Abstract
Objective
Related transcriptional enhancer factor 1 (RTEF-1) is a key transcriptional regulator in endothelial function. In this study, we investigated a possible role for RTEF-1 in the regulation of microvascular relaxation and the underlying mechanism involved. Activation of fibroblast growth factor receptor 1 (FGFR1) by FGFs increases vasodilation, although transcriptional control of the molecular mechanisms underlying FGFR1 is still unclear. Materials and Methods: We demonstrated that RTEF-1 stimulated FGFR1 expression at the transcriptional level, specifically an area including Sp1 elements, as evidenced by promoter assays. Additionally, RTEF-1 increased FGFR1 mRNA and protein expression in vitro and in VE-cadherin-promoted RTEF-1 (VE-Cad/RTEF-1) transgenic mice, whereas RTEF-1 siRNA blocked the upregulation of FGFR1 expression. Furthermore, increased endothelial-dependent microvessel relaxation was observed in the coronary arteries of VE-Cad/RTEF-1 mice, and increased proliferation was observed in RTEF-1-overexpressing cells, both of which correlated to increased FGF/FGFR1 signaling and endothelial nitric oxide synthase (eNOS) upregulation. Our results indicate that RTEF-1 acts as a transcriptional stimulator of FGFR1 and is involved in FGF pathways by increasing microvessel dilatation via eNOS.
Conclusions
These findings suggest that RTEF-1 plays an important role in FGFR1- stimulated vasodilatation. Understanding the effect of RTEF-1 in microvessel relaxation may provide beneficial knowledge in improving treatments in regards to ischemic vascular disorders.
Key Words: Fibroblast growth factor receptor 1, Related transcriptional enhancer factor 1, Endothelial nitric oxide synthase, Endothelial function, Microvascular relaxation
Introduction
The fibroblast growth factor/receptor (FGF/FGFR) system has been implicated in a variety of physiological and pathological conditions, including embryonic development, tissue growth and repair, nervous system control, inflammation, tumor growth and vascularization. FGF directly influences the migratory and proliferative activity of human endothelial cells and is recognized as a mediator of human angiogenesis. FGF/FGFR1 is expressed in the mesenchyme, including endothelial cells, smooth muscle cells and hemopoietic progenitor cells [1]. Many studies have demonstrated the importance of FGF2/FGFR in vessel dilatation; mainly that FGF2/FGFR increases endothelial nitric oxide synthase (eNOS) expression in endothelial cells. Nitric oxide (NO) released from the endothelium plays a major role in the determination of coronary microvascular reactivity and the angiogenic process [2, 3]. However, the underlying molecular mechanisms of transcriptional regulation are less well-defined.
Related transcriptional enhancer factor 1 (RTEF-1) belongs to the TEF-1 multigene family that binds to M-CAT elements in the promoters of many genes expressed in cardiac, smooth and skeletal muscle cells [4, 5, 6, 7, 8, 9]. Previous studies from our laboratory, as well as others, have demonstrated that the expression of RTEF-1 is increased in hypoxic endothelial cells as a result of enhanced proangiogenic activity via targeting Sp1 elements within the VEGF promoter [10, 11]. Recently, we also demonstrated that transcriptional regulation by RTEF-1 in endothelial cells is involved in the development of cardiomyocyte hypertrophy. We found that VEGF-B is a target gene of RTEF-1 that acts as a bridge between endothelial cells and cardiomyocytes during the development of cardiac hypertrophy [12].
The biological activities of FGFs are mediated through high-affinity membrane-bound tyrosine kinase receptors, which have cardioprotective and angiogenic effects in the ischemic myocardium, although the means of regulating transcriptional control of the FGF receptor are unclear. In this study, we demonstrated that RTEF-1 stimulates FGFR1 expression by regulating FGFR1 promoter activity. RTEF-1-stimulated FGFR1 expression is involved in the increase of microvascular relaxation via eNOS upregulation, indicating that RTEF-1 plays an important role in endothelial cell behavior.
Materials and Methods
Cell Culture and Transfection
Human microvascular endothelial cells (HMEC-1) were cultured in MCDB-131 (Center for Disease Control). Bovine aortic endothelial cells (BAEC), including both transfected and untransfected, were cultured in Dulbecco's modified Eagle's medium (DMEM). Both cell lines were supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin and 100 units/ml penicillin, at 37°C in an atmosphere of 95% air and 5% CO2. Cells were transfected with DNA (1 μg/105 cells) using the Lipofectamine method according to the manufacturer's protocol (Invitrogen, Carlsbad, Calif., USA). FGFR1 promoter luciferase construct, control vector, PXJ40 and/or an increasing amount of RTEF-1 expression vector (generous gifts from Dr. Stewart) were transfected into BAEC cells. PXJ40/lacZ (0.01 μg) was cotransfected to determine the transfection efficiency of approximately 70–80%. After transfection, cells were incubated for 48 h before analysis.
Cloning and Promoter Activity Assays
Briefly, the FGFR1 promoter sequences were inserted into pGL3-Basic vector (Promega, Madison, Wisc., USA) and named according to the length of the fragment from the transcription start-site in the 5′ and 3′ directions: FGFR1–865 (–865/+109), FGFR1–426 (–426/+109), FGFR1–217 (–217/+96), FGFR1–100 (–100/+96), FGFR1–48 (–48/+96) and FGFR1–20 (–20/+96). The sequences for the FGFR1–217, 100, 48 and 20 constructs were amplified by PCR with flanking 5′-KpnI and 3′-SacI enzyme restriction sites to facilitate directional cloning into the pGL3-Basic vector. The amount of control vector PJX40 was used for compensatory total volume of DNA. After 24-h transfection, luciferase activity was determined using the Dual-Luciferase assay system (Promega).
