Abstract
Protein phosphorylation is a dynamic post-translational modification that plays a critical role in the regulation of a wide spectrum of biological events and cellular functions including signal transduction, gene expression, cell proliferation, and apoptosis. Determination of the sites and magnitudes of protein phosphorylation has been an essential step in the analysis of the control of many biological systems. A high throughput analysis of phosphorylation of proteins would provide a simple, logical, and useful tool for a functional dissection and prediction of biological functions and signaling pathways in association with these important molecular events. We have developed a functional proteomics technique using the ProteinChip array-based SELDI-TOF-MS analysis for high throughput profiling of phosphoproteins/phosphopeptides in human serum for the early detection and diagnosis as well as for the molecular staging of human cancer. The methodology and experimental approach consists of five steps: (1) generation of a total peptide pool of serum proteins by a global trypsin digestion; (2) rapid isolation of phosphopeptides from the total serum peptide pool by an affinity selection, purification, and enrichment using a novel automated micro-bioprocessing system with phospho-antibody-conjugated paramagnetic beads and a hybrid magnet plate; (3) high throughput phosphopeptide analysis on ProteinChip arrays by automated SELDI-TOF-MS; and (4) bioinformatics and statistical methods for data analysis. This method with appropriate modifications may be equally applicable to serine-, threonine- and tyrosine-phosphorylated proteins and for selectively isolating, profiling, and identifying phosphopeptides present in a highly complex phosphor-peptide mixture prepared from various human specimens such as cells, tissue samples, and serum and other body fluids.
Keywords: Phosphoprotein, Phosphopeptide, Phosphoproteome, High throughput Phospoprotein/Peptide Profiling, ProteinChip Arrays, SELDI-TOF-MS
1. Introduction
The reversible protein phosphorylation is a key regulating switch that controls a wide range of biological functions and activities (1–8). Particularly, phosphorylation of protein kinases plays a critical role in signaling pathways involved in oncogenesis and pathogenesis of various human cancers (6, 8–10). It is clear that the abnormal protein phosphorylation is associated with many major diseases, such as diabetes, rheumatoid arthritis, cardiovascular disease, and cancers (1, 10, 11). Therefore it is vitally important to understand the intracellular signaling events that control protein phosphorylation. Many toxins that are known to cause cancers act by affecting the functions of kinases or phosphatases (5, 9–12). Although protein kinases are now one of the major groups of proteins being targeted by drug discovery and therapeutics programs, the substrates, or proteins that these kinases and phosphatases affect, are largely unknown. Thus, determination of the sites and magnitudes of protein phosphorylation has been an essential step in the analysis of the control of many biological systems. A high throughput analysis of phosphorylation of serum proteins would provide a simple, logical, and useful tool for a functional diagnosis and prediction of human cancers in association with these important molecular events (5, 8, 9, 13–16). Moreover, the current advanced cancer treatment with anti-angiogenesis agents and protein kinase inhibitors showed profound impact on phosphorylation/dephosphorylation of proteins involved in cell proliferation, apoptosis, angiogenesis, tumor progression, and metastasis in laboratory and in preclinical and clinical settings (10, 11, 17). These phospho-modified proteins are secreted from cells and into the circulation system and are easily available in the serum. Thus, a high-throughput proteomic profiling of phosphopeptides in serum samples exposed to these agents will allow the identification of specific serological biomarkers associated with their biological action (3, 8, 14, 17–20). However, direct determination of phosphorylation of individual proteins in a biological system has been difficult to date (5, 9, 14, 15, 21). It typically requires the purification to homogeneity of the phosphoprotein of interest before analysis and there is currently no method available to study this aspect in detail and consistently. Thus, there has been a substantial need for a more rapid, global, and general method for the analysis of protein phosphorylation in complex protein mixtures (3, 14, 17–20, 22). In addition, a large-scale global phosphoproteome analysis poses challenges in several fronts including to simultaneously isolate and enrich phosphopeptides in several hundred parallel samples without introduction of significant experimental errors and to maintain consistent integrity of proteome among all the samples (2, 3, 14, 15, 18, 19, 21, 23). To address these challenges, we developed an innovative, rapid, and simplified method for serum phoshopeptide separation and enrichment using a one-step affinity capture of phosphopeptides on phospho-antibody-conjugated paramagnetic beads or nanoparticles and separation by a hybrid magnet. This method offers the advantage of automation to avoid human errors and enable a high throughput serum phosphopeptide preparation, which is readily coupled with an automated peptide profiling and analysis on ProteinChip arrays by SELDI-TOF-MS.
2. Materials
2.1. Reagents
Phospho-tyrosine antibody: the phospho-tyrosine mouse mAbs (P-Tyr-100) developed by Cell Signaling Technology (Danvers, MA) is a high affinity mouse monoclonal antibody and provides an exceptionally sensitive new tool with increased utility for studying tyrosine phosphorylation and monitoring tyrosine kinase activity in high throughput tyrosine phosphor-protein/peptide analysis. The antibody is supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 μBSA and 50% glycerol. Store at −20°C.
Surface Activated Dynabeads: the Dynabeads® MyOne™ Tosylactivated (Invitrogen, Carlsbad, CA) are superparamagnetic and uniformpolystyrene beads (1.0 μ in diameter) coated with a polyurethane layer. The dynabeads are used for conjugation with phosphor-antibodies and for biomagnetic separation and enrichment of phosphor-antibodycaptured phosphor-peptides.
Sequencing Grade Modified Trypsin: Sequencing Grade Modified Trypsin (Promega, Madison, WI) is a porcine trypsin modified by reductive methylation, rendering it resistant to proteolytic digestion (24). Sequencing Grade Modified Trypsin is supplied as lyophilized powder and can be reconstituted in 50 mM acetic acid. The substrate is dissolved in 50 mM Tris–HCl (pH 7.6), 1 mM CaCl2, and the enzyme is diluted in 50 mM acetic acid.
