Abstract
Here we present a database, WikiCell, as a portal for a unified view of the human transcriptome. At present, WikiCell consists of Expressed Sequenced Tags (ESTs), and users can access, curate, and submit database data by interactive mode, and also can browse, query, upload, and download sequences. Researchers can utilize the transcriptome model based on a human taxonomy graph. The sequences in each model are sorted by attributes such as physiological and pathological samples. The Genbank EST data format are conserved. Gene information is provided, including housekeeping genes, taxonomy location, and gene ontology (GO) description. We believe that WikiCell provides a useful resource for defining expression pattern and tissue differentiation based on human taxonomy mode. It can be accessed at http://www.wikicell.org/.
Introduction
“Transcriptome” is a term that describes the set of all transcripts or messenger RNA (mRNA) molecules produced in cells (Gomas and Tagore, 2008; Subramanian et al., 2005). Transcriptomes of stem cells and cancer cells are of particular interest to researchers who seek to understand the processes of cellular differentiation and carcinogenesis. Transcriptome may also be applied to the specific subset of transcripts present in a particular cell, or the total set of transcripts in a given organism (Gomas and Tagore, 2008). Many transcriptome experiments make use of high-throughput technologies to yield multi-dimensional datasets. These datasets are often quite large, are usually not published in their entirety, or are published as supplementary information that is not easily searchable. Without a system for standardizing and sharing such data, it is less useful for the biomedical community to contribute these types of data to centralized repositories. The annotation and display of pertinent information in the context of the corresponding transcriptome is even more difficult, as shown in Figure 1. Expressed Sequence Tags (ESTs) are one of the most widely applied transcriptome technologies (Lee et al., 2005; Nagaraj et al., 2007). ESTs are used to identify gene transcripts, and are instrumental in gene discovery and gene sequence determination (Adams et al., 1991; Campagne and Skrabanek, 2006; Stanton et al., 2003).
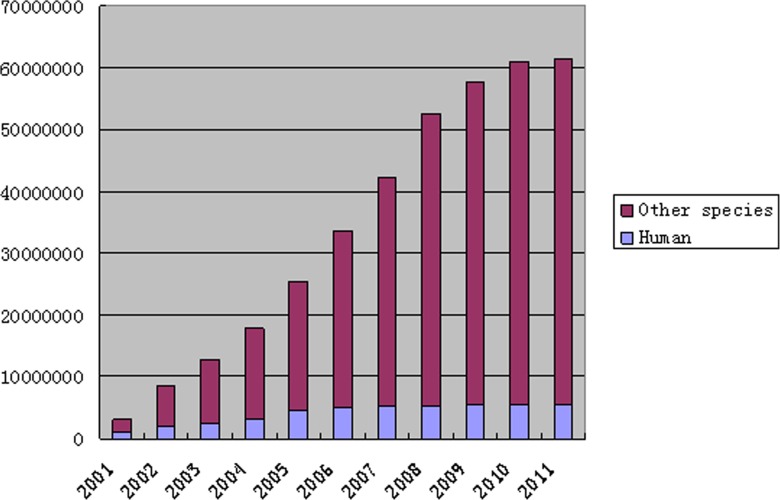
FIG. 1.
Expressed Sequenced Tag (EST) annual cumulative rising rate. There are few EST data available before 2001. The statistics span nearly a decade. The quality of EST sequences is increasing every year, but the rate trend is steady.
Fortunately, the development of Mediawiki (Mediawiki, 2007) software resolves the deficiency described above (www.mediawiki.org). Current biological databases provide users with data submission forms, tools, and access to the compiled data via websites, FTP sites, or programmatic interfaces. Internal curation teams organize and update the data. A wiki model for biological databases, such as WikiPathways (Kelder et al., 2009; Pico et al., 2008), Proteopedia (Eran et al., 2008), and Genewiki (Jon et al., 2008, 2009), provides a single, intuitive interface for submitting, updating, organizing, and accessing data. This allows users to participate in the curation process and keep up with the influx of new data.
To exploit the features of these databases, we have developed WikiCell (www.wikicell.org), a portal that provides a unified view of the human transcriptome. WikiCell is an open and public platform dedicated to the annotation of the human transcriptome. Researchers can contribute transcriptome data, including ESTs and annotations. The wiki format allows authors to create and edit any number of interlinked webpages. Based on the anatomy of the human body, the logical structure traces out an image of refined classification from nine major systems to cell level, and includes both physiological and pathological transcriptome data.