Generation of RTEF-1 Transgenic Mice
RTEF-1 transgenic mice were generated on the FVB background at the Beth Israel Deaconess Medical Center Transgenic Core Facility using the vascular endothelial-cadherin (VE)-cadherin promoter to drive endothelial-specific expression of human RTEF-1, as described previously [12, 13]. The investigation conformed to the Guide for the Care and Use of Laboratory Animals (NIH publication No. 85-23, 1996) and was approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center. RTEF-1 transgenic mice were genotyped by PCR, using the following primers: forward 5′-CTGCAGGCAGCTCACAAAGGAACAAA-3′ and reverse 5′-TCGATGGGCTTGTCCAGTGCCTGACT-3′.
siRNA Transfection
siRNAs targeting human RTEF-1 were synthesized by Genepharma, Inc. (ZhangJiang). Knockdown efficiency of the two duplexes of RTEF-1 siRNAs (siRNA-894: 5′-GGG CAG ACC UCA ACA CCA ATT-3′, 5′-UUG GUG UUG AGG UCU GCC CAG-3′ and siRNA-1748: 5′-ACC CAA GAU GCU GUG UAU UTT-3′, 5′-AAU ACA CAG CAU CUU GGG UTT-3′) or a nontarget control (5′-UUC UCC GAA CGU GUC ACG UTT-3′, 5′- ACG UGA CAC GUU CGG AGA ATT-3′) were determined by transfection into BAEC cells or HMEC-1 cells at a final concentration of 50 nM according to the manufacturer's protocol. Briefly, a master mix of Lipofectamine 2000 was diluted with 1 ml of OPTI-MEM (Invitrogen) and incubated for 5 min. Lipofectamine 2000 dilution was added to the DNA/siRNA dilution, incubated for 20 min and added drop-wise to the cells. Five hours after transfection, the media was changed and the cells were allowed to recover overnight.
RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR Analysis
Total RNA was extracted from the apex of mouse hearts from wild-type (WT) and RTEF-1 transgenic mice as well as from both endothelial cell lines using Trizol (Invitrogen) according to the manufacturer's instructions. A total of 2.0 μg of RNA from both endothelial cell lines transfected with RTEF-1 or isolated from the apex of WT and RTEF-1 transgenic mouse hearts were reverse-transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, Calif., USA) with random primers according to the manufacturer's protocol. Quantitative real-time PCR (QPCR) amplification was done using SYBR Green master mix (Applied Biosystems) according to the manufacturer's protocol with the following primers: hRTEF-1 5′-CCACGAAGGTCTGCTCTTTC-3′ and 5′-AAGTTCTCCAGCACGCTGTT-3′; hFGFR1 5′-GTGGTGTTGGCAGAGGCTATC-3′ and 5′-TCTGTTGCGTCCGACTTCAA-3′ [14]; hENOS 5′-GGGTCCTGTGTATGGATGAGT-3′ and 5′-CAGCGTCTCGTGTTCGAGG-3′; mRTEF-1 5′-GCACCATTACCTCCAACGAG-3′ and 5′-GATCAGCTCATTCCGACCAT-3′; mFGFR1 5′-GCAGAGCATCAACTGGCTG-3′ and 5′-GGAGAAGTAGGTGGTATCGCTG-3′; mGAPDH 5′-TGGTGAAGCAGGCATCTGAG-3′ and 5′-CTCCTGCGACTTCAACAGCA-3′; mENOS 5′-TCAGCCATCACAGTGTTCCC-3′ and 5′-ATAGCCCGCATAGCGTATCAG-3′. Real time quantitative PCR was performed in an SDS 7000 System (Applied Biosystems). For all individual cDNAs, amplification of each specific mRNA sequence was performed in at least 2 independently performed PCR experiments. For each reaction, expression was calculated as 2−ΔCt, where ΔCt is the difference between the Ct for the gene of interest and the Ct for the housekeeping gene GAPDH.
In vitro Assessment of Coronary Microvessel Reactivity
The coronary arteriole branch (80–180 μm internal diameters) was dissected from the left ventricle free wall tissue of isolated hearts from male WT and RTEF-1 transgenic mice (aged 10 weeks) using a ×10–60 dissecting microscope (Olympus Optical, Tokyo, Japan) as described previously [15, 16]. Microvessels were placed in a Plexiglas microvessel chamber, cannulated with dual glass micropipettes measuring 40- to 80-μm in diameter and secured with a 10-0 nylon monofilament suture (Ethicon, Somerville, N.J., USA) as previously described. For all measurements, the vessels were washed 3 times with a cold Krebs buffer solution (118 mM sodium chloride, 25 mM sodium bicarbonate, 4.5 mM potassium chloride, 2.5 mM calcium chloride, 1.0 mM magnesium sulfate and 6.0 mM glucose) and allowed to equilibrate in the buffer solution for 15–30 min between interventions. Vessels were bathed in MOPS buffer maintained at 37°C and aerated with room air. The microvessels were imaged with the use of an inverted microscope connected to a video camera and the internal lumen diameter was measured with an electronic imaging apparatus (Living Systems, Burlington, Vt., USA). Vessels were precontracted with the thromboxane A2 analog U46619 by 30% of the baseline diameter prior to the application of a vasodilator agent. After the constriction reached a steady state level, vascular responses to FGF2 (10−9−10−4M; R&D Systems, Minneapolis, Minn., or Sigma, St. Louis, Mo., USA) endothelium-dependent vasodilator A23187 (10−9−10−4M; Sigma), and sodium nitroprusside (SNP; 10−9−10−4M; Sigma) were examined.
Immunoblot Analysis
BAEC or HMEC-1 cells were washed twice with cold phosphate-buffered saline and lysed in cold RIPA buffer (Boston Bio-Products, Inc.) containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche). Protein concentrations were determined with the DC protein standard assay (Bio-Rad, Munich, Germany). Samples were subjected to 10% SDS-PAGE, transferred to nitrocellulose membranes (Whatman, Springfield Mill, UK) and subsequently blocked in TBS-Tween 20 containing 5% nonfat milk. The membranes were incubated with the indicated primary antibodies: polyclonal anti-FGFR1 antibody with 1:500 dilution, (Santa Cruz, Calif., USA); polyclonal anti-RTEF-1 antibody with 1:10,000 dilution, (Genemed Synthesis, Inc., South San Francisco, Calif., USA), monoclonal anti-vinculin with 1:65,000 dilution, monoclonal anti-phospho-eNOS with 1:500 dilution and anti-eNOS with 1:200 dilution (Cell Signaling Technology, Danvers, Mass., USA) followed by incubation with horseradish peroxidase-conjugated secondary antibodies anti-rabbit IgG with 1:3,000 dilution, (Calbiochem, La Jolla, Calif., USA) or anti-mouse IgG with 1:2,000 dilution (Vector Labs, Burlingame, Calif., USA). Blots were developed using the chemiluminescence detection system according to the instructions of the manufacturer (Thermo Fisher, Pittsburgh, Pa., USA). Densitometric analysis was done using the NIH software program, Image J.
