACATATGAAAGTCAACTTGCCAGATTTGAAGTGGGATTTCGG-3 |
| YSOD-4 | 5′-GGCGGATCCATTAGATCTTGCCAGCGTCAAATCTTCTAGATGCTTCTTTCC-3′ |
| YSODMTS | 5′-GCACGTACGCCTTGCTGTGGTGGAGAGCAATGAC-3′ |
| hMnSOD-1 | 5′-GCACATATGAAGCACAGCCTCCCCGACCTGC-3′ |
| hMnSOD-2 | 5′-GACAAGCTTACTTTTTGCAAGCCATGTATCTTTCAGTTACATTCTCCC-3′ |
| hMnSOD-3 | 5′-GCACGTACGAAGCACAGCCTCCCCGACCTGC-3′ |
| hMnSOD-4 | 5′-GACTCGAGTTACTTTTTGCAAGCCATGTATCTTTCAGTTACATTCTCCC-3′ |
2.2.2 Expression of recombinant yeast MnSOD in E. coli
DNA encoding the mature yMnSOD polypeptide was amplified from genomic template using primers YSOD-3 and YSOD-4 (Table 2). The PCR product was digested with NdeI and BamHI and ligated into a similarly digested pBAD2 E. coli expression vector (15). After nucleotide sequence verification the expression plasmid pBAD2YSOD was electrotransformed into electrocompetent BW25113ΔsodA designer deletion E. coli strain (15).
2.2.3 Expression of human MnSOD in S. cerevisiae
DNA encoding the mature human mitochondrial MnSOD (hMnSOD) polypeptide lacking the human mitochondrial targeting sequence was amplified from pHMNSOD-4 cDNA clone (ATCC, Manassas, VA), using primers hMnSOD-3 and hMnSOD-4 (Table 2). The PCR product was digested with BsiWI and XhoI. The S. cerevisiae yMTS together with the PSOD2 promoter was amplified from genomic template using primers PSOD-1 and YSODMTS (Table 2). A unique BsiWI restriction site was designed into the YSODMTS primer by silent mutagenesis. The PCR product was digested with AgeI and BsiWI. The two digested PCR products were ligated with AgeI/XhoI digested pYES2. The triple ligation product was isolated and the insert verified by nucleotide sequence analysis, yielding pYEPSOD-HSOD.
2.2.4 Expression of human MnSOD isoform B in E. coli
DNA encoding the human mitochondrial MnSOD isoform B was amplified from a cDNA clone (OriGene, Rockville,MD) using the primers hMnSOD-1 and hMnSOD-2 (Table 2). The PCR product was digested with NdeI and HindIII and ligated into similarly digested pET23a vector (Novagen, Madison, WI). pET23HSOD(isoform B) was verified and electrotransformed into BL21-AI and C41(DE3) E. coli strains for expression.
2.2.5 Expression of human hMnSOD isoform B in S. cerevisiae
DNA encoding hMnSOD isoform B was amplified from the cDNA clone using primers hMnSOD-3 and hMnSOD-4 (Table 2). The PCR product was digested with BsiWI and XhoI and ligated together with PSOD2-yMTS into the AgeI/XhoI-digested pYES2 vector yielding the pYEPSOD-HSOD(isoform B) vector. The insert was verified by nucleotide sequence analysis.
2.3. Protein Expression
Recombinant yeast MnSOD with high Mn content was prepared by growing E. coli BW25113 ΔsodA | pBAD2YSOD in Terrific Broth supplemented with 0.15% glucose and 125 μg/ml carbenicillin for 20 hr at 37 °C. The cells were collected, resuspended in induction medium (MOPS minimal medium containing 0.2% glycerol, 10 μM L-arabinose, 5 mM MnCl2 and carbenicillin), and incubated for 24 hr at 37 °C with shaking. E. coli C41(DE3| pET23HSOD(isoform B) was cultivated in LB/glucose media containing carbenicillin for 24 hr at 37 °C and the expression of hMnSOD isoform B was induced with 40 μM of IPTG 4 hr after inoculation. Yeast expression strains were cultivated in lactate medium (25) supplemented with 20 μM MnCl2 at the specified temperature for 24 hr.
2.4. Isolation of mitochondria from S. cerevisiae
Mitochondria were purified from yeast by a modification of standard methods (26). Freshly collected cells (6 to 8 g) from a 24 hour lactate culture (2 L) were treated with Zymolyase (Seikagaku, East Falmouth, MA) and spheroplasts were homogenized with 40 strokes of the tight fitting pestle in a 40 ml Dounce Tissue Grinder (Wheaton Glass, Millville, NJ). Unbroken spheroplasts and subcellular debris were pooled and rehomogenized a total of 4 times. The partially purified mitochondria were further purified with sucrose density gradient using Beckman L8-M ultracentrifuge (113,000×g, 25,000 rpm, 1 h). The mitochondrial fraction suspended at the interface between 60 and 32 % sucrose layers was diluted by adding 2–3 volumes of SEM buffer (26) slowly with gentle swirling, and debris clumps were removed. Purified mitochondria were collected by centrifugation at 14,000 rpm in an Eppendorf centrifuge for 30 min at 4 °C. The mitochondrial pellet was resuspended in 1 ml of SEM and stored in −80 °C.