Human Serum: human clinical serum samples are collected and prepared using approved clinical protocols and standard methods. Serum samples are stored at −80°C.
2.2. Magnetic Plate
A new class of hybrid magnet plates has recently been developed at the Joint Genome Institute and Lawrence Berkeley National Laboratroy (JGI/LBNL, Berkeley, CA) for high throughput purification of biological samples for functional genomics and proteomics and for affinity drug screening due to its superior capability of selectively separating proteins and DNA from complex biological mixtures based on a magnetic field (25, 26). These magnet plates are ideal for any process that requires automated bead manipulation in high-density microtiter plates containing sample volumes in a range of 3–300 μl. The novel hybrid magnetic structure combines a permanent magnet with ferromagnetic materials that produces magnetic fields significantly higher than those of any commercially available magnetic plate. More importantly, the fields at a distance of 1 cm above the magnet are more than 1,000-fold stronger than those of the commercial 96-well magnet. This feature allows for more vigorous washing and sample recovering. The second generation 96-well hybrid magnet plate has been designed and constructed for our proteomics platform by physicists and engineers at JGI/LBNL, producing fields well above 10,000.0 G, which allows a more efficient separation of affinity-captured phosphopeptides from crude serum peptide mixtures and thus improves reproducibility and sensitivity of proteomic analysis by reducing processing loss and increasing peptide recovery while retaining a high peptide-proteome integrity. Alternatively, the commercially available 96-well magnetic plates can also be used.
2.3. ProteinChip Arrays and Peptide Standards
SEND-ID ProteinChip arrays (Bio-Rad, Hercules, CA) have C18 as a functional group and are used for phosphopeptide profiling and fingerprinting on SELDI-TOF-MS. IMAC30 ProteinChip arrays (Bio-Rad) can also be used for phospho-peptide analysis.
All-in-1 Peptide standards (Bio-Rad) for SELDI-TOF-MS is supplied as a dry powder in a glass vial with rubber stopper and is freshly reconstituted in Reconstitution Solution.
2.4. Buffers and Solutions
Trypsin Resuspension Buffer: 50 mM acetic acid.
Protein Denaturation Solution: 6 M guanidine HCl (or 6–8 M urea), 50 mM Tris–HCl (pH 8), 2–4 mM DTT (or β-mercaptoethanol).
1× PBS (phosphate buffered saline) (pH 7.4): 0.26 g NaH2PO4×H2O (MW 137.99), 1.44 g Na2HPO4×2H2O (MW 177. 99), 8.78 g NaCl (MW 58.5). Dissolve in 900 ml dH2O. Adjust pH to 7.4 and the volume to 1,000 ml.
10× Phosphate Buffered Saline (PBS): To prepare 1 L add 80 g sodium chloride (NaCl), 2 g potassium chloride (KCl), 14.4 g sodium phosphate, dibasic (Na2HPO4), and 2.4 g potassium phosphate, monobasic (KH2PO4) to 1 L dH2O. Adjust pH to 7.4.
Coating buffer: 0.1 M sodium borate buffer (pH 9.5): 6.183 g H3BO3 (MW 61.83). Dissolve in 800 ml distilled water. Adjust pH to 9.5 using 5 M NaOH and adjust volume to 1,000 ml with distilled water. The coating buffer is used for pre-washing and coating of Dynabeads.
3 M ammonium sulphate stock solution: 39.6 g (NH4)2SO4 (MW 132.1). Dissolve in 0.1 M sodium borate buffer (pH 9.5), adjust pH to 9.5 and adjust volume to 100 ml.
Blocking buffer: PBS pH 7.4 with 0.5% (w/v) BSA and 0.05% Tween 20 in 100 ml PBS. Blocking buffer is used for blocking of all precoated Dynabeads. Do not use this buffer or any buffer containing protein or amino-groups (glycine, Tris etc.) for pre-washing or coating of Dynabeads.
Washing buffer: PBS pH 7.4 with 0.1% (w/v) BSA and 0.05% Tween-20 in 100 ml PBS. If a preservative is needed in the coated product, a final concentration of 0.02% (w/v) sodium azide (NaN3) may be added to washing buffer. This preservative is cytotoxic and must be carefully removed before use by washing.
Elution buffers: Any conventional method for protein elution can be used, e.g., 0.1 M citrate pH 3, 0.1 M glycine–HCl pH 2.5, or 0.1 M glycine–NaOH pH 10. All reagents used should be analytical grade. Organic solvent containing 50% acetonitrile (CAN) and 0.1% Triflouroaacetic acid (TFA) (Sigma, St. Louis, MO) is best for the SEND-ID chip.
Peptide Standards Reconstitution Solution: 10 mM ammonium acetate, 25% acetonitrile, and 1.25% trifluoroacetic acid.
3. Methods
An important goal of clinical proteomics is to develop sensitive, specific, and robust proteomic platforms to simultaneously measure the human proteome in clinically relevant specimens and to establish protein signature profiles for discriminating between the normal and disease states (27–35). Serum potentially carries a rich archive of histological and biological information and is attracting increasing interest in clinical proteomics. A throughput profiling and an accurate measurement of these serum proteome would serve to improve early detection, diagnosis, and prognosis of cancers and identify new therapeutic targets (27–35). While the importance of studying the serum proteome is obvious, the characterization and analysis of serum proteins, however, are analytically challenging due to their extremely high dynamic range of concentration that spans more than 9–10 orders of magnitude and to the complexity that is composed of biomolecules ranging from large proteins and lipids to small metabolite hormones, peptides, amino acids, and electrolytes. Particularly, the serum protein contents are dominated by a handful of abundant proteins such as albumin, immunoglobulins, haptoglobin, transferrin, and lipoproteins that account for more than 99% of serum protein masses and overwhelmingly shadow the detection of those low abundant but biologically important molecules (30). The reduction of sample complexity and depletion of the level of these abundant proteins are essential first steps for a successful and efficient analysis of the serum proteome. Affinity depletion methods have therefore been developed to remove abundant serum proteins such as albumin and immunoglobulin from serum prior to mass spectrometric analysis (36–38). One of the major pitfalls of these protein depletion methods, however, is that many important low molecular weight proteins or peptides can be concomitantly removed during the sample processing as well (27, 28, 30). Classical methods such as sample fractionation and purification by liquid chromatography with various media, separation by gel electrophoresis, sample desalting and concentration by dialysis, centrifugation, and immuno-precipitation, are often labor-intensive and demand large quantities of sample, suffer from attendant analyte loss due to non-specific binding and dilution effects, and easily introduce experimental errors during multi-task and multi-step sample preparation thus hampering sample quantification and parallel comparison (30, 38, 39). A simple, direct, and efficient mass spectrometric sample preparation and protein/peptide detection in heterogeneous samples is much needed.
