Abstract
Protein–protein interactions are key elements in the assembly of cellular regulatory and signaling protein complexes that integrate and transmit signals and information in controlling and regulating various cellular processes and functions. Many conventional methods of studying protein–protein interaction, such as the immuno-precipitation and immuno-blotting assay and the affinity-column pull-down and chromatographic analysis, are very time-consuming and labor intensive and lack accuracy and sensitivity. We have developed a simple, rapid, and sensitive assay using a ProteinChip array and SELDI-TOF mass spectrometry to analyze protein–protein interactions and map the crucial elements that are directly involved in these interactions. First, a purified “bait” protein or a synthetic peptide of interest is immobilized onto the pre-activated surface of a PS10 or PS20 ProteinChip and the unoccupied surfaces on the chip are protected by application of a layer ethanolamine to prevent them from binding to other non-interactive proteins. Then, the target-containing cellular protein lysate or synthetic peptide containing the predicted amino acid sequence of protein-interaction motif is applied to the protected array with immobilized bait protein/ peptide. The nonspecific proteins/peptides are washed off under various stringent conditions and only the proteins specifically interacting with the bait protein/peptide remain on the chip. Last, the captured interacting protein/peptide complexes are then analyzed by SELDI-TOF mass spectrometry and their identities are confirmed by their predicted distinctive masses. This method can be used to unambiguously detect the specific protein–protein interaction of known proteins/peptides, to easily identify potential cellular targets of proteins of interest, and to accurately analyze and map the structural elements of a given protein and its target proteins using synthetic peptides with the predicted potential protein interaction motifs.
Keywords: SELDI-TOF-MS, ProteinChip, Peptide interactions, SEND-ID, Fus1, Apaf1
1. Introduction
Signal transduction events in eukaryotic cells involve the reversible assembly of large multi-protein complexes which integrate and transmit the information that controls various cellular processes such as ion fluxes, cytoskeletal rearrangements, gene expression, cell cycle progression, and apoptosis (1, 2). Protein–protein interactions are key elements in the assembly of functional cellular regulatory and signaling protein complexes. Many proteins involved in intracellular signaling contain multiple distinct sequence modules that direct their constitutive and signal-regulated interaction with other proteins in a signaling network (3, 4). These interaction domains can target proteins to a specific subcellular location, provide a means for recognition of protein post-translational modifications such phosphorylation, mediate the formation of multi-protein signaling complexes, maintain functional conformations, and control substrate specificity of enzymes (5, 6). Some of these interactions are mediated by structurally conserved protein domains which recognize specific short peptide motifs such as the well characterized Src-homology SH2 and SH3 domains and the phospho-tyrosine-binding PTB domains (7) in protein tyrosine kinases (PTKs). The most recently identified PDZ domains are modular protein-binding domains that can bind to specific recognition sequences at the C-termini of unrelated proteins or dimerize with other PDZ domains or bind to internal peptide motifs to create networks associated with the plasma membrane (8, 9). Hence, deciphering protein–protein interaction cascades is essential for understanding cellular functions of proteins.
Analysis of protein–protein interaction and determination of the precise mechanism and communication among key components in an interactive protein complex such as apoptosome is a very complicated and time-consuming process. The conventional methods currently used to screen potential interacting protein partners, such as the yeast two hybrid assay or phage display systems, are notorious in generating excessive false positives. The Tandem Affinity Purification (TAP) method developed at EMBL (Heidelberg, Germany) has been shown to be a powerful tool for capturing and analyzing cellular proteins potentially interacting with each other under physiological conditions without prior knowledge of the protein composition, activity, or function (10). The TAP assay when combined with mass spectrometry analysis allows the identification of cellular protein targets interacting with a given protein. However, this process is very complex, needs to clone the gene of interest in the TAP-tagged vectors, and requires stringent and lengthy purification efforts (10). Moreover, the TAP system requires validation of targets by means of classical co-immu-nopurification/immuno-bloting experiments and or gel filtration chromatography. The biochemical approach with FPLC or HPLC for studying protein–protein interaction requires not only expensive equipment, but also highly purified proteins and substantial expertise of the user. To circumvent these operational difficulties, a rapid and sensitive approach is needed for accurately and effectively analyzing protein–protein interaction. Mass spectrometry can serve as a powerful and sensitive tool to attain that goal (11).