Materials and Methods
The overall ESTs statistics were identified from human tissues, as shown in Table 1. Data were collected from two sites, with the majority from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov), and the remainder from the Beijing Institute of Genomics (www.big.cas.cn). The second set represents our independent procreant data. The sequences were downloaded from the dbEST database, which contained 8,444,018 reported EST entries from Homo sapiens as of October 2011. ESTs in groups of 100 or more were considered members of a library, which resulted in 5,943,083 EST sequences. These sequences were sorted by physiological and pathological origin based on library information of sequence source, which expatiated on human taxonomy pathologic and medical information. There were more than 4000 libraries in human EST data, and several libraries belonged to the same tissue or cell. Accordingly, these libraries were assigned 231 clusters. Physiological sequences from different tissues or cells were used for researching gene expression relationships. Some specific EST sequences of pathologic tissue can be helpful for researching causes of morbidity.
Table 1.
The Statistics of Expressed Sequenced Tag Information
| System | Numbers of data nodes | Numbers of non-data nodes | Physiology | Pathology | Total |
|---|---|---|---|---|---|
| Circulatory system | 30 | 201 | 93,426 | 391,278 | 484,704 |
| Digestive system | 46 | 229 | 341,245 | 767,429 | 1,108,674 |
| Endocrine system | 21 | 62 | 51,345 | 218,332 | 269,677 |
| Motion system | 25 | 42 | 90,323 | 370,148 | 460,471 |
| Nervous system | 38 | 203 | 23,005 | 1,194,775 | 1,217,780 |
| Reproductive system | 35 | 165 | 550,098 | 697,718 | 1,247,816 |
| Respiratory system | 14 | 126 | 73,981 | 421,045 | 495,026 |
| Sensory system | 8 | 177 | 325,452 | 90,851 | 416,303 |
| Urinary system | 10 | 68 | 122,427 | 120,205 | 242,632 |
The WikiCell online database (www.wikicell.org) was implemented using a mediawiki engine, mySQL relational database, and PHP technology, and provides a simple way to access the EST data and their annotations.
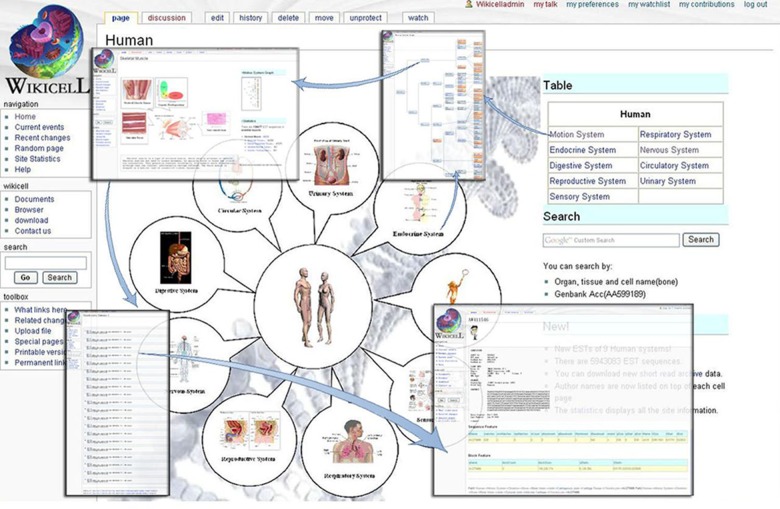
WikiCell presents a new model for EST databases that enhances and complements ongoing efforts. Each node of taxonomy at WikiCell has a dedicated wiki page displaying dynamic pictures, descriptions, references, a system tree diagram, statistics, and a path. The statistics section provides the number of current nodes and sub-nodes owned by the EST. Users may only enter the summary page from the statistic section of a leaf node page, which is shown in Figure 2.
FIG. 2.
The flow chart of browsing each page. Users can browse each of nine systems from the main page. There is a correlative taxonomy graph in each system, in which the link nodes denote that there are Expressed Sequenced Tag data in the pages. The entrance of summary pages is at the statistics region in the note page.
All pages of WikiCell are freely accessible and do not require registration. However, the contribution of information is only possible for registered users who are logged in. Our policy with respect to registrations is based on balancing two conflicting aims: encouraging users to contribute to the wikis by making it simple while ensuring the reliability of the information provided. Thus a mandatory but liberal registration policy seemed to be the best way to balance these two aims.
Results
WikiCell has three main page types: taxonomy, summary, and EST data. Taxonomy pages include human systems, organs, tissues, and cells, as shown in Figure 3. Summary and EST pages are located a level below tissue and cell pages. Taxonomy pages are more like a tree trunk, and summary and EST pages represent the tree branches. The majority of WikiCell is devoted to ESTs, with each page displaying GenBank, information on the EST location, and gene annotation data.