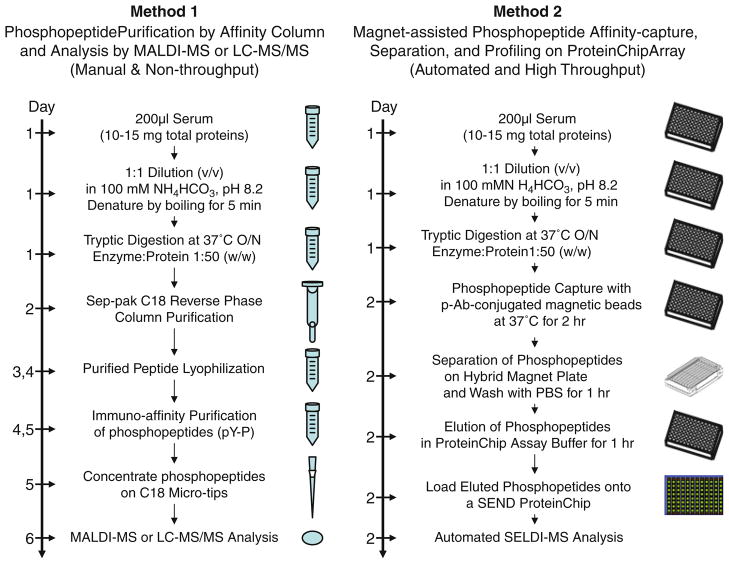
In this method, we use innovative antibody-conjugated magnetic beads to specifically capture a subset of biologically important phosphopeptides from the trypsin-digested serum peptide mixtures. The captured peptides are rapidly and efficiently separated and purified by a novel hybrid magnet specifically developed for our proteomic analysis. This method will significantly reduce the complexity of serum proteins, completely eliminate the interference of abundant serum proteins but not affect the integrity of serum proteome, and avoid multi-steps of serum sample fractionation and purification, thus, allowing a high throughput sample preparation, quantification, and parallel comparison. A comparison of technologies and applications between the existing proteomic approaches (Method 1) and our new serum phosphopeptide proteomic platform (Method 2) is shown in Fig. 1.
Fig. 1.
Comparison of methods and platforms for phosphoprotein/phosphopeptide proteomics analysis by mass spectrometry. Method 1, the conventional phosphopeptide purification by affinity column and analysis by MALDI-MS or LC-MS/MS. Method 2, the magnet-assisted phosphopeptide affinity-capture, separation, enrichment, and profiling on ProteinChip arrays by SELDI-TOF-MS.
The ProteinChip-based SELDI technology is currently being used to successfully detect disease-associated proteins in complex biological specimens and has primarily been applied to search for the cancer-relevant biomarkers in clinical serum samples (33, 34, 40–45). These studies emphasize the capability and potential of SELDI for the detection and characterization of differentially expressed proteins or proteomic patterns for detection and prediction of diseases. For serum proteomics to realize its full potential, however, several potential problems and controversy in sensitivity and reproducibility concerning the SELDI profiling approach needs to be addressed (32, 34, 46, 47). Semmes et al. (48) has recently assessed the platform reproducibility using SELDI-mediated serum protein profiling for the detection of prostate cancer and demonstrated the reproducibility of SELDI serum profiling between laboratories and suggested that this approach could provide a reproducible diagnostic assay platform. The most severe limitation for the reproducibility may be a result of loss of the majority of proteins and peptides present in the sample while using SELDI for the protein profiling (27, 28, 43, 48). This in turns leads to the rather low-resolution pattern that represents only a minority of proteins and peptides in the serum. Other limitations for SELDI protein profiling are due to variability such as in instrumental laser desorption energy level and ProteinChip array quality and in sample collection, processing, and storage (32, 49–53). These changes resulted in reproducible changes in serum proteome, and can sometimes overshadow the biological changes in the serum samples (32). In this study, we use SELDI-TOF-MS to profile serum phosphopeptide sub-proteome instead of total proteins and, thus, will partially circumvent these problems and enhance reproducibility because it concentrates all measurements in a small region of low mass peptides within 200–5,000 Da with an extremely high mass resolution by SELDI-TOF-MS spectrometry. Furthermore, because the affinity capture is performed after a complete global proteolytic digestion of serum proteins, phosphopeptides cleaved from larger proteins are equally captured together with other phosphopeptides in the pool and can be easily and precisely detected on MS spectroscopy, therefore circumventing the difficulty and inefficiency of using mass spectrometry such as SELDI or MALDI in detection of large protein species (>30–50 kDa), improve the measurement sensitivity, and gain a higher coverage of serum proteome.
3.1. Preparation of Phospho-Antibody-Conjugated Magnetic Beads
The Tosylactivated Dynabeads® (1–2 μm in diameter) (Invitrogen, Carlsbad, CA) is used as a solid phase to conjugate phosphor-antibodies for a biomagnetic separation of phosphopeptides from the trypsin-digested serum peptide mixtures. The general protocols given below are based on experience with either phosphotyrosine (pY)-specific antibodies (Cell Signaling Technology) or phospho-Serine/Threonine (pS/pT)-specific antibodies (BD Biosciences). When incubating the beads with the ligand of choice, it will be physically adsorbed onto the surface of Dynabeads MyOne™ Tosylactivated first and followed by the formation of covalent bonds over time (see Note 1).