Proliferation Assays
GFP and RTEF-1 o/e BAEC cells were seeded in 24-well plates (1.7 × 104 cells per well) and incubated for 5 h counting from day 0. At the indicated time point, culture was stopped by fixing with 100% ethanol for 15 min and subsequently staining with 0.1% crystal violet dissolved in 10% ethanol for 20 min. After washing, 10% acetic acid was added to the cells and absorbance was measured at 590-nm wavelength.
Statistical Analysis
Data were obtained from at least 3 independent cell cultures or animals, as denoted in the figure legends. For statistical analysis, if differences were established, the values were compared using a Student t test. The values were expressed as mean ± SD. The results were considered significant if p ≤ 0.05. The relaxation responses were expressed as percent relaxation of the SNP or A23187-induced vascular reactivity of the microvessels.
Results
RTEF-1 Increases FGFR1 Protein Expression in Endothelial Cells
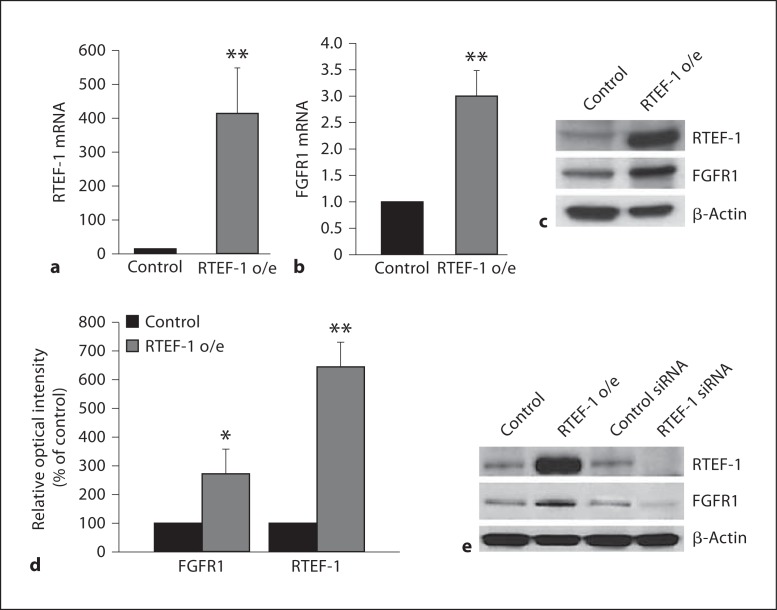
To confirm our previous DNA microarray data that showed a marked upregulation of FGFR1 expression in endothelial cells overexpressing RTEF-1, we performed QPCR analysis. Stably expressing RTEF-1 HMEC-1 cells (RTEF-1 o/e) showed significantly increased RTEF-1 mRNA (fig. 1a), as well as increased FGFR1 mRNA expression (fig. 1b). To determine whether increased FGFR1 mRNA levels by RTEF-1 resulted in altered FGFR1 protein expression, and to confirm that FGFR1 upregulation also occurred in other endothelial cells, RTEF-1 overexpression in BAEC cells was generated by transfecting RTEF-1 cDNA. BAEC cells overexpressing RTEF-1 showed an increase of FGFR1 mRNA levels by approximately 2.7-fold (fig. 1c, d). Furthermore, the knockdown of RTEF-1 by RTEF-1 siRNA in endothelial cells indicated the subsequent inhibition of FGFR1 expression (fig. 1e). These results imply that RTEF-1 regulates FGFR1 expression.
Fig. 1.
RTEF-1 upregulates FGFR1 expression in endothelial cells. a qRT-PCR assay in stably transfected RTEF-1 in HMEC-1 cells analyzing RTEF-1 mRNA levels compared with control, n = 3. ** p ≤ 0.01. b qRT-PCR assay in stably transfected RTEF-1 in HMEC-1 cells analyzing FGFR1 mRNA levels compared with control, n = 3. ** p ≤ 0.01. c Expression of RTEF-1, FGFR and a loading control, β-actin was assessed by immunoblotting. Representative blot is shown, n = 3. d Densitometric analysis of the immunoblot in c. * p ≤ 0.05; ** p ≤ 0.01. e RTEF-1 expression vector and RTEF-1 siRNA were transfected into BAEC cells. Immunoblotting shows expression of RTEF-1, FGFR and a loading control, β-actin after siRNA transfection. Representative blot is shown, n = 3.
RTEF-1 Regulates FGFR1 Promoter Activity in Endothelial Cells
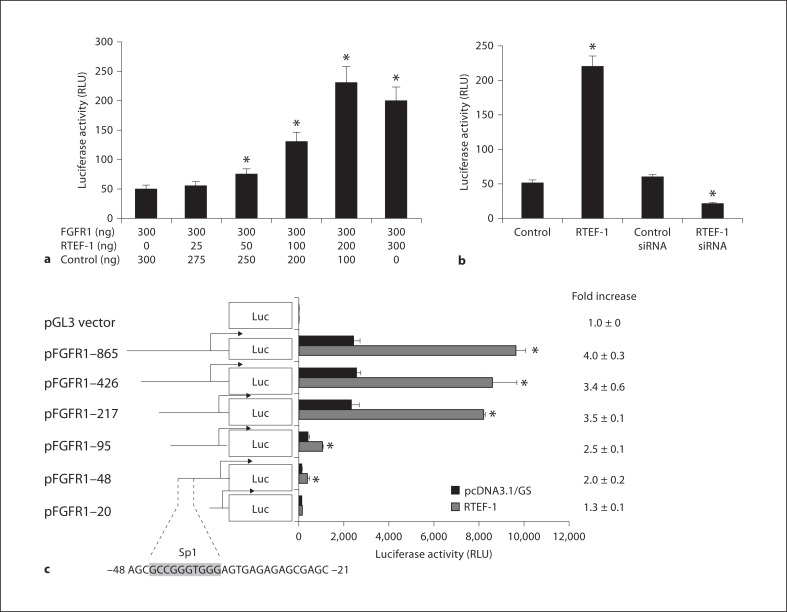
To elucidate whether RTEF-1 stimulates FGFR1 at the transcriptional level, the activities of a luciferase construct under the control of a FGFR1 promoter were measured. BAEC cells were cotransfected with an FGFR1 promoter-luciferase reporter construct with various combinations of PXJ40 (empty vector as a control) and RTEF-1 totaling 600 ng. Luciferase activity was measured 48 h after transfection. Figure 2a shows that FGFR1 promoter activity increased as a function of RTEF-1, indicating that RTEF-1 stimulates the FGFR1 promoter in a dose-dependent manner. Conversely, in BAEC cells transfected with RTEF-1 siRNA, FGFR1 promoter activity decreased significantly (fig. 2b), demonstrating that endogenous levels of RTEF-1 are important for transcriptional regulation of FGFR1 expression.