2.5. Protein Purification
2.5.1 Purification of yeast MnSOD from E. coli
25 g of cells in 50 ml of 50 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA were broken by sonication and the cell free extract was loaded on a DE-52 column (5 × 45 cm) equilibrated with 10 mM Tris/HCl buffer (pH 8.0) containing 1 mM EDTA. The column was washed with 1 L of the same buffer and the protein was eluted with 4 L of 0 to 0.25 M of NaCl gradient in the same buffer. The fractions contained yMnSOD were pooled, concentrated and then dialyzed against 2 L of Tris/EDTA buffer. The dialysate was clarified by centrifugation, and the light pink supernatant was further concentrated by YM 10 ultrafiltration.
2.5.2 Purification of yeast MnSOD from isolated yeast mitochondria
0.8 ml of purified mitochondria in SEM was pelleted by centrifugation at 14000 rpm for 15 min with Eppendorf centrifuge. The mitochondrial pellet was resuspended in 0.8 ml of B-PER reagent (Pierce, Rockford, IL) containing 1 mM of EDTA and vortexed for 2 min. The B-PER supernatant was directly loaded on a DE-52 column (1.5 × 8 cm, equilibrated in 10 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA) and the column was washed with 20 ml of the same buffer. Protein was eluted with a sequential NaCl step gradient (20 ml of 0.05 to 0.3 M NaCl, 0.05 M interval) in the same buffer. yMnSOD fractions were pooled, concentrated and dialyzed against 20 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA,.
2.5.3 Purification of human MnSOD from isolated yeast mitochondria
The B-PER supernatant containing recombinant human MnSOD (rhMnSOD) was loaded onto a DE-52 column as described above for yMnSOD. The column was washed with the same buffer and the flow through fractions contained rhMnSOD were pooled and concentrated as described early. Mn reconstitution of purified protein was done as described previously (13).
2.5.4 Purification of rhMnSOD isoform B from E. coli inclusion bodies
10 g of E. coli cells in 50 ml of 50 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA were broken by sonication and the cell free extract was discarded. The precipitate containing insoluble rhMnSOD isoform B was resuspended in the same buffer by repeated sonication and centrifugation. The gray densely packed pellet was dissolved in 50 mM Tris/HCl buffer (pH 8.5) containing 6 M guanidinium HCl and 100 mM DTT. The mixture was centrifuged and the supernatant was loaded on a Sepharose CL-6B column (1.5 × 98 cm) equilibrated with the same buffer, containing 6 M guanidinium HCl and 5 mM DTT. The fractions containing rhMnSOD isoform B were identified by SDS-PAGE. The fractions containing purified rhMnSOD isoform B were pooled and dialyzed against 4 L of 10 mM Tris/HCl buffer (pH 8.5). The protein precipitate was collected by centrifugation and redissolved in 10 mM Tris/HCl (pH 8.0) containing 3.5 M guanidinium HCl, 10 mM β-mercaptoethanol and 1 mM MnCl2 for refolding. The protein solution was dialyzed against 2.5 M guanidinium HCl in the same buffer. The refolding process was continued by dialysis against several changes of buffer without denaturants. Soluble protein was clarified by centrifugation.
2.6. Effect of Superoxide Stress on the Yeast Cultures
BY4700 and BY4230 yeast strains containing the empty pYES2 vector, pYEPSOD-YSOD, pYEPSOD-HSOD, or pYEPSOD-HSOD(isoform B) were cultured in lactate media at 30 °C. Replicate cultures were inoculated at low cell density in fresh medium with or without 0.3 mM paraquat. The optical density at 600 nm was measured spectrophotometrically after 24 h, and the results analyzed as described (27).
2.7. Biochemical Characterization
The concentration of the purified protein was determined by optical absorption measurements, using the published molar extinction coefficient (ε280, nm = 1.86 × 105 M−1 cm−1 (hMnSOD) (28); 1.87 × 105 M−1 cm−1 (yMnSOD)(29). Superoxide dismutase activity was measured with xanthine oxidase/cytochrome c inhibition assay (30). Metal analyses were performed using atomic absorption spectrometer (Varian Instruments SpectrAA Model 20B equipped with a GTA96 graphite furnace). Whole protein mass analysis was performed by the Proteomics Shared Resource at OHSU.