We have developed a simple, rapid, and sensitive assay using a ProteinChip array and SELDI-TOF mass spectrometry to analyze protein–protein interactions and map the crucial elements that are directly involved in these interactions. First, a purified “bait” protein or a synthetic peptide of interest is immobilized onto the pre-activated surface of a ProteinChip array and the unoccupied surfaces on the array are protected by application of ethanolamine to prevent them from binding to other non-interactive proteins. Then, the target-containing cellular protein lysate or synthetic peptide containing the predicted amino acid sequence of protein-interaction motif is applied to the protected array with immobilized bait protein/peptide. The nonspecific proteins/peptides are washed off under various stringent conditions and only the proteins specifically interacting with the bait protein/peptide remain on the chip. Last, the captured interacting protein/peptide complexes are then analyzed by SELDI-TOF mass spectrometry and their identities are confirmed by their predicted distinctive masses. This method can be used to unambiguously detect the specific protein–protein interaction of known proteins/peptides, to easily identify potential cellular targets of proteins of interest, and to accurately analyze and map the structural elements of a given protein and its target proteins using synthetic peptides with the predicted potential protein interaction motifs. This simplified assay system with ProteinChip array-based SELDI-TOF-MS platform can also ideally be used in the confirmatory studies following exploratory studies with the yeast two-hybrid system, the TAP-tag assay, and the computer-aided protein interaction modeling (see Note 1). Several databases and bioinformatics tools with a multitude of algorithms, such as BOND (Biomolecular Object Network Databank), PIP (Potential Interactions of Proteins) (12), and PDB (Protein Data Bank), are currently available for the prediction and visualization of interacting protein partners. “Interweaver” is one other versatile program developed by the Institute for InfoComm Research in Singapore that uses BIND, DIP (Database of Interacting proteins) and PDB collectively for generating experimental data. The hit candidates obtained from these databases can be applied in this SELDI-TOF-based protocol to confirm not just direct protein interactions but also the exact interacting motifs of the individual protein partners.
2. Materials
2.1. Protein Chips
Several ProteinChip arrays with chemically modified surfaces (pre-activated) that form covalent bonds with free amine groups are available from Bio-Rad Laboratories (Hercules, CA) (see Note 2). They are developed for diverse biological applications including biomarker discovery, protein profiling, protein–protein interaction studies, peptide mapping for protein ID, immunoassays, and receptor-ligand binding studies. These arrays have eight 2-mm diameter spots (A–H format) that correlate with the spacing of wells in a single column of a standard 96-well microplate and are therefore amenable for high-throughput applications in varied robotics. Here, we describe two types of arrays that are particularly amenable to covalently immobilize biomolecules for subsequent capture of targeted proteins from complex biological samples. The arrays differ in their surface chemistry and hence both should be tested to determine the most suitable array for the application envisioned. Other considerations include lower non-specific binding and higher sensitivity.
PS10: The pre-activated surface is composed of carbonyl diim-idazole moities to capture amine groups on the protein.
PS20: The pre-activated surface is composed of epoxy moities to capture amine groups on the protein.
2.2. Reagents
EAM (energy absorbing molecule): CHCA (α-Cyano-4-hydroxycinnamic acid) was purchased from Ciphergen Biosystems (Freemont, CA). These molecules can now be procured from Bio-Rad Laboratories (Hercules, CA).
Molecular weight standards (All-in-One Peptide) were purchased from Ciphergen Biosystems (Freemont, CA). They can now be procured from BioRad Laboratories (Hercules, CA).
Ethanolamine was purchased from Sigma (St. Louis, MO). Ethanolamine (ETA), also called 2-aminoethanol or mono-ethanolamine (MEA), is an organic chemical compound that is both a primary amine (due to an amino group in its molecule) and a primary alcohol (due to a hydroxyl group).
2.3. Peptide Synthesis
The peptides used in this study were formulated in the Peptide Synthesis Facility at The University of Texas M. D. Anderson Cancer Center.
FUS1: KLRRVHKNLIPQGIVKLDHER (2,418 Da).
Stearated FUS1: Stearate-KLRRVHKNLIPQGIVKLDHER (2,767 Da).
Mutant FUS1: KREHLRKNRPQPGPKPREDHR (2,386 Da).
Apaf1: LGILYILQTLE (1,272 Da).
3. Methods
A schematic representation of the overall methodology is depicted in Fig. 1. PS10 arrays are processed as described by Liu and coworkers (13) (see Note 3).