Fig. 2.
RTEF-1 increases FGFR1 promoter activity. a BAECs were cotransfected with an FGFR1 promoter-luciferase reporter construct and RTEF-1 expression vector and/or PXJ40 (empty vector) with various combinations of the latter 2 vectors totaling 600 ng. Luciferase activity was assessed 48 h after transfection, n = 3. * p ≤ 0.05. b BAECs were cotransfected with an FGFR1 promoter-luciferase reporter construct, RTEF-1 expression vector, control vector, siRNA against RTEF-1 and control siRNA. Luciferase activity was assessed 48 h after transfection, n = 3. * p ≤ 0.05. c BAEC cells were cotransfected with RTEF-1 and progressively smaller regions of the FGFR1 promoter-luciferase reporter construct. Luciferase activity was assessed 48 h after transfection, n = 3. * p ≤ 0.05.
Our laboratory has previously demonstrated that RTEF-1 stimulates VEGF expression by upregulating VEGF promoter activity through an Sp1-like motif in endothelial cells, indicating that RTEF-1 may be able to regulate different target genes through individual promoter elements in different cells [11]. A series of truncated portions of the promoter were created extending up to 0.86 kb from the transcription start site of the human FGFR-1 gene. These constructs were inserted upstream of a luciferase reporter gene in the pGL2 basic vector and designated: FGFR1–865 (–865/+109), FGFR1–426 (–426/+109), FGFR1–217 (–217/+96), FGFR1–100 (–100/+96), FGFR1–48 (–48/+96), and FGFR1–20 (–20/+96). Each construct was transiently transfected into BAEC cells and subsequently subjected to a luciferase activity assay. RTEF-1 over-expression in BAEC cells resulted in over 3-fold activation of the FGFR1 promoter activities in 5 of the 6 truncated promoter sequences: FGFR1–865, FGFR1–426, FGFR1–217, FGFR1–100, FGFR1–48, and FGFR1–20. Deletion of the FGFR1 promoter sequence from FGFR1-48 to FGFR1-865 did not abolish the ability of RTEF-1 to stimulate transcription. However, luciferase activity was abolished in the promoter construct with deletion of the region between FGFR1–48 and FGFR1–20. This truncation inhibited the RTEF-1-mediated effect on the FGFR1 promoter (fig. 2c). Interestingly, sequence analysis of the proximal upstream DNA revealed 4 potential Sp transcription factor binding sites located at the start of transcription and at −23, −42, and −54 bp. As previously mentioned, studies by our laboratory reported that the Sp1 element is involved in the RTEF-1-mediated transcription of VEGF.
RTEF-1 Induces FGFR1 Expression in vivo
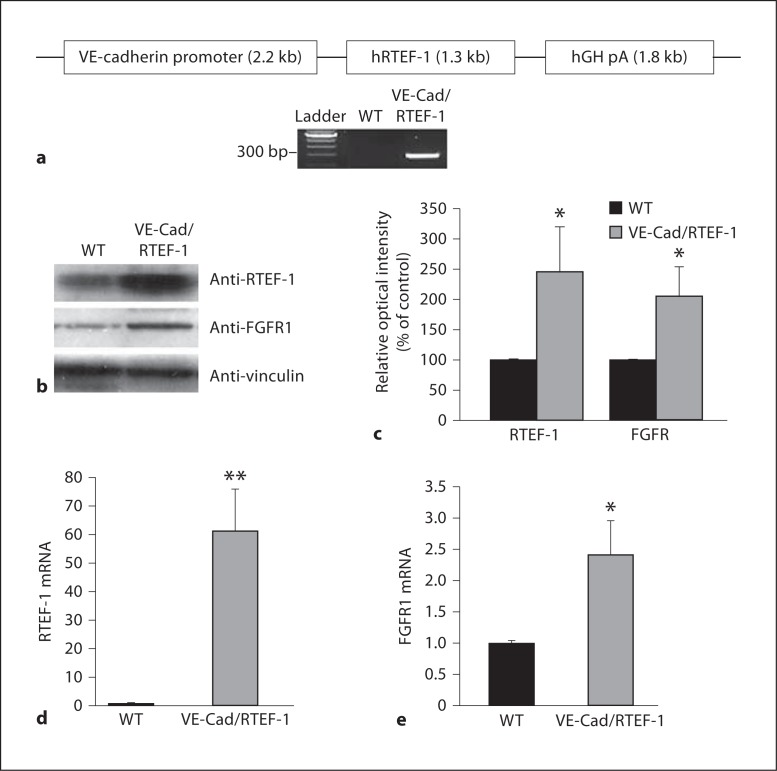
To study the effect of RTEF-1 expression on endothelial function, we generated a conditional transgenic mouse line which expresses the full-length human RTEF-1 cDNA in the endothelium under control of the VE-cadherin promoter (fig. 3a). All transgenic lines grew and bred normally when compared with WT mice. The RTEF-1 transgene expression was confirmed by an immunoblot that showed an increase of RTEF-1 expression in the heart of VE-Cad/RTEF-1 mice (fig. 3b, c). In addition, FGFR1 expression was also increased over 2-fold in VE-Cad/RTEF-1 mice, as shown by the densitometric analysis (fig. 3b, c). Moreover, QPCR using hearts from VE-Cad/RTEF-1 transgenic mice compared to littermate controls showed that levels of RTEF-1 and FGFR1 were significantly elevated in the transgenic mice (fig. 3d, e, n = 3 each).