In vitro respiratory assays of isolated mitochondria were performed as described (31). The measurement of mitochondrial oxygen consumption was performed using a YSI (Yellow Springs, OH) Clark oxygen electrode and a high-accuracy oxygen polarograph circuit (32). The temperature of the reaction mixture was maintained at 30 °C by a circulating water bath connected to the water-jacketed reaction chamber. The response of the electrode was calibrated using the protocatechuic acid/protocatechuate dioxygenase reaction (33). The respiration medium (1.8 ml) containing 10 mM potassium phosphate and 20 mM HEPES buffer (pH 7.2), 0.5 mM EGTA, 3 mM MgCl2, 20 mM taurine, 60 mM potassium lactobionate, 110 mM sucrose, and 1 mg/ml fatty acid free BSA (all stock solutions were adjusted to neutral pH) was air saturated with constant stirring for 20 min. A suspension of purified mitochondria in SEM (~20 μl) was added and the reaction chamber was sealed with a glass stopper containing a sample addition capillary channel. The reaction was initiated by sequential addition (at 2–4 min interval) of 10 mM succinate, 2.5 mM ADP and 10 μM cytochrome c. For evaluation the P/O ratio (34) 1 μM of ADP was added after addition of succinate and the reaction was continued until oxygen was depleted. Alternatively 0.5 mM NADH was used instead of succinate to evaluate the P/O ratio for mitochondria complex I.
Lumi-Light western blotting kit (Roche Applied Science, Indianapolis, IN) and rabbit polyclonal anti-HSOD antibodies (Santa Cruz Biotechnology) were used to detect rhMnSOD. Total protein from yeast was prepared for SDS-PAGE analysis by the quantitative proteomics method (35). Briefly, the cell pellet from 5 ml of culture was resuspended in 135 μl of yeast lysis buffer (0.1 M NaOH, 0.05 M EDTA, and 2 % SDS, pH 8.0) and 2.7 μl of β-mercaptoethanol was added to the mixture. The sample was incubated at 90 °C for 10 min, then 13.5 μl of 1 M acetic acid was added and the sample vortexed for 30 seconds and further incubated for 10 min at 90 °C.
3. Results and Discussion
3.1. Temperature dependent metallation of rhMnSOD in yeast mitochondria
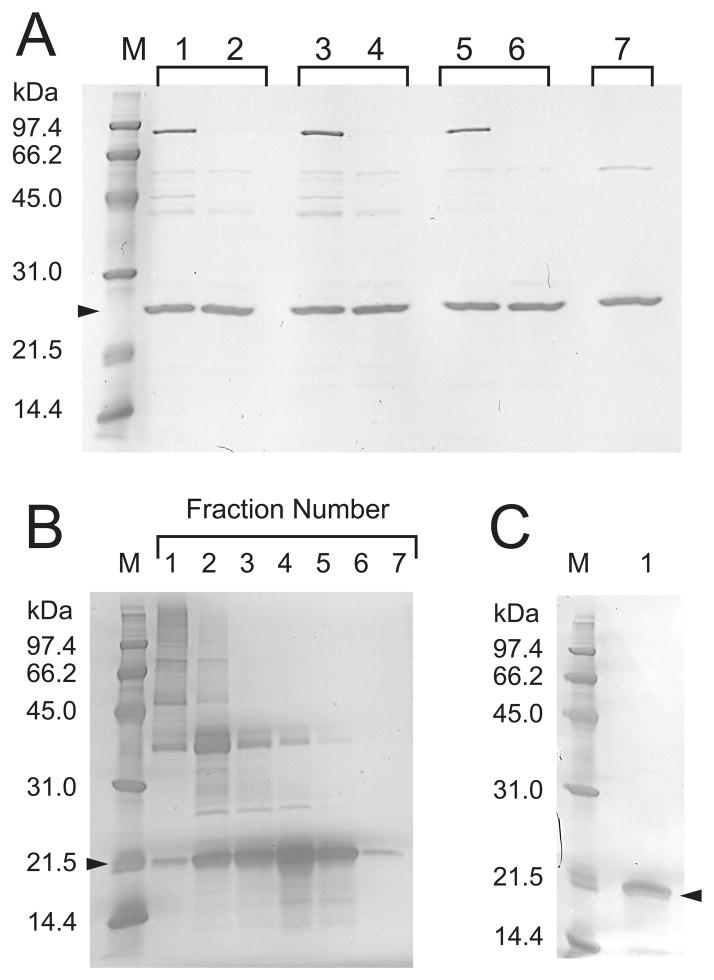
Recombinant human rhMnSOD was expressed in a Δsod2 knock-out strain of S. cerevisiae containing an episomal expression vector (pYEPSOD-HSOD) designed to provide on-demand expression of MnSOD controlled by the native yeast PSOD2 promoter. Using sucrose gradient purified yeast mitochondria as the source, we were able to isolate essentially pure rhMnSOD following a single DE-52 chromatography purification step (Fig. 2A). The purified protein has low Mn content and SOD specific activities (Table 3) compared to purified recombinant rhMnSOD from E. coli grown in Mn supplemented medium (13). In contrast, yeast ryMnSOD purified from an equivalent expression system has the same high Mn content and enzymatic activity as ryMnSOD from E. coli grown on Mn enriched minimal media. In vitro reconstitution of the purified rhMnSOD with Mn significantly increased both the Mn content and specific activity, implying that at 30 °C the majority of rhMnSOD expressed in yeast mitochondria is the apoprotein. Consistent with previous observations on the instability of apo-hMnSOD compared to the metallated forms (13, 36), attempts to remove the minor contaminants (Fig. 2A, lane 3) from rhMnSOD by CM-52 chromatography resulted in loss of most of the protein. The chimeric rhMnSOD protein is targeted to the yeast mitochondria by the yMTS signal sequence, which is properly processed yielding the correct protein mass (rhMnSOD: 22203 Da, predicted: 22192.1 Da).