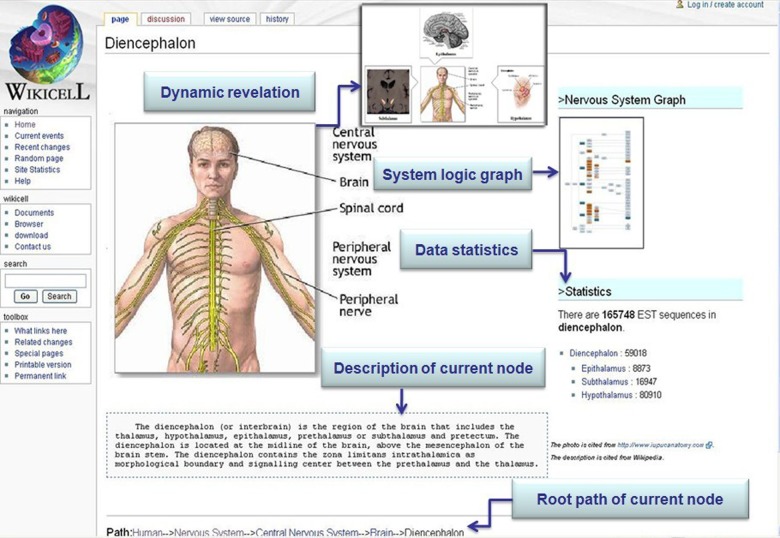
FIG. 3.
Sample of a human taxonomy page. This page on the WikiCell website is one note page in a human taxonomy graph. It introduces some information about human local composition.
WikiCell can be searched by organ, tissue, and cell type, as well as GenBank accession number. In addition, one can browse available human gene information, including chromosome location, housekeeping (HK) genes, gene description, and gene ontology (GO) function. Gene information may also be sorted into three categories: chromosome, HK, and GO function. We have chosen to utilize the definition of Zhu's group of HK genes (Jiang et al., 2008a, 2008b). Through the analysis of public expression data (from ESTs and microarray data) from 18 human tissues, these researchers found that 40% of the currently annotated human genes were constitutively expressed in at least 16 of 18 tissues. We have adopted the most rigorous definition of an HK gene, in which a gene must be expressed in all 18 tissues examined. According to this definition, 3182 HK genes are compiled in each chromosome using this method.
Features of WikiCell
WikiCell is a portal for querying, browsing, communicating, uploading, and downloading contributed datasets. One important character of this database compared to a traditional database is that users can access, curate, and submit database by interactive mode, which speeds up the maintenance and renewal of WikiCell. First, WikiCell provides EST data relative to spatial expression in the human body. You can access the “body path” of the data, such that it identifies in which system, organ, tissue, and/or cell the transcript is expressed. For example, there are 185 EST sequences for the intraglomerular mesangial cell. WikiCell shows the following taxonomy path: Human -;-> Urinary System -;-> Kidney -;-> Renal Parenchyma -;-> Uriniparous Tubule -;-> Renal Corpuscle -;-> Glomerulus -;-> Intraglomerular Mesangial Cell. It also indicates the number and type of EST data that are from the same library. This kind of structure of the database is useful for further research on defining expression patterns and tissue differentiation. Second, WikiCell allows searches for multiple page types, as shown in Figure 4. A user may query with general key words for things such as taxonomy, or with specific terms like GenBank accession numbers. Third, there are three parts in each EST page. The GenBank EST reporting format is preserved to display identifiers such as clone information, primers, sequence, comments, library, submitter, and citations. In addition, there is detailed information about the EST chromosomal location (e.g., matches, strand, sequence size, chromosome number, and start and end location), and block feature (block count and block size). The gene name where the EST is located and the taxonomy path are also provided at the bottom of the page. Fourth, the links of some relevant human integrative resources are added to the front page, such as elements of the human gene compendium, including: GeneCards (http://www.genecards.org/), a member of the biocyc database collection; MetaCyc (http://metacyc.org/), a curated knowledgebase of biological pathways in humans; Reactome (http://www.reactome.org/ReactomeGWT/entrypoint.html), a model organism protein expression database; MOPED (http://moped.proteinspire.org), pathways for the people; and WikiPathways (http://wikipathways.org/index.php/WikiPathways) (Rebhan et al., 1997; Ron et al., 2008; Joshi-Tope et al., 2005; Kolker et al., 2012; Kelder et al., 2009).
FIG. 4.
Search function. Two search tools are on the WikiCell website: Mediawiki-owned and Google custom search extension. You can input keywords to find correlative matches.
Finally, WikiCell offers a bulk data download option, which increases database utility for computational biologists, who may use WikiCell data to conduct in silico analyses of transcriptome activity.