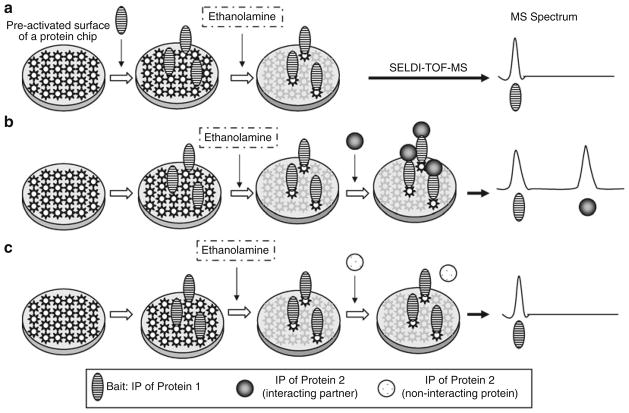
Fig. 1.
Schematic presentation of analysis of the protein–protein interaction by an ethanolamine protection on a ProteinChip array with SELDI-TOF-MS. (a ) a single peak will be detected in the mass spectrum when a single protein/peptide is immobilized onto the array surface: (b ) two peaks will be detected when the target protein/peptide specifically interacts with the bait; and (c) failure of the second peak to show up in the mass spectrum data when the protein/peptide applied subsequently does not interact with the bait protein/peptide.
Apply 10 μl (1 ng/μl) of bait protein or peptide to individual spots on PS10 chips as bait molecules. The arrays are incubated at room temperature in a humidified chamber for 1 h.
Add 10 μl of 1 M ethanolamine onto each spot of the array to block the unoccupied functional surface and incubate at room temperature for 30 min.
Wash individual spots three times with (10 μl) of 1× PBS, pH 7.5, containing 0.25% Triton X100 (see Note 4).
Apply the second target protein or peptide (10 μl: concentration to be determined empirically) onto the spots containing the immobilized bait protein/peptide and incubated in a humidity chamber for 5–8 h at room temperature or overnight at 4°C.
Wash the captured targets on each spot three times in (10 μl) of PBS, pH 7.5, containing 0.25% Triton X100 (see Note 4).
Quickly wash each spot two times in distilled water without detergent and air-dry the surface.
Add 1 μl of 25× diluted EAM solution (CHCA) onto each spot and air-dry.
Acquire mass spectrum using the ProteinChip reader.
The molecular mass of the resultant peptide peaks are analyzed and confirmed by externally calibrating the spectra with All-in-One peptide standards with pre-determined molecular weights (Bio-Rad Laboratories, Hercules, CA).
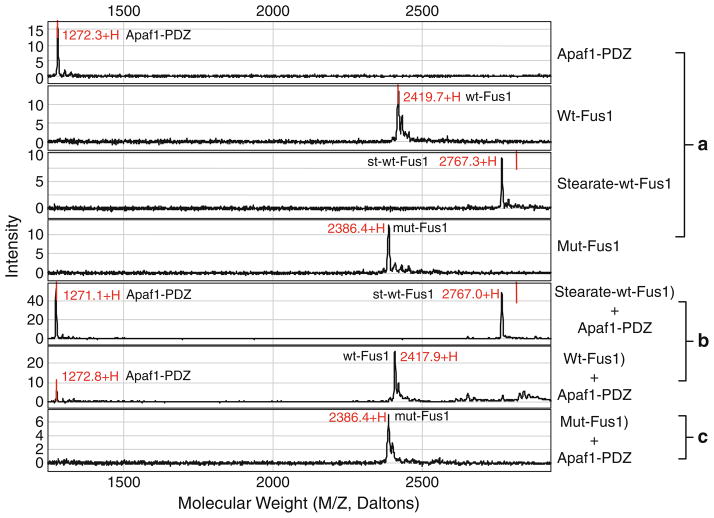
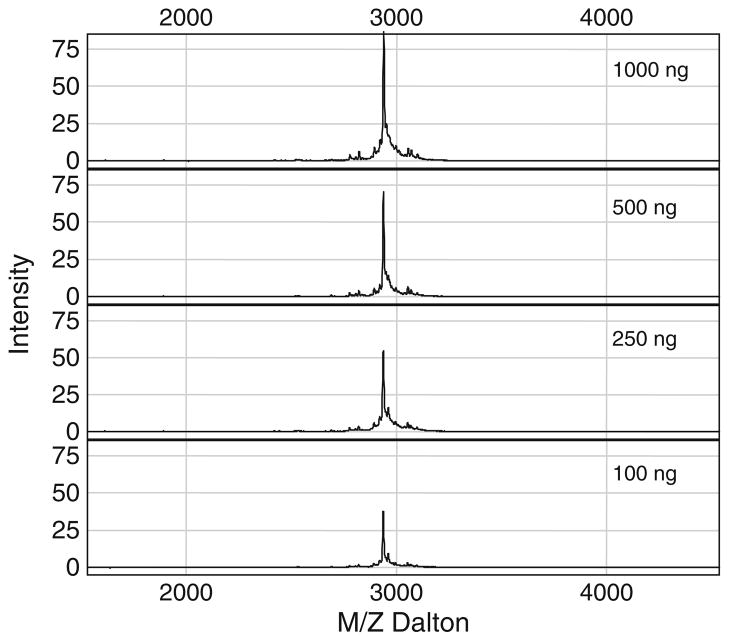
We used this method to study the molecular events in the mitochondria-dependent intrinsic apoptosis pathway mediated by the tumor suppressor FUS1-Apaf-1 protein–protein interaction in lung cancer cells. One of the FUS1-mediated tumor suppressing activities is its ability to induce apoptosis when ectopically expressed by wt-FUS1 gene transfer in FUS1-deficient tumor cells in vitro and in vivo (14, 15). However, the exact molecular mechanism in FUS1-induced apoptosis is unknown. To determine the mechanism of the FUS1-mediated tumor suppression and apoptosis induction, we carried out immuno-precipitations using rabbit anti-FUS1 polyclonal antibodies to pull down FUS1 and its interacting cellular protein partners in crude protein lysates prepared from FUS1-transfected lung cancer cells. We identified one of the potential cellular targets of FUS1 to be the Apaf-1 protein, which has been extensively studied and implicated to be a major component in the apoptosome found in the intrinsic apoptosis pathway (16). We applied this technique to confirm the FUS1-Apaf-1 protein interaction and identify the interacting motifs between two