Fig. 2.
SDS-PAGE of recombinant human MnSOD. M, molecular weight standards. (A) rhMnSOD purified from yeast mitochondria. Lanes 1–6: rhMnSOD prepared from cultures grown at 25°C, (lanes 1 and 2); 30°C (lanes 3 and 4); and 37°C (lanes 5 and 6). Before Mn reconstitution: lanes 1, 3, and 5. After Mn reconstitution: lanes 2, 4, and 6. Lane 7: Recombinant yeast yMnSOD purified from yeast mitochondria. (B) rhMnSOD isoform B purification. Sepharose CL-6B chromatography fractions eluted in 50 mM Tris/HCl buffer pH 8.5, containing 6 M guanidinium HCl and 5 mM DTT. (C) Refolded rhMnSOD isoform B (2 μg soluble protein).
Table 3.
Metal Content and SOD Activity of recombinant MnSOD.
| Protein | T (°C)c | Protein Property | Before Mn reconstitution | After Mn reconstitution | Ref. |
|---|---|---|---|---|---|
| hMnSODa | 25 | Mn/monomer | 0.07 ± 0.01 | 0.70 ± 0.06 | This work |
| Fe/monomer | 0.15 ± 0.01 | 0.11 ± 0.02 | |||
| SOD activity (U/mg) | 925 ± 25 | 7115 ± 566 | |||
| 30 | Mn/monomer | 0.17 ± 0.01 | 0.71 ± 0.02 | This work | |
| Fe/monomer | 0.27 ± 0.05 | 0.13 ± 0.02 | |||
| SOD activity (U/mg) | 1406 ± 18 | 6639 ± 107 | |||
| 37 | Mn/monomer | 0.38 ± 0.01 | 0.71 ± 0.02 | This work | |
| Fe/monomer | 0.21 ± 0.03 | 0.15 ± 0.01 | |||
| SOD activity (U/mg) | 4734 ± 193 | 6900 ± 63 | |||
| hMnSOD | 37 | Mn/monomer | 0.84 ± 0.06 | n. d.d | 13 |
| Fe/monomer | n. d.c | n. d.d | |||
| SOD activity (U/mg) | 7164 ± 48 | n. d.d | |||
| yMnSODa | 30 | Mn/monomer | 0.87 ± 0.04 | n. d.d | This work |
| Fe/monomer | 0.20 ± 0.01 | n. d.d | |||
| SOD activity (U/mg) | 3823 ± 34 | n. d.d | |||
| yMnSODb | 37 | Mn/monomer | 0.83 ± 0.02 | n. d.d | This work |
| Fe/monomer | 0.03 ± 0.02 | n. d.d | |||
| SOD activity (U/mg) | 4353 ± 38 | n. d.d |
mitochondrial MnSOD isolated from yeast grown at different temperatures.
recombinant yMnSOD purified from E. coli grown at 37 °C in Fe-limited/Mn-supplemented media.
growth temperature.
n. d., not determined.