The volume of beads used is based on the number of serum samples to be analyzed (1 mg beads per 200 μl serum sample). The conjugation is at a ratio of 40 μg antibodies to 1 mg beads (w/w). These conditions are for the coating of 1.0 mg phosphor-antibody to 25 mg Dynabeads MyOne™ Tosylactivated (250 μl at 100 mg beads/ml).
Resuspend Dynabeads thoroughly by vortexing for 30 min. Transfer 250 μl of beads into a test tube. Place the tube on a magnet (Dynal MPC, Invitrogen) for 2 min or until the beads have migrated to the side of the tube and the liquid is clear.
Pipette the supernatant off carefully, leaving beads undisturbed.
Remove the test tube from the magnet and resuspend the beads thoroughly in 1 ml of Coating buffer (Solution 5) by vortexing.
Repeat steps 3–4.
Resuspend the washed beads in 100 μl volume coating buffer and the beads are ready for conjugation with antibodies.
Dilute 1.0 mg of antibodies in coating buffer to a total of 600 μl. For optimized coating, the antibodies may be pre-treated and acidified (see Note 2)
Add 970 μl of coating buffer to the above washed beads (100) and mix properly.
Add the diluted antibodies (1.0 mg/600 μl) to the suspended beads and mix properly.
Add 830 μl of 3 M ammonium sulphate stock solution to the antibody/beads mixture to a total of 2,500 μl of conjugation reaction.
Incubate the conjugation reaction for 16–24 h at 37°C with slow tilt rotation. Do not let the beads settle during the incubation period (see Note 3 for optimized coating time, temperature, and pH).
After incubation, place the tube on the magnet for 2 min, or until the beads have migrated to the side of the tube, and remove the supernatant.
Add the same total volume (2,500 μl) of PBS with 0.5% BSA and 0.05% Tween-20 and incubate at 37°C over night.
Wash three times with PBS with 0.1% BSA and 0.05% Tween-20, and resuspend the washed conjugates to the desired volume or concentration. The Dynabeads are now coated and ready for use (see Note 4).
For storage, the desired preservative, e.g., 0.02% sodium azide may be added and store at 2–8°C. The coated beads can usually be stored for several months at 2–8°C, depending on the stability of your immobilized antibodies.
3.2. Protease Digestion of Serum Proteins
Serum Protein Denaturation: Serum proteins require denaturation and disulfide bond cleavage before enzymatic digestion can go to completion. 200 μl (10–15 mg of total proteins) of serum is used for each individual assay. Dilute serum sample 1:1 (v/v) in 100 mM NH4HCO3, pH 8.2, in each well of a 96-well plate.
Boil for 5 min in a heating-block that fits a 96-well plate. If smaller amounts of protein are to be digested, the recommended conditions given can be scaled down proportionally. However, under no conditions should less than 25 μl of dissolving agent be used.
After denaturation, allow the reaction to cool to room temperature.
Protease Digestion: add sequencing grade modified trypsin (Promega) to a final protease:serum proteins ratio of 1:50 (w/w). Incubate at 37°C overnight.
Remove a small aliquot and chill the reaction on ice or freeze. Add an inhibitor to the aliquot to terminate the protease activity or precipitate the sample by the addition of TCA to a 10% final concentration.
Determine the extent of digestion by subjecting a portion of the digestion products to reverse phase HPLC or SDS-PAGE. If further proteolysis is required, return the reaction tube to 37°C and continue incubating until the desired digestion is obtained.
The reaction can be terminated by freezing or by the addition of specific inhibitors (see Note 5).
3.3. Capture, Separation, and Enrichment of Serum Phosphopeptides
For affinity capture and enrichment of serum phosphopeptides, wash the phosphor-antibody-conjugated magnetic beads with two volumes of 1× PBS three times to remove any unbound antibodies.
Resuspend the phosphor-antibody (p-Ab)-beads conjugates in PBS at a desired concentration (1 mg per 200 μl of serum sample).
Transfer the p-Ab/beads conjugates at a ratio of 200 μl trypsin-digested serum proteins to 1 mg p-Ab/beads into each well of the 96-well plates containing digested serum proteins.
Incubate the plate for 1 h at room temperature with gentle shaking.
Place the plate on a 96-well magnet plate and allow phosphorpeptide-captured beads to settle down.
Gently remove the supernatant and the unbound peptides by washing three times with 400 μl of PBS.
The captured phosphor-peptides are then eluted in 15 μl of elution buffer comprising 50% acetonitrile (CAN) and 0.1% Trifluoroacetic acid (TFA).
Place the plate on a 96-well hybrid magnet plate. After the beads settle down, carefully collect the eluted phosphorpeptides into a fresh 96-well plate.
All these steps of process are performed automatically using a Biomek-2000 Laboratory Automation Workstation (Beckman, Fullerton, CA).
3.4. Phosphopeptide Profiling on ProteinChip Arrays by SELDI-TOF-MS
The enriched phosphopeptides are further processed automatically using a micro bio-processor (Bio-Rad) and a Biomek-2000 Laboratory Automation Workstation (Beckman, Fullerton, CA) and loaded onto a SEND (Surface Enhanced Neat Desorption) or IMAC proteinchip array purchased from (Bio-Rad). SEND arrays are unique compared to other arrays by having the energy-absorbing molecule (EAM) incorporated into the array chemistry and is added after sample addition. They are best suited for small molecule analysis, which in our case are phosphopeptides. Pooled serum samples are used as quality assurance (QA) controls and samples are randomly loaded onto each ProteinChip array with duplicates. One spot is loaded on each array with peptide standards for peptide mass calibration. The chips are automatically loaded and analyzed by SELDI-TOF-MS spectrometer (Model PSII or new model PCS 4000, Bio-Rad).
