Abstract
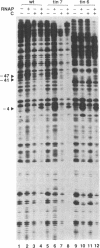
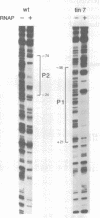
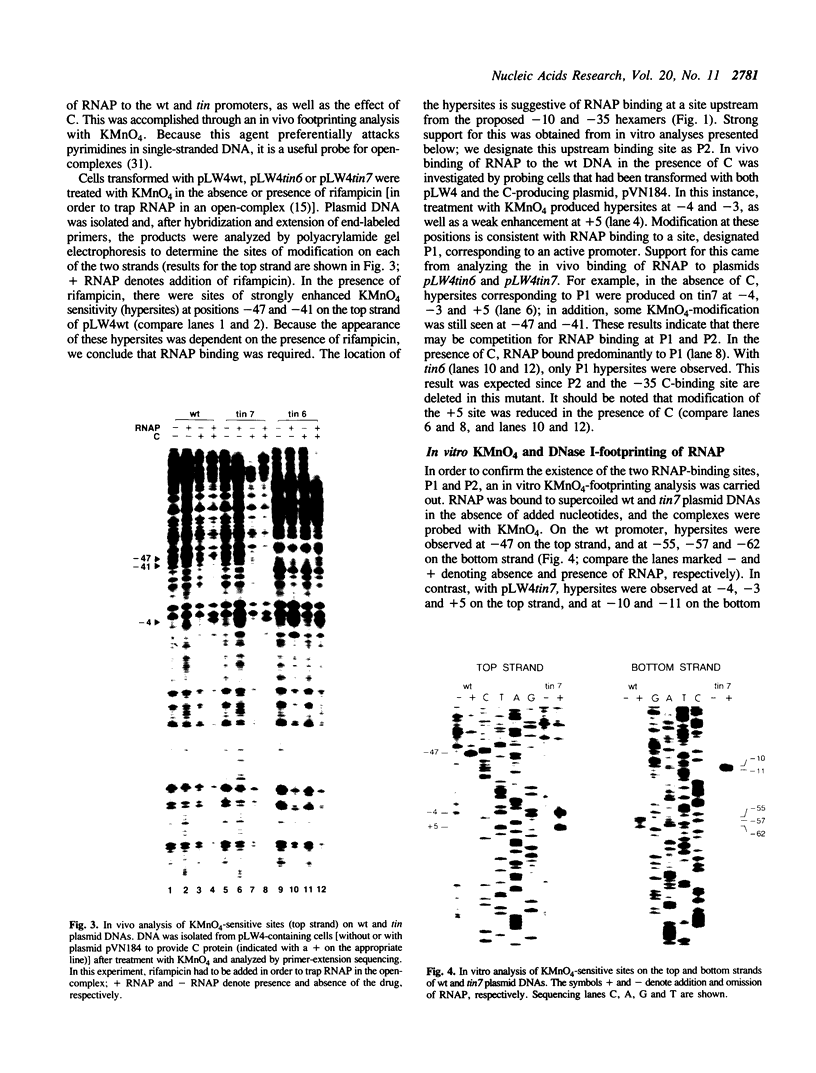
Transcription of the phage Mu com/mom operon is trans-activated by another phage gene product, C, a site-specific DNA binding protein. To gain insight into the mechanism by which C activates transcription, we carried out footprinting analyses of Escherichia coli RNA polymerase (= RNAP) binding to various com-lacZ fusion plasmids. KMnO4-sensitive sites (diagnostic of the melted regions in open-complexes) and DNase I-sensitive sites were located by primer-extension analysis. The results are summarized as follows: (i) in vivo, in the absence of C, RNAP bound in the wild-type (wt) promoter region at a site designated P2; in vitro DNase I-footprinting showed that P2 extends from -74 to -24 with respect to transcription initiation. This overlaps a known strong C-binding site (at -35 to -54). RNAP bound at P2 appeared to be in an open-complex, as evidenced by the presence of KMnO4-hypersensitive sites. (ii) In contrast, when C was present in vivo, RNAP bound in the wt promoter region at a different site, designated P1, located downstream and partially overlapping P2. RNAP bound at P1 also appeared to be in an open-complex, as evidenced by the presence of KMnO4-hypersensitive sites. (iii) Two C-independent mutants, which initiate transcription at the same position as the wt, were also analyzed. In vivo, in the absence of C, RNAP bound mutant tin7 (contains a T to G substitution at -14) predominantly at P1; in vitro DNase I-footprinting showed that P1 extends from -56 to +21. With mutant tin6 (a 63 base-pair deletion removing P2, as well as part of P1 and the C-binding site from -35 to -54), RNAP bound to P1 independent of C. We conclude that P1 is the 'functional' RNAP binding site for mom-transcription initiation, and that C activates transcription by promoting binding at P1, while blocking binding at P2.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama T., Oka A. A common mechanism of transcriptional activation by the three positive regulators, VirG, PhoB, and OmpR. FEBS Lett. 1990 Apr 9;263(1):1–4. doi: 10.1016/0014-5793(90)80691-b. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989 Aug;8(8):2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Wulczyn F. G., Kahmann R. Role of bacteriophage Mu C protein in activation of the mom gene promoter. J Bacteriol. 1989 Apr;171(4):2019–2027. doi: 10.1128/jb.171.4.2019-2027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. R., Morrissey L. M., Foster L. M., Geiduschek E. P. DNA binding by the bacteriophage SPO1-encoded type II DNA-binding protein, transcription factor 1. Formation of nested complexes at a selective binding site. J Biol Chem. 1986 Sep 25;261(27):12820–12827. [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Goradia M., Monaghan C., Bukhari A. I. Regulation of the DNA-modification function of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):647–653. doi: 10.1101/sqb.1983.047.01.076. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J. S1 nuclease mapping of the phage Mu mom gene promoter: a model for the regulation of mom expression. Gene. 1984 Jul-Aug;29(1-2):185–198. doi: 10.1016/0378-1119(84)90179-3. [DOI] [PubMed] [Google Scholar]