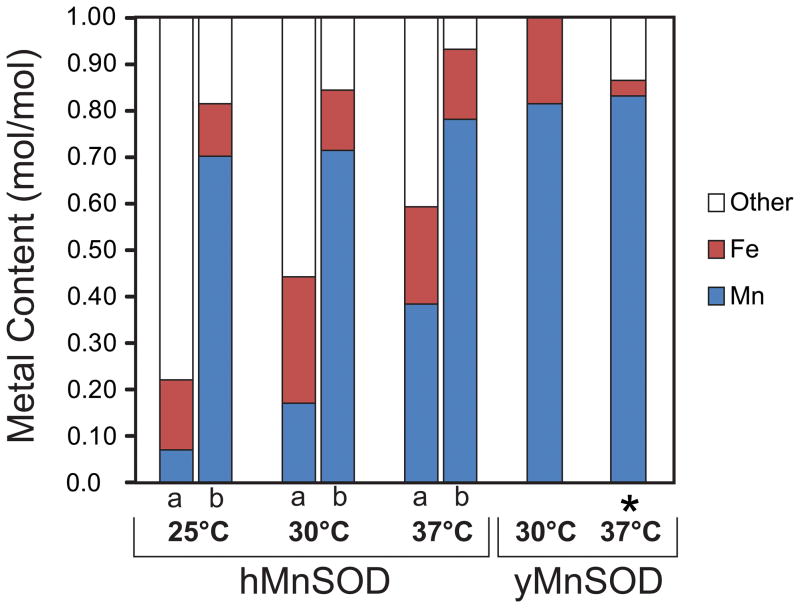
Previous studies (37) have shown that recombinant thermophilic MnSOD from Thermus thermophilus is expressed as soluble metal-free apoprotein in a mesophilic host (E. coli), and that in vitro incubation of purified apoprotein with Mn salt at the physiological growth temperature of T. thermophilus (65 °C) fully restores enzymatic activity. In the present study, the metal content and SOD activity of rhMnSOD isolated from yeast grown at various temperatures shows a clear trend (Table 3, Fig. 3). The as-isolated rhMnSOD protein has low Mn content and SOD activity, particularly at the lowest growth temperature (25 °C), but high Mn content and SOD activity can be achieved by reconstitution (Table 3, Fig. 3), confirming that a significant fraction of apoprotein was present in the purified SOD even at the highest temperature (37 °C). The presence of a small amount of Fe in rhMnSOD is not surprising, given the low metal selectivity of the protein and the abundance of Fe in the mitochondrial matrix. Prokaryotic E. coli FeSOD targeted to the yeast mitochondrion has been shown to be expressed in a functional (Fe-containing) form (38), and Fe is incorporated into the endogenous yMnSOD when mitochondrial Fe homeostasis is disrupted (4). In fact, isolation of apo-rhMnSOD from yeast mitochondria demonstrates that the majority of the Fe in healthy yeast mitochondria is not readily available for binding to apo-rhMnSOD, and that rhMnSOD protein can fold in vivo in the absence of any metal cofactor. It is not clear whether rhMnSOD apoprotein is also produced in E. coli during expression of the recombinant protein, since any apoprotein present would be lost during the multi-step purification.
Fig. 3.
Metallation state of MnSOD. Metal content of protein samples was measured before (a) and after (b) Mn reconstitution as described in the Methods for MnSOD from cultures grown at different temperatures: mitochondrial rhMnSOD isolated from yeast grown at 25, 30 and 37 °C; mitochondrial yMnSOD isolated from yeast grown at 30 °C; and yMnSOD isolated from E. coli grown at 37 °C in medium supplemented with MnCl2 (*).
The rate of metal uptake during in vitro reconstitution of apo-rhMnSOD prepared by denaturation/chelation/refolding methods is temperature-, pH-, and metal ion concentration-dependent (13). In vitro metal uptake by rhMnSOD exhibits a simple Arrhenius temperature dependence, described by (equation 1):
| (1) |
where κ is the rate, Ea the activation energy (110 kcal/mol)(13), R the gas constant, T the absolute temperature, and A a constant. From this equation, rate ratios for in vitro metal uptake at different temperatures can be calculated: κ(37 °C)/κ(30 °C) and κ(37 °C)/κ(25 °C) can be estimated as 2.1 and 5.6, respectively. Interestingly, the ratios of Mn content found for rhMnSOD isolated from yeast grown at 25 °C, 30 °C, and 37 °C (Table 3) show a similar trend, with Mn(37 °C)/Mn(30 °C) = 2.2 and Mn(37 °C)/Mn(25 °C) = 5.4. This suggests that the in vivo metal uptake process for rhMnSOD in yeast mitochondria may be associated with a similar activation barrier as the in vitro reaction, although the possibility that in yeast the metal was bound before protein folding was completed cannot be ruled out.
The contrasting metallation behavior exhibited by rhMnSOD and the yeast homolog (ryMnSOD) was rather unexpected, given that both proteins share a similar tetrameric structure (17,29) and both presumably acquire their metal cofactors by similar mechanisms within mitochondria. The difference in metal binding by the two proteins in vivo may reflect differences in temperature adaptation of the source organisms (yeast, Topt = 30°C; human, Topt = 37°C), which might result in more rapid folding and a more rigid structure for the human enzyme at lower temperatures. Alternatively, the results may reveal the specificity of metal delivery systems within the yeast mitochondrion for the yeast enzyme, and an inability to efficiently recognize hMnSOD as a partner.
3.2. No thermal activation was observed for hMnSOD isoform B
Unlike the wildtype (wt) hMnSOD isoform A, hMnSOD isoform B is expressed as inclusion bodies in E. coli. Insoluble rhMnSOD isoform B was isolated from E. coli by repeated sonication and centrifugation of the cell pellet and the rhMnSOD isoform B protein was purified by gel filtration under denaturating conditions with Sepharose CL-6B (Fig. 2B). Attempts to refold the purified protein by slowly removing the denaturant in the presence of MnCl2 met with only limited success (Fig. 2C). The soluble rhMnSOD isoform B recovered from the refolding mixture has a mass of 18306.5 Da (predicted: 18309.6 Da, with N-terminal methionine) and lacks any detectable SOD activity. The soluble protein precipitated when the MnCl2 was removed by dialysis.