Add 5 μl of 0.1% TFA to each spot of a SEND-ID array and then quickly remove.
Add 10 μl of the eluted phosphorpeptides onto each spot of the SEND-ID array that is placed in a humidity chamber and incubate for 30 min.
Remove samples from the array surface and wash bound phospho-peptides quickly once with 5 μl of 0.1% TFA.
Then add 2 μl of 25% ACN and 0.1% TFA to each spot. Air-dry.
Read arrays with desired and optimized instrumental parameters and settings in a ProteinChip reader, according to manufacturer’s instruction.
3.5. Data Processing and Analysis
The SELDI-MS spectral data collected from the mass spectrometer are calibrated and subjected to further analysis using bioinformatics tools and statistical methods developed by Dr. Coombes (52, 54). Advanced proteomic data processing and analysis methodologies and bioinformatics algorithms are needed to address concerns regarding reliability, sensitivity, and reproducibility of peak detection, quantification, and identification in clinical serum proteomics (32). Some advanced methodologies and algorithms for spectral alignment, baseline correction and normalization, peak detection and quantification, and statistical analysis of peaks are also needed for evaluating clinical significance of cancer diagnostic peptides (32, 49–55). These methods will improve the reproducibility of peak quantifications and provide tools for evaluating the variations in this phosphor-peptide proteomics platform and more accurately interpret serum phosphor-peptide profiling results.
To overcome the technology barriers and circumvent the potential problems and limitations facing MS spectrometry-based serum proteomics and as demonstrated in the above serum protein-profiling experiments, we have developed an innovative and integrated functional proteomics technique using the ProteinChip array-based SELDI-MS analysis for a high throughput profiling of phospho-peptides in human serum. To demonstrate the feasibility of this proteomic platform for clinical serum sample processing and analysis, we performed phosphopeptide (phosphor-Tyrosine) proteomic profiling on serum samples from human normal and lung cancer patients in different stages and with different smoking histories. We analyzed three groups of samples: (1) 20 serum samples from lung cancer patients collected by Dr. Roth in the Department of Thoracic & Cardiovascular Surgery (Cancer Group 1, C-G1), (2) 20 lung cancer serum samples collected by Dr. Spitz in the Department of Epidemiology (Cancer Group 2, C-G2), and (3) 20 serum samples from normal controls. Tyrosin-phosphopeptides (pYPs) were prepared and selectively isolated from these serum samples as described in the Subheading 3. The purified pYPs were randomly loaded onto SEND ProteinChip arrays in duplicate. Phosphopeptide MS-spectra and data were analyzed using bioinformatics tools and statistical methods developed by Dr. Coombes (52, 54).
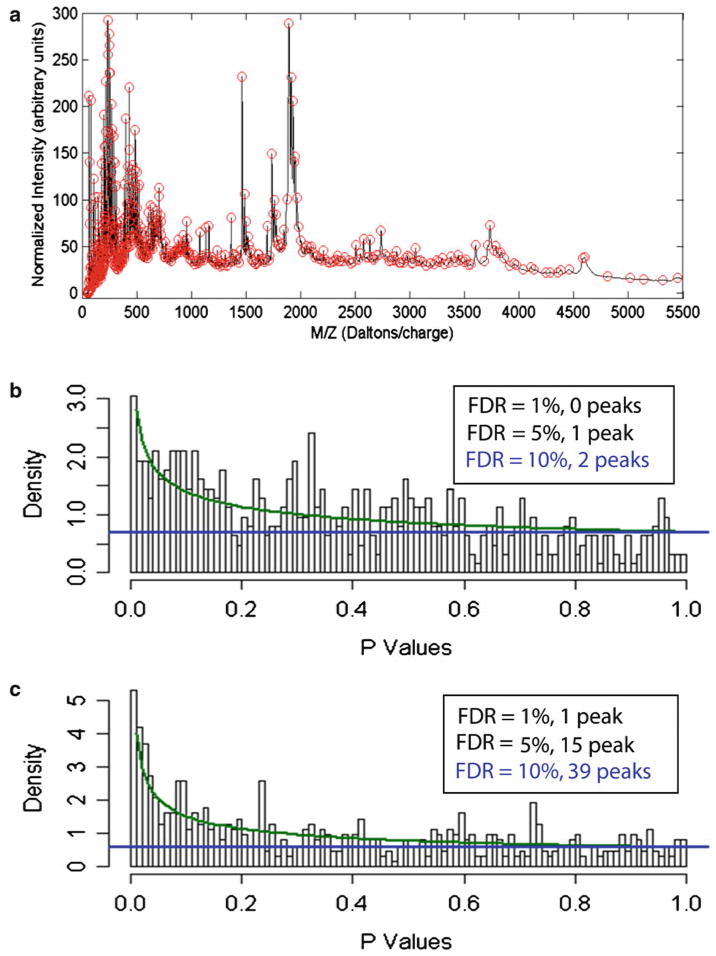
We used wavelets and the mean spectrum for peak detection. Briefly, we first computed the mean of the aligned, baseline-corrected, normalized spectra. We used an undecimated discrete wavelet transform (UDWT) to denoise the mean spectrum by hard thresholding the wavelet coefficients that were less than ten times the standard deviation. Peaks were defined as local maxima in the denoised mean spectrum. Along with the location of each peak, we also recorded an interval that contained the peak by finding the nearest local minimum on either side of the peak. Using this procedure, we detected 622 distinct peaks spanning a m/z range from 50 to 5,500 Da (Fig. 2a). The smallest signal-to-noise ratio (S/N) of any of these peaks was 4.92; the median S/N was 416. In order to quantify the peaks in the individual spectra, we began by locating the time interval containing the peak. We then took the maximum value of the spectrum in that interval and subtracted the three minimum values in the interval to define the peak height. Peak quantification was performed on the aligned, baseline-corrected, normalized spectra. Implicitly, the minimum value was used as a local estimate of the baseline in the interval. Because no smoothing was performed, the peak heights might be slightly biased on the high side. However, this is a reasonable trade-off because it decreases the variance.