Fig. 3.
RTEF-1 induces FGFR1 expression in VE-Cad/RTEF-1 transgenic mice. a Schematic structure of the VE-cadherin/RTEF-1 transgene construct in which the human RTEF-1 transgene is driven by the promoter sequence from the VE-cadherin gene that drives endothelial-specific expression. hGH pA indicates human growth hormone polyadenylation signal. Genotyping results via PCR for the transgenic mice. b Expression of RTEF-1, FGFR and a loading control, vinculin was analyzed by immunoblotting in the hearts of VE-Cad/RTEF-1 mice compared with those of littermate WT mice. Representative blot is shown, n = 3. c Relative optical intensity of both FGFR1 and RTEF-1 protein levels in vivo, calculated using Image J software, n = 3. * p ≤ 0.05. d qRT-PCR assay in hearts isolated from VE-Cad/RTEF-1 mice compared with control mice analyzing RTEF-1 mRNA levels, n = 6. ** p ≤ 0.01. e qRT-PCR assay in hearts isolated from VE-Cad/RTEF-1 mice compared with control mice analyzing FGFR1 mRNA levels, n = 6. * p ≤ 0.05.
RTEF-1 Increases Cardiac Microvascular Relaxation
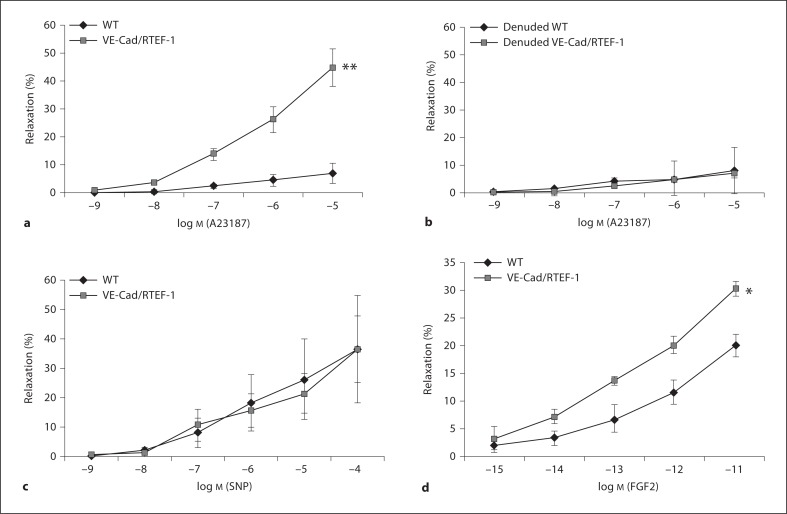
Previous data indicate that increased expression of proangiogenic genes such as VEGFR and FGFR1 have augmented the relaxation response in the endothelium [3, 17]. In addition, impaired microvascular relaxation was improved by FGF-2 treatment, and molecular analysis in these animals revealed increased FGFR1 expression [18]. To evaluate whether microvascular relaxation function is attenuated in RTEF-1 transgenic mice compared with WT mice, coronary artery branches (microvessels) at the epicardial surface were isolated microscopically from male WT and RTEF-1 transgenic mice and relaxation (diameter) was measured after preconstriction with thromboxane A2 analog U46619. After the constriction reached a steady state, an endothelium-dependent vasodilator (A23187), or a smooth muscle-dependent vasodilator (SNP), was added and relaxation was measured. Figure 4a demonstrates that microvascular relaxation was increased in VE-Cad/RTEF-1 transgenic mice compared with control mice (44.7 ± 6.7% vs. 6.8 ± 3.5%). In contrast, the addition of A23187 to endothelial denuded WT or VE-Cad/RTEF-1 mice did not show any significant differences in relaxation (fig. 4b), as A23187 is an endothelium-dependent vasodilator. In addition, when SNP was added, relaxation was similar between WT and the transgenic VE-Cad/RTEF-1 mice (fig. 4c), suggesting that the RTEF-1-mediated increases in microvascular relaxation are endothelial-cell-specific. FGF-2 is known as an inducer of angiogenesis as well an enhancer of endothelium-dependent vessel relaxation (Sellke et al. 1994). We next examined whether FGF-2-stimulated microvascular relaxation could be stimulated by RTEF-1, as FGFR1 expression was enhanced by RTEF-1. Figure 4d indicates that RTEF-1 o/e mice showed significantly increased microvascular relaxation in response to FGF-2 compared with WT mice.
Fig. 4.
RTEF-1 increases endothelial cell-specific coronary microvascular relaxation. a Plot of the in vitro response of the coronary artery branches (microvessels) to A23187 in the transgenic mice compared to WT mice. Relaxation (diameter) was measured after preconstriction with thromboxane A2 analog U46619. b Plot of the in vitro response of the denuded endothelium to A23187 in the transgenic mice compared to WT mice. Relaxation (diameter) was measured after preconstriction with thromboxane A2 analog U46619. c Plot of the in vitro relaxation response of the microvessels to SNP in the transgenic mice compared to WT mice. d Plot of the in vitro relaxation response of the microvessels to FGF-2 in the transgenic mice compared to WT mice. * p≤ 0.05, ** p ≤ 0.01. WT mice: n = 6, RTEF-1 o/e mice: n = 8.