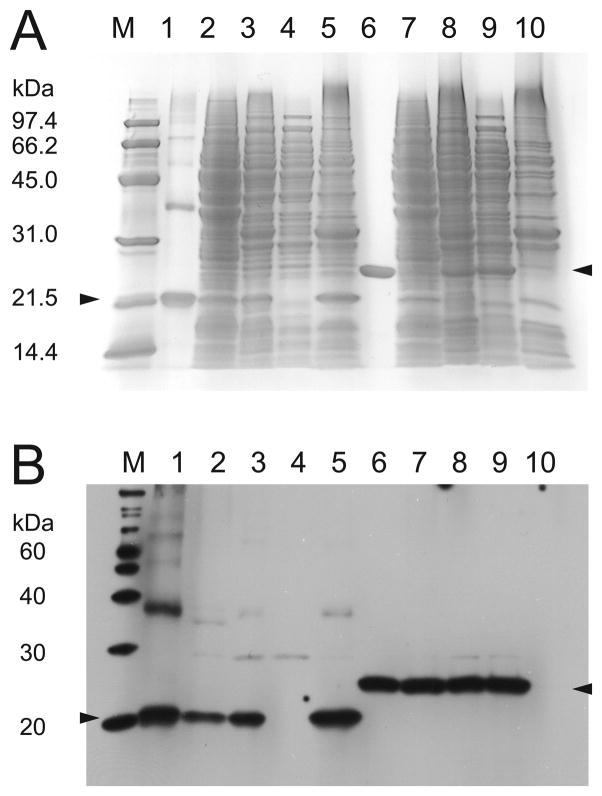
rhMnSOD isoform B expressed in yeast at 30° C also produced an insoluble protein product (Fig. 4, lane 5) that is exclusively found in the insoluble B-PER fraction of purified mitochondria, while wt rhMnSOD is exclusively in the soluble fraction (Fig. 4, lane 10). rhMnSOD isoform B fused to the yMnSOD yMTS appears to be efficiently imported into yeast mitochondria, and the signal sequence is correctly cleaved yielding a polypeptide of mass 18175.3 Da (predicted 18177.6 Da). The N-terminal yMTS may direct the precursor protein to the mitochondrial import machinery as the nascent polypeptide emerges from the ribosome, so that the full-length protein is never released into the cytosol (39), preventing aggregation of the insoluble preprotein in that space. Identical expression results (data not shown) were observed for yeast grown at 37° C, thus there is no thermal activation behavior for rhMnSOD isoform B. Under the growth conditions used in this study the absence of soluble MnSOD or fully metallated functional MnSOD in the mitochondria of Δsod2 knock-out yeast did not appreciably affect the in vitro P/O ratio of purified mitochondria, which was consistently ~1.5, even when hMnSOD isoform B was expressed. Thus, expression of hMnSOD isoform B did not appear to produce any significant deleterious effects in yeast. However, it is possible that the effects of hMnSOD isoform B expression may be different in mammalian cells, particularly if a soluble protein product is formed in the latter.
Fig. 4.
SDS-PAGE of rhMnSOD and rhMnSOD isoform B. Upper panel, GelCode blue stain detection. Lower panel, western blot detection. M, molecular weight standards; (1) rhMnSOD isoform B purified from E. coli, (2) rhMnSOD isoform B in yeast cell lysis, (3) rhMnSOD isoform B in purified yeast mitochondria, (4) mitochondrial B-PER supernatant, (5) rhMnSOD isoform B in mitochondrial B-PER precipitate, (6) rhMnSOD purified from E. coli, (7) rhMnSOD in yeast cell lysis, (8) rhMnSOD in purified yeast mitochondria, (9) rhMnSOD in mitochondrial B-PER supernatant, (10) mitochondrial B-PER precipitate.