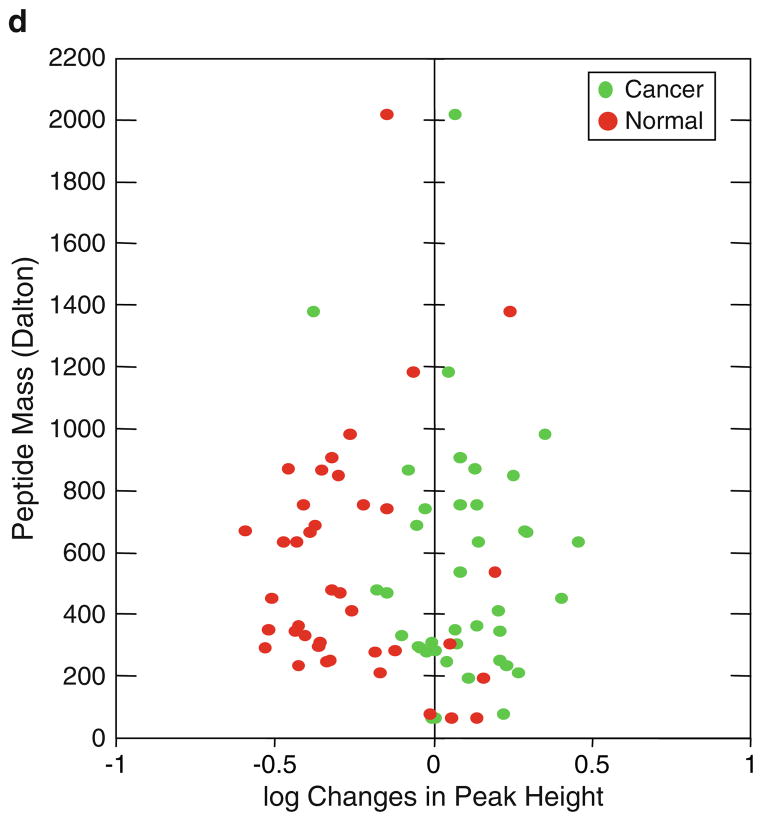
Fig. 2.
Detection, quantification, and statistical analysis of specific human serum phosphopeptide peaks determined by ProteinChip array-based SELDI-TOF-MS. (a ) Phosphopeptide peaks are detected using wavelets and determined by the mean spectrum of the aligned, base-line-corrected, and normalized spectra. More than 600 phospho-tyrosine peptide peaks are detected from the magnet-assisted and affinity-enriched human serum phosphopetide pools. Statistical analysis of phosphor-peptide modulations between the normal and lung cancer serum samples using a beta-uniform mixture (BUM) Fig. 2. (continued) model to estimate the false discovery rate (FDR). (b) BUM analysis of peaks that are different between the three groups of samples using one-way NOVA, and (c) BUM analysis of p-values from an F -test of the significance of group effects after accounting for other technological factors, including laser intensity, pressure, and spot position. Peaks with significant changes (p<0.05) between the normal and the cancer groups are determined at FDR = 1, 5, and 10%, respectively. (d) Scattered plot of 39 pairs of phosphor-tyrosine peptides with significant changes between the normal and lung cancer serum samples, as determined by BUM in c.
Information on the peak locations and heights was further exported from MATLAB and imported to the software program R for statistical analysis. To answer the primary question whether there are any peaks that are different between cancer samples and normal samples, we performed a one-way analysis of variance (ANOVA) using a single factor that takes on three levels (Cancer-G1 Cancer-G2, verses Normal). We performed a separate ANOVA for each of the 622 peaks, using the base-two logarithm of the peak height to try to separate the three sample groups. For each peak, we recorded the p-value from an F-test of the model; small p-values suggest that the peak height is different between at least two of the three groups in the study. In order to account for multiple testing, we modeled the set of p-values using a beta-uniform mixture (BUM) model to estimate the false discovery rate (FDR) (56–59). Setting FDR at 1, 5, and 10%, we found 0, 1, and 2 significant peaks, respectively (Fig. 2b). However, using a BUM analysis after accounting with other technological factors including laser intensity, vacuum chamber pressure, and spot positions on the ProteinChip arrays, we found 1, 15, and 39 signifiant peaks with FDR = 1, 5, and 10%, respectively (Fig. 2c). The fold changes in intensity detected on MS profiles between the normal and cancer serum samples are plotted in Fig. 2d. Using this phosphopeptide proteomic profiling technology and data analysis we are able to find peaks that significantly differ between the three groups. The data generated from these phosphopeptide profiles are also highly reproducible, as shown by the consistent mass spectra among each sample group (Fig. 3). Differences in sample handling explain some of the peaks that are found to be differentially expressed. Nevertheless, many of the changes can clearly be attributed to differences between normal samples and cancer samples, regardless of who collected them. These pilot experiments clearly demonstrate the feasibility of our serum phosphopetide proteomic platform in detecting temporal changes of phosphopeptide proteome in clinically relevant serum samples. Our proteomics platform also demonstrated the capability of overcoming and circumventing a number of technological problems and barriers facing the current MS spectrometry-based proteomics technologies, including reduction of proteome complexity, enhancement of specificity and sensitivity of detection of low abundance proteins and peptides, increase of throughput rate of sample process and analysis, and improvement of identification and quantification of specific peptides and proteins on MS profiles and proteomic data processing and analysis.