RTEF-1 Enhances Proliferation via the FGFR1 Signaling Pathway
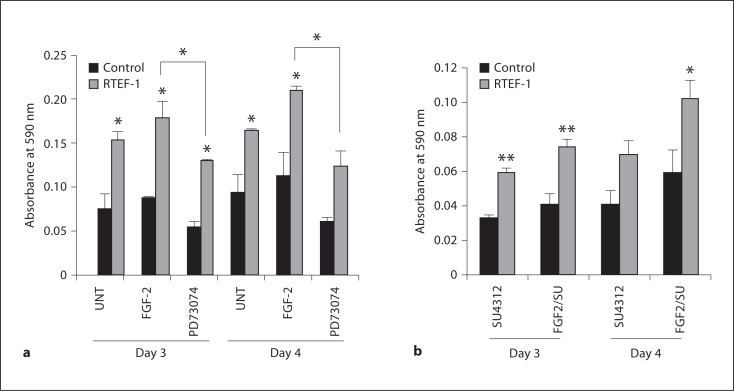
During neovascularization, rapidly proliferating endothelial cells are incorporated into new vessels. Therefore, we assessed whether proliferation was enhanced in RTEF-1 o/e cells in response to FGF-2 stimulation. As our laboratory has previously shown, overexpression of RTEF-1 increases proliferation in endothelial cells when compared with control. Figure 5a indicates that proliferation in RTEF-1 o/e cells treated with FGF-2 was significantly enhanced. Interestingly, the FGFR1 tyrosine kinase inhibitor, PD173074, did not hinder proliferation in control cells, yet blocked RTEF-1-induced proliferation (fig. 5a). To determine if VEGF is responsible for the RTEF-1-mediated increase in proliferation, cells were treated with FGF-2 and/or a selective inhibitor of VEGF receptor tyrosine kinases, SU4312. RTEF-1 o/e cell proliferation was partially inhibited when treated with SU4312. However, RTEF-1 o/e cells pretreated with SU4312 demonstrated increased proliferation when FGF-2 was added when compared with control cells. Additionally, FGF-2 did not restore proliferation in control cells pretreated with SU4312 (fig. 5b). While VEGF is surely involved in RTEF-1-related endothelial proliferation, these data suggest that the FGFR1/FGF-2 signaling pathway is necessary for RTEF-1-mediated proliferation.
Fig. 5.
RTEF-1 increases proliferation via FGFR1 signaling pathway. a Cells were treated with or without 25 ng/ml FGF-2 or 200 nM PD73074 and growth was examined by crystal violet growth assay over 4 days, n = 3. * p ≤ 0.05. b Cells were treated with or without 25 ng/ml FGF-2 and/or 100 μM SU4312 and growth was examined by crystal violet growth assay over 4 days, n = 3. * p ≤ 0.05, ** p ≤ 0.01.
RTEF-1 Overexpression Attenuates Microvascular Relaxation via Increased eNOS Levels
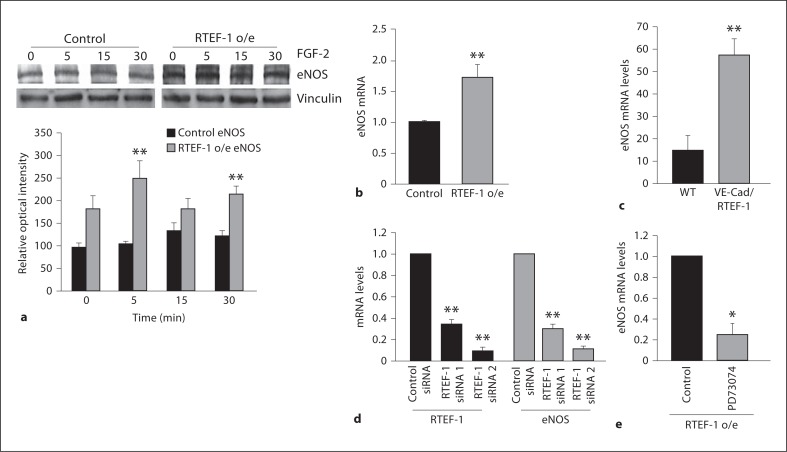
The endothelium plays an essential role in maintaining vascular tone by releasing various biochemical factors that modulate contraction and relaxation of the vasculature. The endothelium regulates vascular reactivity by activating eNOS, which is responsible for NO production. eNOS is constitutively expressed in ECs and its activity is regulated mainly by protein-protein interactions and multisite phosphorylation. eNOS activity, in part, is regulated by the phosphorylation state of specific serines, threonines and tyrosines including Serine 1177. To examine if RTEF-1-induced upregulation of FGFR1 increased microvascular relaxation through increasing eNOS activation, we examined HMEC-1 cells treated with FGF-2 for eNOS phosphorylation at serine 1177. There were no significant changes in eNOS phosphorylation in RTEF-1 o/e HMEC-1 cells (data not shown). Additionally, QPCR analysis demonstrated that eNOS mRNA levels were increased in RTEF-1/HMEC (fig. 6b) as well as in the hearts of VE-Cad/RTEF-1 mice (fig. 6c). Knockdown of endogenous RTEF-1 by siRNA significantly inhibited eNOS mRNA expression (fig. 6d). Finally, RTEF-1 o/e cells treated with the FGFR1 inhibitor PD73074 showed dramatically reduced eNOS expression (fig. 6e). These data indicate that RTEF-1 may increase microvascular relaxation via the regulation of eNOS expression.
Fig. 6.
RTEF-1 overexpression increases eNOS signaling. a Control and RTEF-1 o/e HMEC-1 cells were treated with FGF-2 (25 ng/ml) for the times indicated and immunoblotted with total eNOS antibodies. Relative optical intensity of the total eNOS of the immunoblot is shown on the bottom panel. b Assessment of eNOS levels by QPCR in stably transfected RTEF-1 o/e HMEC-1 cells and control, n = 3. ** p ≤ 0.01. c Assessment of eNOS levels by QPCR in the hearts of VE-Cad/RTEF-1 and WT mice, n = 3. ** p ≤ 0.01. d qRT-PCR assay of HMEC-1 cells transfected with nontarget control, RTEF-1 and eNOS siRNAs. e qRT-PCR assay of HMEC-1 cells analyzing eNOS mRNA levels after treatment with 200 nMPD73074.