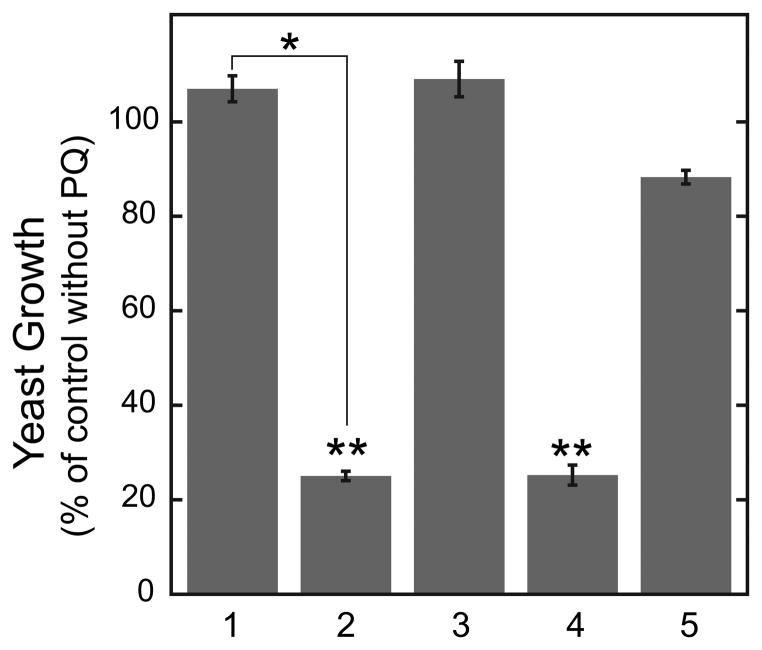
3.3. hMnSOD in yeast mitochondria is fully active against superoxide stress produced by paraquat
Paraquat (PQ) induces oxidative stress in yeast by generating superoxide in the mitochondrial matrix through redox cycling coupled to electron transport. MnSOD protects mitochondria against PQ toxicity (27), and the growth phenotype of yeast in the presence of PQ can serve as a stress test for MnSOD function. Yeast expressing rhMnSOD or ryMnSOD were both insensitive to the presence of 0.3 mM PQ (Fig. 5, 1&3), demonstrating that even partially metallated rhMnSOD can fully protect yeast lacking endogenous yMnSOD. Both of these overexpression strains were slightly more resistant to the effects of PQ than the yeast strain containing a single chromosomal copy of the sod2 gene (Fig. 5, 5). On the other hand, expression of insoluble rhMnSOD isoform B had no effect on the growth of the yeast under these conditions, neither enhancing protection against PQ toxicity, nor producing any significant additional deleterious effect (Fig. 5, 2) compared to the Δsod2 control culture (Fig. 5, 4). These results suggest that expression of the hMnSOD isoform B polypeptide does not itself produce deleterious effects, but in human tissue its expression may correlate with a quantitative decrease in normal hMnSOD (isoform A) expression, which is known to result in severe metabolic disorders. Association of hMnSOD isoform B with cancer may simply reflect an increase in aberrant splicing of transcripts in the disease state (40–42). As noted above, there may be differences in the behavior of hMnSOD isoform B in yeast and mammalian cells.
Fig. 5.
Effect of paraquat superoxide stress on yeast cultures. Yeast sensitivity to paraquat is expressed as the ratio of culture density at 24 h for cells in lactate medium with and without paraquat. (1) BY4230 (Δsod2) yeast containing the hMnSOD expression plasmid; (2) BY4230 (Δsod2) yeast containing containing the isoform B hMnSOD expression plasmid; (3) BY4230 (Δsod2) yeast containing the yeast yMnSOD expression plasmid; (4) Δ BY4230 (Δsod2) yeast containing the empty pYES2 vector; (5) BY4700 (Δura3) strain yeast containing the empty pYES2 vector. Data are the mean ± S.D. of two independent experiments. Statistical significance was calculated with a Student’s two-tailed t-test: *, p = .008, (very significantly different); **, p = 0.925 (not significantly different).
Acknowledgments
Support for this project from National Institutes of Health (GM 42680 to J.W.W.) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCord JM. Oxygen-derived free radicals. New Horiz (Baltimore) 1993;1:70–76. [PubMed] [Google Scholar]
- 2.Fridovich I. Superoxide radical and superoxide dismutase. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 3.Halliwell B. Reactive oxygen species in living systems: source, biochemistry, and role in human disease. Am J Med. 1991;91:14S–22S. doi: 10.1016/0002-9343(91)90279-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. The EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer WF, Jr, Fridovich I. In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J Biol Chem. 1991;266:303–308. [PubMed] [Google Scholar]
- 6.Whittaker MM, Whittaker JW. Mutagenesis of a proton linkage pathway in Escherichia coli manganese superoxide dismutase. Biochemistry. 1997;36:8923–8931. doi: 10.1021/bi9704212. [DOI] [PubMed] [Google Scholar]
- 7.Culotta VC, Yang M, O’Halloran TV. Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 9.Holley AK, Bakthavatchalu V, Velez-Roman JM, St Clair DK. Manganese superoxide dismutase: guardian of the powerhouse. Int J Mol Sci. 2011;12:7114–7162. doi: 10.3390/ijms12107114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Huang HT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Saavedra D, McCord JM. Association of a new intronic polymorphism of the SOD2 gene (G1677T) with cancer. Cell Biochem Funct. 2009;27:223–227. doi: 10.1002/cbf.1560. [DOI] [PubMed] [Google Scholar]