proteins. A computer-aided prediction of the functional motifs in FUS1 and Apaf1 protein sequence revealed a Class I and Class II PDZ protein binding motif in FUS1 and Apaf-1 proteins, respectively. We synthesized peptides containing wild-type or mutated amino acid sequences of the predicted PDZ motifs in both proteins. As demonstrated in Fig. 2, we alternatively used the peptide derived from the PDZ domain of Apaf1 protein or from the PDZ or other functional motifs of FUS1 protein as a bait to test against different peptides derived from both proteins with a wild-type or a mutated amino acid sequence within their PDZ motifs, using PS10 array (see Note 5) by SELDI-TOF-MS analysis. The specific interaction between the FUS1 and Apaf-1 proteins is clearly demonstrated by the interaction between the peptides derived from wild type amino acid sequences containing the PDZ protein-binding motifs from both proteins. However, no interaction in the non-PDZ sequence containing peptide and the mutated peptides can be detected (Fig. 2). The other attractive feature of this technique is the sensitivity of detection. As demonstrated in Fig. 3, even a very low concentration of peptides and their interactions can be detected. Protein/peptide quantity as low as 10 ng can easily be detected using this method with several modifications and different arrays.
Fig. 2.
Analysis of Fus1-Apaf-1 interaction using synthetic peptides designed from predicted protein interaction motifs by an ethanolamine protection assay on a ProteinChip array with SELDI-MS. The specific interaction of the Fus1 PDZ domain with Apaf-1 c-terminal peptide was detected as indicated by accurate mass of each peptide and by comparison with negative mutant and nonspecific control peptides. (a) Panels show individual peaks of various Fus1 or Apaf1 peptides when single peptides were loaded onto proteinchip arrays; (b ) two distinctive peaks are detected only when the peptides specifically interact with each other; and (c) when a mutation is introduced in the PDZ binding motif of Fus1, no Apaf1 peptide peak appears in the spectrum.
Fig. 3.
Quantification of peptide bound to the surface and determination of detection sensitivity on PS10 ProteinChip array by SELDI-TOF-MS. A serial dilution of known concentrations of purified synthetic peptides is loaded onto PS10 chip individually and the amount of peptide is quantified based on the peak intensity.
Acknowledgments
This work was partially supported by grants from NIH/NCI SPORE P50CA070907, RO1CA116322, DOD PROSPECT W81XWH-0710306; and the M. D. Anderson Cancer Center Support Core Grant (CA16672) for using the Peptide Synthesis Facility to synthesize all the peptides used in this study.
Footnotes
For the characterization of protein-interacting motifs, this technique requires prior knowledge of candidate molecules to be tested and hence cannot function as a screening strategy. It can be useful for validating and confirming the observation and knowledge obtained from screening procedures or database searches.