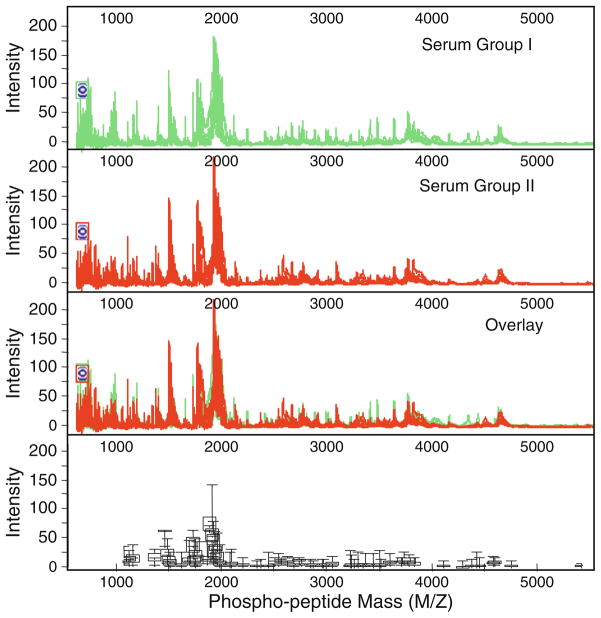
Fig. 3.
Profiles and analysis of tyrosine-specific phosphopeptides in normal and lung cancer serum samples on SEND ProteinChip arrays by SELDI-TOF-MS. The variations of phosphopetide levels as defined by peak intensity on the mass spectra among serum samples are shown by the overlapping spectra (top three panels ) and by the box plots (bottom panel ) with error bars indicating the ranges of each paired peptide peak among three serum sample groups.
Acknowledgments
The authors would like to thank Drs. Xifeng Wu and Margaret Spitz for providing lung cancer serum samples for serum phosphopeptide profiling and analysis and Dr. Kevin Coombes for bioinformatics and statistical analysis, at The University of Texas M. D. Anderson Cancer Center, Houston, TX, and David Humphries at Lawrence Berkeley National Laboratory, Oak Land, CA for developing hybrid magnetic plates for serum phosphopeptide enrichment. This work was partially supported by grants from NIH/NCI SPORE P50CA070907, RO1CA116322, and MMHCC U01CA105352; DOD PROSPECT W81XWH-0710306; The University of Texas M. D. Anderson Cancer Center Support Core Grant (CA16672).
Footnotes
The efficacy of immunomagnetic separation is critically dependant on the specificity and avidity of the antibody or other ligand applied. A concentration of 40 μg antibody/mg Dynabeads is generally optimal for coating. Antibody/protein to be coated directly onto the surface of Dynabeads must be purified, since all proteins will bind to the bead surface. Sugars or stabilizers may disturb the binding and should be removed from the antibody preparation.
For antibody pre-treatment and acidification, in general, lowering pH to 2.5 for 15 min at room temperature or 1 h at 1–4°C, and then raising the pH to approximately neutral prior to addition of the beads, will increase binding and function of antibodies, but this must be optimized for your specific antibodies.
The physical adsorption to the bead surface is rapid, while the formation of covalent bonds will need more time. After the recommended 16–24 h at 37°C, a maximal chemical binding is achieved. Coating at 20°C will require an extended incubation time to 48 h and longer to obtain the same degree of chemical binding. At 4°C the chemical binding is very slow (>48 h). Both higher temperatures and a higher pH will speed up the formation of covalent bonds, provided that the antibodies in question are stable and functional under these conditions. Sodium borate buffer pH 9.5 is recommended. Molarities between 0.1 and 0.5 are optimal.
If the presence of BSA will interfere with your downstream application, this protein can be omitted from the buffer. Detergent may similarly be omitted.
Trypsin can also be inactivated by lowering the pH of the reaction to below 4. Trypsin will regain activity as the pH is raised above 4. Reducing the temperature will decrease the digestion rate. Longer incubation periods, up to 24 h, may be required depending on the nature of the protein.
References
- 1.Hunter T. Signaling–2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 4.Peters EC, Brock A, Ficarro SB. Exploring the phosphoproteome with mass spectrometry. Mini-Rev Med Chem. 2004;4:313–324. doi: 10.2174/1389557043487330. [DOI] [PubMed] [Google Scholar]
- 5.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 6.York JD, Hunter T. Signal transduction. Unexpected mediators of protein phosphorylation. Science. 2004;306:2053–2055. doi: 10.1126/science.1107225. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SA, Hunter T. Phosphoproteomics finds its timing. Nat Biotechnol. 2004;22:1093–1094. doi: 10.1038/nbt0904-1093. [DOI] [PubMed] [Google Scholar]
- 8.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 9.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 10.Hunter T. Tyrosine phosphorylation in cell signaling and disease. Keio J Med. 2002;51:61–71. doi: 10.2302/kjm.51.61. [DOI] [PubMed] [Google Scholar]
- 11.Hunter T. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lectures. 1998;94:81–119. [PubMed] [Google Scholar]
- 12.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 14.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Pan S, Zhang H, Rush J, Eng J, Zhang N, Patterson D, et al. High throughput proteome screening for biomarker detection. Mol Cell Proteomics. 2005;4:182–190. doi: 10.1074/mcp.M400161-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 18.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 19.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc Natl Acad Sci USA. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 22.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 23.Mann M. Quantitative proteomics? Nat Biotechnol. 1999;17:954–955. doi: 10.1038/13646. [DOI] [PubMed] [Google Scholar]
- 24.Rice RH, Means GE, Brown WD. Stabilization of Bovine Trypsin by Reductive Methylation. Biochim Biophys Acta. 1977;492:316–321. doi: 10.1016/0005-2795(77)90082-4. [DOI] [PubMed] [Google Scholar]
- 25.Elkin H, Kapur T, Humphries D, Pollard M, Nammon N, Hawkins T. Magnetic bead purification of labeled DNA fragments for high throughput capillary electorphoresis sequencing. Biotechniques. 2002;32:1296–1307. doi: 10.2144/02326st05. [DOI] [PubMed] [Google Scholar]
- 26.Humphries DE, Pollard MJ, Elkin CJ. The Regents of the University of California; 10/305658 (6954128B2)2002. California, USA: High Performance Hybrid Magnetic Structure for Biotechnology Applications. (Patent)