- 12.Aguirre JD, Culotta VC. Battles with iron: manganese in oxidative stress protection. J Biol Chem. 2012;287:0000–0000. doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittaker MM, Whittaker JW. In vitro metal uptake by recombinant human manganese superoxide dismutase. Arch Biochem Biophys. 2009;491:69–74. doi: 10.1016/j.abb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittaker MM, Mizuno K, Bächinger HP, Whittaker JW. Kinetic analysis of the metal binding mechanism of Escherichia coli Manganese superoxide dismutase. Biophys J. 2006;90:598–607. doi: 10.1529/biophysj.105.071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittaker MM, Whittaker JW. Conformationally gated metal uptake by apomanganese superoxide dismutase. Biochemistry. 2008;47:11625–11636. doi: 10.1021/bi8015636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittaker MM, Lerch TF, Kirillova O, Chapman MS, Whittaker JW. Subunit dissociation and metal binding by Escherichia coli apo-manganese superoxide dismutase. Arch Biochem Biophys. 2011;505:213–225. doi: 10.1016/j.abb.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borgstahl GE, Parge HE, Hickey MJ, Johnson MJ, Boissinot M, Hallewell RA, Lepock JR, Cabelli DE, Tainer JA. Human mitochondrial manganese superoxide dismutase polymorphic variant Ile58Thr reduces activity by destabilizing the tetrameric interface. Biochemistry. 1996;35:4287–4297. doi: 10.1021/bi951892w. [DOI] [PubMed] [Google Scholar]
- 18.Wan XS, Devalaraja MN, St Clair DK. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol. 1994;113:1127–1136. doi: 10.1089/dna.1994.13.1127. [DOI] [PubMed] [Google Scholar]
- 19.St Clair DK, Holland JC. Complementary DNA encoding human colon cancer manganese superoxide dismutase and the expression of its gene in human cells. Cancer Res. 1991;51:939–943. [PubMed] [Google Scholar]
- 20.Anziano PQ. Manganese superoxide dismutase exon 3-deleted isoforms and nucleic acid molecules encoding the isoforms. US7, 277, 001. Patent No. 2007;B2:1–30.
- 21.Wang F, Huang L. Study on expression of human manganese superoxide dismutase gene splicing isoforms. Biotechnology Bulletin. 2010;3:39. CNKI:SUN:SWJT.0.2010-03-039. [Google Scholar]
- 22.AceView. URL: www.ncbi.nlm.nih.gov/IEB/Research/Acembly.
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 25.Tatsuta T, Langer T. Studying proteolysis within mitochondria. Methods Mol Biol. 2007;372:343–360. doi: 10.1007/978-1-59745-365-3_25. [DOI] [PubMed] [Google Scholar]
- 26.Gregg C, Kyryakov P, Titorenko VI. Purification of Mitochondria from Yeast Cells. J Vis Exp. 2009;30:e1417. doi: 10.3791/1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cochemé HM, Murphy MP. Complex I is the major site of mitochondria superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 28.Ho YS, Crapo YD. Isolation and characterization of complementary DNAs encoding human manganese-containing superoxide dismutase. FEBS Lett. 1988;229:256–260. doi: 10.1016/0014-5793(88)81136-0. [DOI] [PubMed] [Google Scholar]
- 29.Ravindranath SD, Fridovich I. Isolation and characterization of a Manganese-containing superoxide dismutase from yeast. J Biol Chem. 1975;250:6107–6112. [PubMed] [Google Scholar]
- 30.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 31.Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–372. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meunier PC, Popovic R. High-accuracy oxygen polarograph for photosynthetic systems. Rev Sci Instrum. 1988;59:486–491. [Google Scholar]
- 33.Whittaker MM, Ballou DP, Whittaker JW. Kinetic isotope effects as probe of the mechanism of galactose oxidase. Biochemistry. 1998;37:8426–8436. doi: 10.1021/bi980328t. [DOI] [PubMed] [Google Scholar]
- 34.Guérin B, Labbe P, Somlo M. Preparation of yeast mitochondria (Saccharomyces cerevisiae) with good P/O and respiratory control. Methods Enzymol. 1979;50:149–159. doi: 10.1016/0076-6879(79)55021-6. [DOI] [PubMed] [Google Scholar]
- 35.von der Haar T. Optimized protein extraction for quantitative proteomic of yeasts. PLoS ONE. 2007;10:e1078, 1–8. doi: 10.1371/journal.pone.0001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, Radi R. Reaction of peroxynitrite with Mn-superoxide dismutase. J Biol Chem. 2001;276:11631–11638. doi: 10.1074/jbc.M009429200. [DOI] [PubMed] [Google Scholar]
- 37.Whittaker MM, Whittaker JW. Thermally triggered metal binding by recombinant Thermus thermophilus manganese superoxide dismutase, expressed as the apo-enzyme. J Biol Chem. 1999;274:34751–34757. doi: 10.1074/jbc.274.49.34751. [DOI] [PubMed] [Google Scholar]
- 38.Balzan R, Bannister WH, Hunter GJ, Bannister JV. Escherichia coli iron superoxide dismutase targeted to the mitochondria of yeast cells protects the cells against oxidative stress. Proc Natl Acad Sci. 1995;92:4219–4223. doi: 10.1073/pnas.92.10.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yogev O, Karniely S, Pines O. Translation-coupled Translocation of Yeast Fumarase into Mitochondria in Vivo. J Biol Chem. 2007;282:29222–29229. doi: 10.1074/jbc.M704201200. [DOI] [PubMed] [Google Scholar]
- 40.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 41.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 42.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]