This study describes the use of SELDI-TOF based proteinchip arrays to capture interacting partners after covalent immobilization of bait protein/peptide molecules onto pre-activated surfaces. According to the manufacturer, these arrays are pre-activated with carbonyldiimidazole that have a high affinity toward amino groups. Hence, the amino groups in the peptides applied onto the spots form durable covalent bonds with carbonyldiimidazole already present on the arrays.
Akin to other mass spectrometry-based proteomic techniques, crude samples are not amenable to this procedure. Quality of the input protein sample determines the quality of the data generated with it. A high degree of purity in the protein/peptide samples loaded onto arrays is desirable to generate best results and avoid ambiguous and erroneous data. The best results are obtained with pure synthetic peptides. In our hands, partially purified protein samples did work to a considerable extent, although they did not generate spectacular data. However, it can be used for partially purified over-expressed protein candidates. Liu and coworkers (16) used this elegant technique to determine the interaction between calbindin-D28k and caspase 3 in their study of glucocorticoid-induced apoptosis in osteocytes and osteoblasts. Their study provides a real-world example of application of this technique to decipher meaningful molecular interactions.
Stringency of the washes (amount of detergent used in the wash buffer) should be experimentally determined for each project based on the sensitivity the assay and specificity of the binding involved.
This procedure can also be accomplished using another proteinchip array with a slightly different chemistry, PS20. It can also be performed using other compatible blocking agents between the two protein-binding steps. As suggested by the manufacturer, Tris–HCl or Glycine (0.1–0.5 M), pH 8.0 can also function as a blocking vehicle.
References
- 1.Yaffe MB, Cantley LC. Signal transduction. Grabbing phosphoproteins. Nature. 1999;402:30–31. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 2.Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can’t pSERious? Structure. 2001;9:R33–R38. doi: 10.1016/s0969-2126(01)00580-9. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J. A solid base for assaying protein kinase activity. Nat Biotechnol. 2002;20:232–233. doi: 10.1038/nbt0302-232. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;2003:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T, Nash P. Protein-protein interactions define specificity in signal trans-duction. Genes Develop. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 6.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 7.Fanning AS, Anderson JM. Protein modules as organizers of membrane structure. Curr Opin Cell Biol. 1999;11:432–439. doi: 10.1016/S0955-0674(99)80062-3. [DOI] [PubMed] [Google Scholar]
- 8.Fanning AS, Anderson JM. PDZ domains and the formation of protein networks at the plasma membrane. Curr Topics Microbiol Immunol. 1998;228:209–233. doi: 10.1007/978-3-642-80481-6_9. [DOI] [PubMed] [Google Scholar]
- 9.Fanning AS, Anderson JM. Protein modules as organizers of membrane structure. Curr Opin Cell Biol. 1999;11:432–439. doi: 10.1016/S0955-0674(99)80062-3. [DOI] [PubMed] [Google Scholar]
- 10.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 11.Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz PA, Ou K, Sanchez JC, Bairoch A, Williams KL, Hochstrasser DF. High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol. 1999;289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 12.Jonsson PF, Cavanna T, Zicha D, Bates PA. Cluster analysis of networks generated through homology: automatic iden-tification of important protein communities involved in cancer metastasis. BMC Bioinformatics. 2006;7:2–14. doi: 10.1186/1471-2105-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Porta A, Peng X, Gengaro K, Cunningham EB, Li H, Dominguez LA, Bellido T, Christakos S. Prevention of glucocorticoid-induced apopto-sis in osteocytes and osteoblasts by calbindin-D-28k. J Bone Miner Res. 2004;19:479–490. doi: 10.1359/JBMR.0301242. [DOI] [PubMed] [Google Scholar]
- 14.Ito I, Ji L, Tanaka F, Saito Y, Gopalan B, Branch CD, Xu K, Atkinson EN, Bekele BN, Stephens LC, Minna JD, Roth JA, Ramesh R. Liposomal vector mediated delivery of the 3p FUS1 gene demonstrates potent antitumor activity against human lung cancer in vivo. Cancer Gene Ther. 2004;11:733–739. doi: 10.1038/sj.cgt.7700756. [DOI] [PubMed] [Google Scholar]
- 15.Kondo M, Ji L, Kamibayashi C, Tomizawa Y, Randle D, Sekido Y, Yokota J, Kashuba V, Zabarovsky E, Kuzmin I, Lerman M, Roth J, Minna JD. Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene. 2001;20:6258–6262. doi: 10.1038/sj.onc.1204832. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Miller CW, Koeffler PH, Berk AJ. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFII-D, and a neighboring p53 domain inhibits transcription. Cell. 1992;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]