Discussion
The principal finding of this study is that RTEF-1 is an important molecule involved in endothelium-dependent vessel dilatation. We demonstrate that the mechanism of RTEF-1-induced FGFR1 expression promotes endothelial cell-dependent microvascular relaxation via attenuation of eNOS expression. Several lines of evidence support our finding. First, FGFR1 expression was induced by RTEF-1 at both the mRNA and protein level in vitro and in vivo. Second, studies indicated that FGFR1 expression is attenuated by RTEF-1, and RTEF-1 is also able to specifically stimulate the Sp1 region on the FGFR1 promoter to increase transcription. Third, we determined that the FGFR1/FGF-2 signaling pathway is integral for RTEF-1-mediated proliferation. Finally, RTEF-1 induced eNOS expression in vitro and increased eNOS levels in our transgenic VE-Cad/RTEF-1 mice, and that interference with RTEF-1 signaling decreased the ability of RTEF-1 to stimulate both FGFR1 and eNOS levels. Collectively, these data suggest that RTEF-1 plays a pivotal role in regulating endothelial function.
Previous reports indicate that RTEF-1 is a muscle-enriched transcriptional factor that forms a sequence-specific complex on M-CAT sites of targeting genes in muscle cells [5, 11]. Our previous studies demonstrated that RTEF-1 is upregulated in hypoxic endothelial cells and that overexpression of RTEF-1 increases VEGF promoter activity [10, 11]. We also confirmed that VEGF is increased in hypoxic endothelial cells overexpressing RTEF-1 [11]. Considering that RTEF-1 is a member of the TEF-1 family, which regulates genes expressed in cardiac and skeletal muscle cells through the M-CAT elements in their promoters, we hypothesized that RTEF-1 regulates FGFR1 expression by activating the FGFR1 promoter. Our data confirm that RTEF-1 upregulates FGFR1 in endothelial cells and that inhibition of RTEF-1 blunts FGFR1 expression. FGF-2 induces the growth of new vasculature in a variety of animal models [19] and FGFRs are mediators of the FGF ligand family which signal the cellular effects of FGFs. Heartless mutants of Drosophila and studies of FGFR-1-deficient embryonic stem cells in chimeric embryos suggest that FGFR-1 may play an important role in cardiogenesis [20, 21]. However, the mechanisms involved in the induction of FGFR-1 expression are not completely understood. We investigated the FGFR-1 gene, as it is a key mediator in forming new blood vessels during ischemia in vivo and stimulating endothelial cell proliferation in vitro. In addition, there are more than 5 M-CAT-like sequences as well as Sp1 sites existing in the FGFR-1 promoter region, indicating that these sites on the FGFR-1 promoter are possible targets for RTEF-1. The studies to determine the direct interactions between RTEF-1 and the FGFR1 promoter are under way.
In this study, we demonstrated for the first time that RTEF-1 activates the FGFR-1 promoter, which, in turn, increases FGFR-1 mRNA and protein expression. It is well characterized in the literature that the FGF/FGFR system can be a potent angiogenic agent and promote the growth of vascular smooth muscle and endothelial cells [22, 23]. Additionally, intracoronary treatment with bFGF increases microvessel numbers in infarcted and noninfarcted myocardium, as well as protecting vascular function in the setting of ischemia; chronic administration was found to increase endothelium-dependent relaxation of the collateral perfused coronary microcirculation in pigs [3, 24]. FGF-2 and other angiogenic growth factors operate to a large extent through the release of endothelial-derived NO via the activation of tyrosine kinase receptors, resulting in vasodilation and elevated blood flow. Additionally, eNOS-derived NO preserves collateral blood flow during ischemia [25]. We have identified that eNOS mRNA levels are increased in RTEF-1 transgenic mice and that eNOS expression is also elevated in vitro in RTEF-1 o/e endothelial cells treated with FGF2, suggesting that RTEF-1 plays a necessary role in FGF/FGFR signaling. This study also demonstrates that endothelial-dependent microvessel relaxation is elevated in VE-Cad/RTEF-1 transgenic mice. Thus, our studies maintain that RTEF-1-stimulated FGFR1 can increase endothelial-dependent vessel dilatation, which coincides with the aforementioned studies, showing a direct relationship with FGF/FGFR and microvascular relaxation.
In summary, we have demonstrated that RTEF-1 is a novel regulator of FGFR-1 expression in vitro and in vivo. Given the strong evidence for the involvement of the FGF/FGFR system in the regulation of eNOS, RTEF-1 could potentially be a therapeutic target in the regulation of both angiogenesis and vasodilation.
Acknowledgements
We would like to thank Dr. Alex Stewart (University of Ottawa, Canada) for providing the RTEF-1 expression vector and Dr. Pablo Quintero Pinzon for helpful discussions regarding the manuscript. This study was supported by NIH R01HL082837 (J.L.), NIH R01HL69024 (F.S.) and by an NIH training grant T32HL007374 (A.F.M.-B.).