- Heisig P., Kahmann R. The sequence and mom-transactivation function of the C gene of bacteriophage Mu. Gene. 1986;43(1-2):59–67. doi: 10.1016/0378-1119(86)90008-9. [DOI] [PubMed] [Google Scholar]
- Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Knaus R., Bujard H. PL of coliphage lambda: an alternative solution for an efficient promoter. EMBO J. 1988 Sep;7(9):2919–2923. doi: 10.1002/j.1460-2075.1988.tb03150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Genetically separable functional elements mediate the optimal expression and stringent regulation of a bacterial tRNA gene. Cell. 1985 Feb;40(2):319–326. doi: 10.1016/0092-8674(85)90146-1. [DOI] [PubMed] [Google Scholar]
- Maeda S., Mizuno T. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J Bacteriol. 1990 Jan;172(1):501–503. doi: 10.1128/jb.172.1.501-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Rao G., Howe M. M. Bacteriophage Mu late promoters: four late transcripts initiate near a conserved sequence. J Bacteriol. 1989 Apr;171(4):2003–2018. doi: 10.1128/jb.171.4.2003-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs C. F., Howe M. M. Kinetics and regulation of transcription of bacteriophage Mu. Virology. 1990 Jan;174(1):192–203. doi: 10.1016/0042-6822(90)90068-3. [DOI] [PubMed] [Google Scholar]
- Minton N. P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984 Nov;31(1-3):269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- Nagaraja V., Hecht G., Hattman S. The phage Mu 'late' gene transcription activator, C, is a site-specific DNA binding protein. Biochem Pharmacol. 1988 May 1;37(9):1809–1810. doi: 10.1016/0006-2952(88)90457-1. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vollering M., Brinkman A., Van de Putte P. Analysis of the methylation-regulated Mu mom transcript. Cell. 1984 Jan;36(1):189–196. doi: 10.1016/0092-8674(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S., Chan B., Busby S. Functional analysis of different sequence elements in the Escherichia coli galactose operon P2 promoter. Mol Microbiol. 1988 Mar;2(2):165–172. doi: 10.1111/j.1365-2958.1988.tb00018.x. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing co-operative DNA-binding in vivo. The lac O1:O3 interaction. J Mol Biol. 1988 Jul 5;202(1):107–119. doi: 10.1016/0022-2836(88)90523-2. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Schumann W., Bade E. G., Forgie R. A., Howe M. M. Cloning of DNA fragments of the right end of phage mu and location of the HindIII, SalI, PstI, and BamHI restriction sites on the genetic map of mu. Virology. 1980 Jul 30;104(2):418–425. doi: 10.1016/0042-6822(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Seiler A., Blöcker H., Frank R., Kahmann R. The mom gene of bacteriophage Mu: the mechanism of methylation-dependent expression. EMBO J. 1986 Oct;5(10):2719–2728. doi: 10.1002/j.1460-2075.1986.tb04556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Tiedeman A. A., Smith J. M. lacZY gene fusion cassettes with KanR resistance. Nucleic Acids Res. 1988 Apr 25;16(8):3587–3587. doi: 10.1093/nar/16.8.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Mueckl D., Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r protein operon. Cell. 1980 Sep;21(2):523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]