- 27.Aebersold R, Rist B, Gygi SP. Quantitative proteome analysis: methods and applications. Annal New York Acad Sci. 2000;919:33–47. doi: 10.1111/j.1749-6632.2000.tb06865.x. [DOI] [PubMed] [Google Scholar]
- 28.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 29.Anderson NL, Matheson AD, Steiner S. Proteomics: applications in basic and applied biology. Curr Opin Biotechnol. 2000;11:408–412. doi: 10.1016/s0958-1669(00)00118-x. [DOI] [PubMed] [Google Scholar]
- 30.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Celis JEG. Proteomics in translational cancer research: Toward an integrated approach. Cancer Cell. 2003;3:9–15. doi: 10.1016/s1535-6108(02)00242-8. [DOI] [PubMed] [Google Scholar]
- 32.Coombes KR, Morris JS, Hu J, Edmonson SR, Baggerly KA. Serum proteomics profiling–a young technology begins to mature. Nat Biotechnol. 2005;23:291–292. doi: 10.1038/nbt0305-291. [DOI] [PubMed] [Google Scholar]
- 33.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 34.Petricoin EF, Liotta LA. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol. 2004;15:24–30. doi: 10.1016/j.copbio.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Sauer S, Lange BM, Gobom J, Nyarsik L, Seitz H, Lehrach H. Miniaturization in functional genomics and proteomics. Nat Rev Genet. 2005;6:465–476. doi: 10.1038/nrg1618. [DOI] [PubMed] [Google Scholar]
- 36.Shrivastava A, von Wronski MA, Sato AK, et al. A distinct strategy to generate high-affinity peptide binders to receptor tyrosine kinases. Prot Engin, Design Select. 2005;18:417–424. doi: 10.1093/protein/gzi049. [DOI] [PubMed] [Google Scholar]
- 37.Sato AK, Sexton DJ, Morganelli LA, et al. Development of mammalian serum albumin affinity purification media by peptide phage display. Biotechnol Prog. 2002;18:182–192. doi: 10.1021/bp010181o. [DOI] [PubMed] [Google Scholar]
- 38.Adkins JN, Varnum SM, Auberry KJ, et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Chan KC, Lucas DA, Hise DGM, et al. Analysis of the human serum proteome. Clin Proteomics J. 2004;1:101–225. [Google Scholar]
- 40.Ressom HW, Varghese RS, Abdel-Hamid M, et al. Analysis of mass spectral serum profiles for biomarker selection. Bioinformatics. 2005;21:4039–4045. doi: 10.1093/bioinformatics/bti670. [DOI] [PubMed] [Google Scholar]
- 41.Veenstra TD, Prieto DA, Conrads TP. Proteomic patterns for early cancer detection. Drug Discov Today. 2004;9:889–897. doi: 10.1016/S1359-6446(04)03246-5. [DOI] [PubMed] [Google Scholar]
- 42.Yu LR, Zhou M, Conrads TP, Veenstra TD. Diagnostic proteomics: serum proteomic patterns for the detection of early stage cancers. Dis Markers. 2003;19:209–218. doi: 10.1155/2004/612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Issaq HJ, Conrads TP, Prieto DA, Tirumalai R, Veenstra TD. SELDI-TOF MS for diagnostic proteomics. Anal Chem. 2003;75:148A–155A. [PubMed] [Google Scholar]
- 44.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. The SELDI-TOF MS approach to proteomics: protein profiling and biomarker identification. Biochem Biophys Res Commun. 2002;292:587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 45.Rosenblatt KP, Bryant-Greenwood P, Killian JK, et al. Serum proteomics in cancer diagnosis and management. Ann Rev Med. 2004;55:97–112. doi: 10.1146/annurev.med.55.091902.105237. [DOI] [PubMed] [Google Scholar]
- 46.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 47.Koomen JM, Li D, Xiao LC, et al. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Prot Res. 2005;4:972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 48.Semmes OJ, Feng Z, Adam BL, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 49.Baggerly KA, Morris JS, Wang J, Gold D, Xiao LC, Coombes KR. A comprehensive approach to the analysis of matrix-assisted laser desorption/ionization-time of flight proteomics spectra from serum samples. Proteomics. 2003;3:1667–1672. doi: 10.1002/pmic.200300522. [DOI] [PubMed] [Google Scholar]
- 50.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 51.Baggerly KA, Morris JS, Edmonson SR, Coombes KR. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97:307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- 52.Coombes KR, Tsavachidis S, Morris JS, Baggerly KA, Hung MC, Kuerer HM. Improved peak detection and quantification of mass spectrometry data acquired from surface-enhanced laser desorption and ionization by denoising spectra with the undecimated discrete wavelet transform. Proteomics. 2005;5:4107–4117. doi: 10.1002/pmic.200401261. [DOI] [PubMed] [Google Scholar]
- 53.Coombes KR. Analysis of mass spectrometry profiles of the serum proteome. Clin Chem. 2005;51:1–2. doi: 10.1373/clinchem.2004.040832. [DOI] [PubMed] [Google Scholar]
- 54.Coombes KR, Fritsche HA, Jr, Clarke C, et al. Quality control and peak finding for proteomics data collected from nipple aspirate fluid by surface-enhanced laser desorption and ionization. Clin Chem. 2003;49:1615–1623. doi: 10.1373/49.10.1615. [DOI] [PubMed] [Google Scholar]
- 55.Baggerly KA, Edmonson SR, Morris JS, Coombes KR. High-resolution serum proteomic patterns for ovarian cancer detection. Endocr Relat Cancer. 2004;11:583–584. doi: 10.1677/erc.1.00868. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 57.Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 58.Pounds S, Cheng C. Improving false discovery rate estimation. Bioinformatics. 2004;20:1737–1745. doi: 10.1093/bioinformatics/bth160. [DOI] [PubMed] [Google Scholar]
- 59.Pounds S, Cheng C. Sample size determination for the false discovery rate. Bioinformatics. 2005;21:4263–4271. doi: 10.1093/bioinformatics/bti699. [DOI] [PubMed] [Google Scholar]