The content management system
Mediawiki was chosen as the software platform for the WikiCell. This interface is identical to that used by Wikipedia, and thus many users will be quickly familiar with the system. In addition, Mediawiki allows the use of extensions. Due to the popularity of software, many extensions are readily available. For WikiCell, we use several extensions: DataInvoker (www.mediawiki.org/wiki/Extension:DataInvoker), which introduces new parser functions allowing database data retrieval; Secure HTML (www.mediawiki.org/wiki/Extension:Secure_HTML), which allows a user to specify an arbitrary HTML when the HTML includes a corresponding hash created by combining the HTML input with an authorized key; and ConfirmEdit (www.mediawiki.org/wiki/Extension:ConfirmEdit), which enables a simple text Captcha that should minimize automated editing.
Discussion and Future Directions
With the exponential growth of biological wikis, it is clear that the wiki model resonates with biological and scientific communities. However, many of these biological wikis appear to suffer from a lack of participation. Establishing a critical mass of users and useful content appears to be the most common obstacle in these efforts.
Active participation by members of the transcriptomics community in sharing data through WikiCell has resulted in a prolific increase in the amount of transcriptome annotations. A great deal of additional transcriptome data, such as SAGE, SRA, GEO, and RNA-Seq, will be added to the WikiCell (Wang et al., 2009). Some useful tools for transcriptomics research, such as DEGSeq, Tophat, and Cufflink, will be integrated for online data analysis (Likun et al., 2010; Roberts et al., 2011; Trapnell et al., 2009). In addition, the further research on expression patterns for different tissue/cell types based on these data is an important resource, as secondary analysis data will be integrated into WikiCell. With the increased participation of the scientific community, we foresee that WikiCell will serve as a unified resource for transcriptome research. It will be an important resource for defining transcriptome models and tissue diversity using the human taxonomy method.
Acknowledgements
We are grateful to Jin Jing, Zhang Ning, and Li Hongjie for their helpful suggestions. This study was supported by a grant from the National Basic Research Program (973 Program, no. 2010CB126604), a grant from the Special Foundation Work Program (no. 2009FY120100), a grant (2012AA020409) from National Programs for High Technology Research and Development (863 Program), support from the Ministry of Science and Technology of the People's Republic of China, and a grant from the National Science Foundation of China (no. 31071163 and 31101063).
Author Disclosure Statement
No competing financial interests exist.
References
- Adams M.D. Kelley J.M. Gocayne J.D., et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Campagne F. Skrabanek L. Mining expressed sequence tags identifies cancer markers of clinical interest. BMC Bioinformatics. 2006;7:481. doi: 10.1186/1471-2105-7-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eran H. Jaime P. Eric M., et al. Proteopedia—a scientific ‘wiki’ bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol. 2008;9:1–10. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomas V.S. Tagore S. Transcriptomics. Curr Drug Metab. 2008;9:245–249. doi: 10.2174/138920008783884759. [DOI] [PubMed] [Google Scholar]
- Jiang Z. Fuhong H. Shuhui S., et al. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008a;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. Fuhong H. Songnian H., et al. On the nature of human housekeeping genes. Trends Genetics. 2008b;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Jon W.H. Camilo O. James G., et al. A gene Wiki for community annotation of gene function. PLOS Biol. 2008;6:1398–1402. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jon W.H. Pierre L. Michael M., et al. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2009;760:1–7. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G. Gillespie M. Vastrik I., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T. Pico A.R. Hanspers K., et al. Mining biological pathways using WikiPathways web services. PLoS One. 2009;4:6447. doi: 10.1371/journal.pone.0006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E. Higdon R. Haynes W., et al. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. 2012;40:1093–1099. doi: 10.1093/nar/gkr1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. Tsai J. Sunkaras J., et al. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likun W. Zhixing F. Xi W., et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Mediawiki. The Free Wiki Engine. 2007. www.mediawiki.org/w/index.php?title=MediaWiki&oldid=65192 www.mediawiki.org/w/index.php?title=MediaWiki&oldid=65192
- Nagaraj S.H. Gasser R.B. Ranganathan S. A hitchhiker's guide to expressed sequence tag (EST) analysis. Brief. Bioinformatics. 2007;8:6–21. doi: 10.1093/bib/bbl015. [DOI] [PubMed] [Google Scholar]
- Pico A.R. Kelder T. Van I.M.P., et al. WikiPathways: Pathway editing for the people. PLoS Biol. 2008;6:60184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M. Chalifa-Caspi V. Prilusky J., et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genetics. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Roberts A. Pimentel H. Trapnell C. Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Ron C. Hartmut F. Carol A.F., et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton J.A. Macgregor A.B. Green D.P. Identifying tissue-enriched gene expression in mouse tissues using the NIH UniGene database. Appl Bioinformatics. 2003;2:S65–S73. [PubMed] [Google Scholar]
- Subramanian A. Tamayo P. Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. Pachter L. Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Gerstein M. Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]