References
- 1.Poole TJ, Finkelstein EB, Cox CM. The role of FGF and VEGF in angioblast induction and migration during vascular development. Dev Dyn. 2001;220:1–17. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1087>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Kuo L, Chilian WM, Davis MJ, Laughlin MH. Endotoxin impairs flow-induced vasodilation of porcine coronary arterioles. Am J Physiol. 1992;262:H1838–H1845. doi: 10.1152/ajpheart.1992.262.6.H1838. [DOI] [PubMed] [Google Scholar]
- 3.Sellke FW, Wang SY, Stamler A, Lopez JJ, Li J, Li J, Simons M. Enhanced microvascular relaxations to VEGF and BFGF in chronically ischemic porcine myocardium. Am J Physiol. 1996;271:H713–H720. doi: 10.1152/ajpheart.1996.271.2.H713. [DOI] [PubMed] [Google Scholar]
- 4.Chen HH, Baty CJ, Maeda T, Brooks S, Baker LC, Ueyama T, Gursoy E, Saba S, Salama G, London B, Stewart AF. Transcription enhancer factor-1-related factor-transgenic mice develop cardiac conduction defects associated with altered connexin phosphorylation. Circulation. 2004;110:2980–2987. doi: 10.1161/01.CIR.0000146902.84099.26. [DOI] [PubMed] [Google Scholar]
- 5.Farrance IK, Ordahl CP. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J Biol Chem. 1996;271:8266–8274. doi: 10.1074/jbc.271.14.8266. [DOI] [PubMed] [Google Scholar]
- 6.Gan Q, Yoshida T, Li J, Owens GK. Smooth muscle cells and myofibroblasts use distinct transcriptional mechanisms for smooth muscle alpha-actin expression. Circ Res. 2007;101:883–892. doi: 10.1161/CIRCRESAHA.107.154831. [DOI] [PubMed] [Google Scholar]
- 7.Maeda T, Maeda M, Stewart AF. TEF-1 transcription factors regulate activity of the mouse mammary tumor virus LTR. Biochem Biophys Res Commun. 2002;296:1279–1285. doi: 10.1016/s0006-291x(02)02085-5. [DOI] [PubMed] [Google Scholar]
- 8.Maeda T, Mazzulli JR, Farrance IK, Stewart AF. Mouse DTEF-1 (ETFR-1, TEF-5) is a transcriptional activator in alpha 1-adrenergic agonist-stimulated cardiac myocytes. J Biol Chem. 2002;277:24346–24352. doi: 10.1074/jbc.M201171200. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appukuttan B, McFarland TJ, Davies MH, Atchaneeyasakul LO, Zhang Y, Babra B, Pan Y, Rosenbaum JT, Acott T, Powers MR, Stout JT. Identification of novel alternatively spliced isoforms of RTEF-1 within human ocular vascular endothelial cells and murine retina. Invest Ophthalmol Vis Sci. 2007;48:3775–3782. doi: 10.1167/iovs.06-1172. [DOI] [PubMed] [Google Scholar]
- 11.Shie JL, Wu G, Wu J, Liu FF, Laham RJ, Oettgen P, Li J. RTEF-1, a novel transcriptional stimulator of vascular endothelial growth factor in hypoxic endothelial cells. J Biol Chem. 2004;279:25010–25016. doi: 10.1074/jbc.M403103200. [DOI] [PubMed] [Google Scholar]
- 12.Xu M, Jin Y, Song Q, Wu J, Philbrick MJ, Cully BL, An X, Guo L, Gao F, Li J. The endothelial-dependent effect of RTEF-1 in pressure overload cardiac hypertrophy – role of VEGF-B. Cardiovasc Res. 2011;90:325–334. doi: 10.1093/cvr/cvq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gory S, Vernet M, Laurent M, Dejana E, Dalmon J, Huber P. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood. 1999;93:184–192. [PubMed] [Google Scholar]
- 14.McCabe CJ, Khaira JS, Boelaert K, Heaney AP, Tannahill LA, Hussain S, Mitchell R, Olliff J, Sheppard MC, Franklyn JA, Gittoes NJ. Expression of pituitary tumour transforming gene (PTTG) and fibroblast growth factor-2 (FGF-2) in human pituitary adenomas: relationships to clinical tumour behaviour. Clin Endocrinol (Oxf) 2003;58:141–150. doi: 10.1046/j.1365-2265.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 15.Feng J, Bianchi C, Sandmeyer JL, Sellke FW. Bradykinin preconditioning improves the profile of cell survival proteins and limits apoptosis after cardioplegic arrest. Circulation. 2005;112:I190–I195. doi: 10.1161/CIRCULATIONAHA.104.524454. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Partovian C, Li J, Hampton TG, Metais C, Tkachenko E, Sellke FW, Simons M. Modulation of microvascular signaling by heparan sulfate matrix: studies in syndecan-4 transgenic mice. Microvasc Res. 2002;64:38–46. doi: 10.1006/mvre.2002.2399. [DOI] [PubMed] [Google Scholar]
- 17.Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Boodhwani M, Voisine P, Ruel M, Sodha NR, Feng J, Xu SH, Bianchi C, Sellke FW. Comparison of vascular endothelial growth factor and fibroblast growth factor-2 in a swine model of endothelial dysfunction. Eur J Cardiothorac Surg. 2008;33:645–650. doi: 10.1016/j.ejcts.2007.12.016. discussion 251–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons M, Laham RJ, Post M, Sellke FW. Therapeutic angiogenesis: potential role of basic fibroblast growth factor in patients with severe ischaemic heart disease. BioDrugs. 2000;14:13–20. doi: 10.2165/00063030-200014010-00002. [DOI] [PubMed] [Google Scholar]
- 20.Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (FGFR1) function: A role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- 21.Dell'Era P, Ronca R, Coco L, Nicoli S, Metra M, Presta M. Fibroblast growth factor receptor-1 is essential for in vitro cardiomyocyte development. Circ Res. 2003;93:414–420. doi: 10.1161/01.RES.0000089460.12061.E1. [DOI] [PubMed] [Google Scholar]
- 22.Scott-Burden T, Schini VB, Elizondo E, Junquero DC, Vanhoutte PM. Platelet-derived growth factor suppresses and fibroblast growth factor enhances cytokine-induced production of nitric oxide by cultured smooth muscle cells. Effects on cell proliferation. Circ Res. 1992;71:1088–1100. doi: 10.1161/01.res.71.5.1088. [DOI] [PubMed] [Google Scholar]
- 23.Vlodavsky I, Folkman J, Sullivan R, Fridman R, Ishai-Michaeli R, Sasse J, Klagsbrun M. Endothelial cell-derived basic fibroblast growth factor: synthesis and deposition into subendothelial extracellular matrix. Proc Natl Acad Sci USA. 1987;84:2292–2296. doi: 10.1073/pnas.84.8.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battler A, Scheinowitz M, Bor A, Hasdai D, Vered Z, Di Segni E, Varda-Bloom N, Nass D, Engelberg S, Eldar M, et al. Intracoronary injection of basic fibroblast growth factor enhances angiogenesis in infarcted swine myocardium. J Am Coll Cardiol. 1993;22:2001–2006. doi: 10.1016/0735-1097(93)90790-8. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava S, Terjung RL, Yang HT. Basic fibroblast growth factor increases collateral blood flow in spontaneously hypertensive rats. Am J Physiol Heart Circ Physiol. 2003;285:H1190–H1197. doi: 10.1152/ajpheart.00280.2003. [DOI] [PubMed] [Google Scholar]